Abstract
The normal function of human intercellular adhesion molecule-1 (ICAM-1) is to provide adhesion between endothelial cells and leukocytes after injury or stress. ICAM-1 binds to leukocyte function-associated antigen (LFA-1) or macrophage-1 antigen (Mac-1). However, ICAM-1 is also used as a receptor by the major group of human rhinoviruses and is a catalyst for the subsequent viral uncoating during cell entry. The three-dimensional atomic structure of the two amino-terminal domains (D1 and D2) of ICAM-1 has been determined to 2.2-Å resolution and fitted into a cryoelectron microscopy reconstruction of a rhinovirus–ICAM-1 complex. Rhinovirus attachment is confined to the BC, CD, DE, and FG loops of the amino-terminal Ig-like domain (D1) at the end distal to the cellular membrane. The loops are considerably different in structure to those of human ICAM-2 or murine ICAM-1, which do not bind rhinoviruses. There are extensive charge interactions between ICAM-1 and human rhinoviruses, which are mostly conserved in both major and minor receptor groups of rhinoviruses. The interaction of ICAMs with LFA-1 is known to be mediated by a divalent cation bound to the insertion (I)-domain on the α chain of LFA-1 and the carboxyl group of a conserved glutamic acid residue on ICAMs. Domain D1 has been docked with the known structure of the I-domain. The resultant model is consistent with mutational data and provides a structural framework for the adhesion between these molecules.
Intercellular adhesion molecule-1 (ICAM-1, CD54) is a cell-surface, transmembrane molecule that is normally expressed at a very low level (1). However, it is rapidly up-regulated by cytokine stimulation, enhancing adhesion of leukocytes to endothelial cells at sites of infection or injury. ICAM-1 is also used by various pathogens, such as common cold human rhinoviruses (HRVs) (2, 3), coxsackievirus A21, and the malarial parasite Plasmodium falciparum (4, 5).
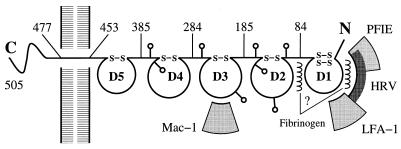
The ICAM-1 molecule consists of five Ig-like domains (D1–D5), a short transmembrane region, and a small carboxyl-terminal cytoplasmic domain (Fig. 1). The second, third, and fourth Ig domains are heavily N-glycosylated with four potential sites in D2, two in D3, and two in D4 (6, 7). The normal adhesive ligands are two integrins, leukocyte function-associated antigen (LFA-1, CD11a/CD18) (7–9), and macrophage-1 antigen (Mac-1, CD11b/CD18) (10). Adhesion between ICAM-1 and LFA-1 is primarily between the D1 domain (11, 12) and the insertion (I)-domain (9, 10) of the respective molecules, whereas adhesion between ICAM-1 and Mac-1 is between the D3 domain and the I-domain (10). Two other molecules, ICAM-2 and ICAM-3, have at least 30% sequence identity with ICAM-1 and have similar adhesive properties. ICAM-1 and ICAM-2 normally have low expression levels, whereas ICAM-3 is more abundant in resting monocytes and lymphocytes. Fibrinogen can also bind to domain D1 of ICAM-1, mediating leukocyte adhesion to vascular endothelium (1). Unlike many other integrin receptors, ICAM-1 does not possess an Arg-Gly-Asp (RGD) motif, but has a larger, more extended binding surface.
Figure 1.
A diagram of an ICAM-1 molecule showing sites of glycosylation (lollipop-shaped structures) and the approximate location of binding sites of LFA-1, Mac-1, human rhinoviruses, fibrinogen, and Plasmodium falciparum-infected erythrocytes (PFIE).
The major group of human rhinoviruses (at least 80% of the approximately 100 identified serotypes) utilizes human ICAM-1 (but not ICAM-2 or ICAM-3) as a cell-surface receptor (2, 3). Mutational and antigenic analyses (5, 12, 13), as well as cryoelectron microscopy (cryo-EM) (14), show that human rhinoviruses attach to domain D1 of ICAM-1. This interaction initiates entry of the virus into the host cell (15, 16). Erythrocytes infected by the malarial parasite P. falciparum gain the ability to bind to domain D1 of ICAM-1 in venular endothelium cells (4, 5). The binding sites for LFA-1, fibrinogen, rhinoviruses, and malaria-infected erythrocytes on ICAM-1 are overlapping, but are not identical (4, 5). However, whereas the binding of LFA-1 to ICAM-1 requires the presence of Ca2+ or Mg2+ (11, 17) and may require a dimeric form of ICAM-1 (18), rhinoviruses bind to monomeric ICAM-1 (3, 5) and do not require a divalent cation.
Electron microscopy studies have shown that the extracellular component of ICAM-1 has a rod-like shape, 190 Å long, with a kink between the second and third domain at about 76 Å from the amino end (11). Cryo-EM reconstructions at 25-Å resolution of complexes between a soluble D1D2 recombinant fragment of ICAM-1 and HRV16 (14) or HRV14 (unpublished results) show a rod-like structure, about 75 Å long, with a width of about 20 Å for the ICAM-1 fragment.
X-ray crystallographic structures have been determined of the homologous adhesion molecules CD4 (19, 20), CD2 (21), vascular adhesion molecule-1 (VCAM-1) (22, 23), and ICAM-2 (24). CD4 and VCAM-1 are utilized by certain viruses as cellular receptors. VCAM-1 and ICAM-2 are members of a subclass of the Ig superfamily that are structurally similar, are expressed on endothelial cells, and are stimulated by inflammatory cytokines. Their structures are characterized by an “Intermediate” domain D1 and a “Constant 2” domain D2 (25). Furthermore, there is an extra disulfide bond in domain D1 compared with the Intermediate Ig fold (25). In spite of these similarities between VCAM-1 and ICAM-2, the amino acid sequence identity between these and the homologous ICAM-1 molecule is less than 31%. Because the specificity of HRVs is strictly for human ICAM-1, even to the exclusion of murine ICAM-1 (12, 13), the sequence and structure of the loops [identified as the site of interaction with HRVs (14)], at the end distal to the transmembrane region, are critical to the understanding of the interaction of the major group of HRVs with their human cellular receptor. Here, we present the structure of the amino-terminal domains D1 and D2 of human ICAM-1 at 2.2-Å resolution.
MATERIALS AND METHODS
Expression and Purification of ICAM-1 D1D2 with Reduced Glycosylation.
Crystals of fully glycosylated ICAM-1 D1D2 expressed in CHO cells (26) exhibited an extremely high degree of variability in their cell dimensions. To avoid these problems, three of the four potential glycosylation sites were removed in D2. A mutant in which all four N-linked glycosylation sites were eliminated by mutating asparagine to glutamine was not secreted by either CHO or sF9 cells. A series of mutants in which glycosylation sites were singly eliminated, and a second series in which only single glycosylation sites were retained, indicated that the site at Asn-175 was both necessary and sufficient for high-level secretion (data not shown). All subsequent studies, including the crystal structure determination, were performed with the D1D2 mutated protein N103Q/N118Q/N156Q (mutICAM-1).
The D1D2 construct (residues 1–185) for baculovirus expression was generated by PCR amplification. The site-directed mutagenesis of the single-stranded template was performed according to standard protocols to delete the four potential glycosylation sites. The desired mutated protein N103Q/N118Q/N156Q was then generated by back-mutation of Gln-175 in the mutated template. All constructs were cloned into the baculovirus transfer vector PVL1393 (PharMingen), and recombinant baculovirus was generated and propagated by the manufacturer’s standard protocols. Fermentation in sF9 cells was carried out in spinner flasks, and the secreted mutICAM-1 was purified by a modification of the protocol described by Kolatkar et al. (26).
Crystallization of mutICAM-1 D1D2.
The mutICAM-1 was used for growing crystals suitable for diffraction experiments by using the hanging drop method. Drops of 5 or 10 μl, containing 10 mg/ml of mutICAM-1 in 10 mM Tris, pH 7.5/25 mM NaCl, were equilibrated against a reservoir containing 1 ml of 20% PEG 4000 in 10 mM Tris as a precipitant. Self-nucleation under these conditions produced showers of microscopic crystals with needle-shaped habits. These needles were then used for macroseeding. Typically, one or two specimens were transferred from needle stocks into freshly prepared drops. Needle-shaped crystals at least 0.5 mm long and up to 0.1 mm thick appeared in 1–2 weeks after seeding.
Data Collection and Structure Determination.
A large number of x-ray diffraction data sets were collected to resolution limits varying between 4 and 2.2 Å (Table 1). Derivatization of the crystals produced considerable changes in cell dimensions and poor isomorphism. The resulting experimental phases failed to produce an interpretable map. In an attempt to overcome the nonisomorphism, crystals were grown of selenomethionine-derivatized (SeMet) mutICAM-1, and multiple wavelength data were collected around the SeK edge (Table 1). The first two domains of ICAM-1 contain only one Met residue. A single peak corresponding to the Se site was observed in an anomalous dispersion Patterson map (27) and confirmed by a difference Bijvoet-difference Patterson map of the Pt compound, K2Pt(NO2)4. The multiple wavelength anomalous dispersion (MAD) (28) phases were, however, unable to produce an interpretable map. Further attempts were made to combine (29) the MAD phases from the SeMet data with multiple isomorphous replacement (MIR) phases from other derivatives (Table 1), or with MAD phases collected at the Pt LIII edge from crystals of the cis-Pt(NH3)2Cl2 derivative.
Table 1.
X-ray diffraction data summary
| Compound* | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| a (Å) | 41.7 | 41.8 | 41.8 | 41.9 | 41.9 | 41.9 |
| b (Å) | 123.7 | 123.6 | 123.5 | 123.3 | 124.1 | 124.7 |
| c (Å) | 82.5 | 82.8 | 82.2 | 81.6 | 83.3 | 83.3 |
| Source | ← Brookhaven→ | — | CHESS | |||
| X12C | X4A | X12C | X12C | R-axis | A1 | |
| Detector | Mar | Fuji | Mar | Mar | CCD | |
| No. of wavelengths | 1 | 4 | 1 | 4 | 1 | 1 |
| Resolution, Å | 3.0 | 2.6 | 3.0 | 3.1 | 3.5 | 2.2 |
| Rmerge, % | 8.8 | 7.6† | 5.4 | 6.4† | 9.0 | 4.5 |
| Completeness, % | 87 | 94 | 85 | 87 | 70 | 90 |
Compound 1 is native (the most isomorphous data set to compounds 3 and 5); compound 2 is SeMet; compound 3 is K2Pt(NO2)4; compound 4 is cis-Pt(NH3)2Cl2; compound 5 is UO2(NO3)2; and compound 6 is Ir3N(SO4)6.
These statistics correspond to merging data from all wavelengths, neglecting anomalous effects. The wavelengths were 0.9879, 0.9793, 0.9791, and 0.9686 Å and 1.0720, 1.0714, 1.0711, and 0.9800 Å for the Se and Pt compounds, respectively.
A number of attempts were made to solve the mutICAM-1 structure by molecular replacement (MR), by using the known homologous structures of CD4, CD2, and VCAM-1, without success. Only the structure of ICAM-2 was sufficiently close to that of mutICAM-1 to allow a successful structure determination. A homology model for mutICAM-1 was built with the program look (version 2.0, Molecular Applications Group, Palo Alto, CA) based on the three-dimensional structure of ICAM-2 (24). Searches with domain D1 by using amore (30) readily produced a clear solution that was confirmed by the consistency in the position of the sulfur atom in the single Met residue of the model and its experimentally determined positions from anomalous dispersion data of the Se compound. Searches with domain D2, by using a model in which the loops had been removed that were likely to be different between mutICAM-1 and ICAM-2, did not produce a clear solution. Electron density maps were calculated by combining the phases derived from the D1-domain MR solution and the MIR experimental phases obtained from the UO2(NO3)2 and K2Pt(NO2)4 derivatives. This map had a protein–solvent boundary for the second domain, consistent with the expected shape of a molecule made of two Ig domains, and also showed the β-strand-like features.
Crystallographic refinement using the program x-plor (31) allowed identification of the missing loops in the resultant electron density map. The current model shows reasonable agreement with both x-ray data and ideal geometry (Table 2). Detailed crystallographic information will be given in a fuller manuscript (unpublished results). The coordinates have been deposited with the Brookhaven Protein Data Bank (accession no. 1iam).
Table 2.
Refinement statistics
| Refinement* | SeMet | Ir3N(S04)6 |
| Resolution, Å | 5-2.6 | 5-2.2 |
| Rworking, % | 20.5 | 21.4 |
| Rfree, % | 30.9 | 30.3 |
| rms deviation from idealized values | ||
| Bonds, Å | 0.008 | 0.009 |
| Angles, ° | 1.65 | 1.69 |
| Average B values | ||
| In backbone (Å2) | 18 | 24 |
| In side chain (Å2) | 21 | 27 |
| Ramachandran plot % of residues | ||
| In most favored region | 88 | 88 |
| In additional allowed region | 12 | 12 |
| In disallowed region | 0 | 0 |
A subset of data (10%) was excluded from refinement and used for the free R-factor calculation (F > 0σ(F)).
RESULTS AND DISCUSSION
The mutICAM-1 Structure.
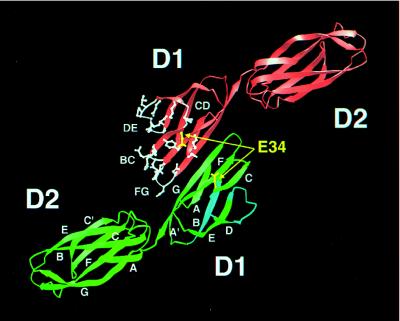
The structure of the two Ig-like domains (Fig. 2) has a total length of ≈75 Å. Each domain has a diameter of about 20 Å, with an elbow angle of about 30° between the major axes of the two domains. The fold of each domain is similar to the corresponding domain of ICAM-2 (24) with an rms difference of 1.2 Å between 82 superimposed Cα atoms in D1 and of 2.1 Å between 100 superimposed Cα atoms in D2. Both mutICAM-1 and ICAM-2 have an additional disulfide bond between Cys-25 and Cys-69 connecting the BC and FG loops in domain D1 compared with the classical Intermediate Ig-type domain (25) (see Fig. 2 for the accepted nomenclature of β-strands A, B, … , G in an Ig fold). The greater similarity between domains D1 versus domains D2, when comparing mutICAM-1 with ICAM-2, reflects the easier recognition of this domain by the molecular replacement search procedure that was used in the structure determination (see Materials and Methods). The similarity of structure between mutICAM-1 and ICAM-2 is not surprising in light of their 30% amino acid sequence identity. However, the angular relationship of the two domains within each molecule differs by 15°. Superposition of mutICAM-1 domain D1 onto D2 shows that D2 is rotated by 170° about an axis roughly parallel to its length relative to the orientation of domain D1, whereas this rotation is only 155° for ICAM-2.
Figure 2.
The crystallographic dimer in which there is extensive antiparallel β-sheet between the G strands. The residues and loops that penetrate into the HRV “canyon” are colored white in one of the two monomers. Glu-34, essential for LFA-1 binding, is shown in yellow. Strands and loops important for binding PFIE are in light blue in one monomer. β-strands in domains D1 and D2 are labeled consistent with the nomenclature of an “Intermediate” and “Constant 2” Ig-like fold, respectively (25).
Three of the potential four glycosylation sites of the baculovirus-expressed ICAM-1 used in the crystal structure determination were mutated. The remaining site was an N-linked sugar at residue Asn-175. Mass spectroscopic studies show that there are two glycosylation variants of mutICAM-1 D1D2 with 6 and 10 sugar units. Although more than one carbohydrate moiety is clearly visible in the electron density map, only the first sugar unit attached to Asn-175 has been modeled as N-acetyl-d-glucosamine.
ICAM-1 as a Receptor for Rhinoviruses.
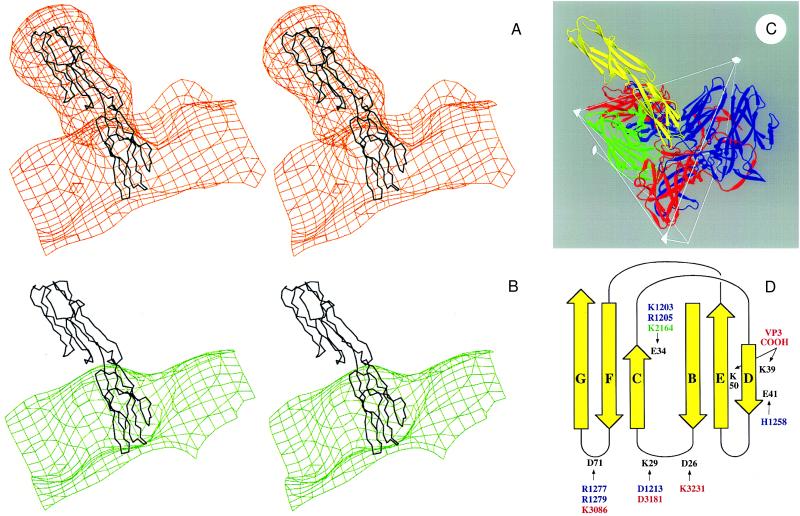
The structure of ICAM-1 was placed into a phase contrast transfer function (32) improved version of the cryo-EM electron density map (unpublished results) of the HRV16:ICAM-1 (D1D2) complex described previously (14). Fitting of the mutICAM-1 molecule into the density was simple and unique (Fig. 3A). It also would have been possible to fit domain D1 into the density when rotated by approximately 180° relative to the best fit, but this would have placed D2 outside the electron density as a consequence of the 30° elbow angle. The fit assumes that the elbow angle between the two domains is the same in the cryo-EM reconstruction as in the crystal structure. This can be partly justified by observing the significant interactions between residues from the opposing domains (see below) and by the fact that the fit is good, without involving additional conformational changes. In the model corresponding to the unique fit, the three loops BC, DE, and FG penetrate deep into the canyon [a surface depression into which the receptor binds (16)] (Fig. 3 B and C), consistent with mutational studies (33). In addition, the short CD loop of ICAM-1 (Fig. 2) lies against VP2 of HRV16 on the “south” side of the canyon. The footprint of ICAM-1 onto the HRV16 surface is essentially as previously published (14), and there are extensive charge interactions between ICAM-1 and HRV16 (Fig. 3D). Lys-29 interacts with HRV16 residue Asp-1213 (see Fig. 3D for nomenclature of amino acid numbering), which is part of the flexible loop that has been postulated to regulate the presence of a stabilizing cellular “pocket factor”—possibly a small fatty acid—by association with receptor (16). Analysis of the conservation of HRV16 residues that interact with ICAM-1 (Fig. 3D) does not show any obvious differentiation between the major and minor groups of rhinoviruses. We conclude that possibly both major and minor groups should be able to bind ICAM-1 to some extent, notwithstanding competition-binding results (c.f. ref. 16).
Figure 3.
Interpretation of the cryo-EM electron density for HRV16 complexed with a D1D2 fragment of ICAM-1 expressed in Chinese hamster ovary cells (14). (A) Stereoview of the cryo-EM electron density of the complex (orange) fitted with the mutICAM-1 Cα backbone. The extra electron density regions around D2 of ICAM-1 correspond to the predicted locations of the four glycosylation sites. Domain D1 is not glycosylated. (B) Stereoview of the cryo-EM density of HRV16 (green) with the ICAM-1 Cα backbone, which can be seen to fit into the canyon depression on the HRV16 surface. (C) Ribbon diagram showing the interaction of ICAM-1 (yellow) and HRV16. HRV16 proteins VP1, VP2, and VP3 are in blue, green, and red, respectively. Two symmetry-related VP1s and VP3s are shown. Some icosahedral symmetry elements and the boundary of an icosahedral asymmetric unit are shown. (D) Diagrammatic figure of the fold of ICAM-1 domain D1 (yellow). ICAM-1 residues that could make salt bridges in the HRV16 complex are in black. The opposing residues on HRV16 are colored blue, green, and red and are numbered starting at 1001, 2001, 3001 according to whether they are in viral proteins VP1, VP2, or VP3, respectively. There are 60 copies of each viral protein in the HRV16 complex.
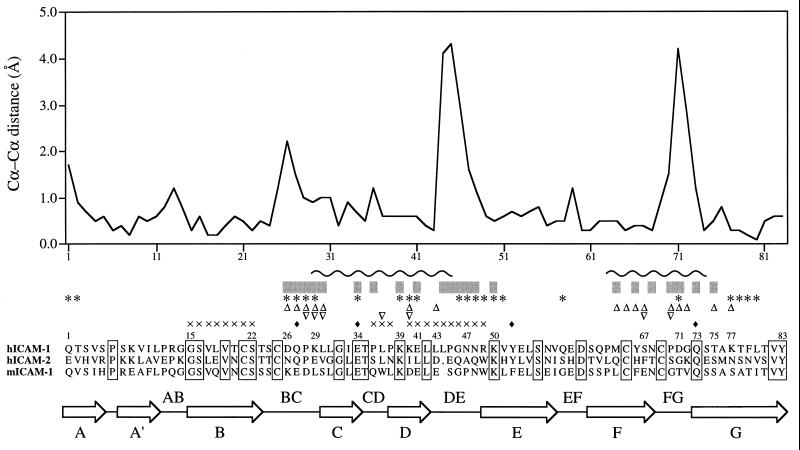
Superposition of domain D1 of ICAM-2 onto domain D1 of mutICAM-1 gave a structure-based amino acid sequence alignment that differs slightly from the predicted alignment (24) in the vicinity of an amino acid insertion in mutICAM-1. A plot of the distance between superimposed, equivalenced Cα atoms between mutICAM-1 and ICAM-2 in domain D1 shows three short regions where there is a substantial and significant difference in structure (Fig. 4); these regions correspond to the BC, DE, and FG loops that are identified by the cryo-EM studies as being the site of binding to HRV16 (14) and HRV14 (unpublished results). Hence, by far the largest conformational changes between mutICAM-1 and ICAM-2 occur at those sites of ICAM-1 that interact with HRVs, suggesting why ICAM-1, but not ICAM-2 or ICAM-3, binds to rhinoviruses. A comparison of the amino acid sequences of human ICAM-1 with murine ICAM-1 and other human ICAMs at the BC, DE, and FG loops (Fig. 4) shows major differences in the disposition of proline and charged residues. These are likely to cause large conformational and functional differences between human ICAM-1 and the other ICAMs.
Figure 4.
Structural alignment of D1 of mutICAM-1 with that of ICAM-2. The plot is of differences in Cα positions for the superimposed structures. By far, the largest differences occur at the BC, DE, and FG loops, all of which are at the amino end of the domain and important for binding of HRVs to human (h) ICAM-1 [but not human (h) ICAM-2 or murine (m) ICAM-1]. Residues below the gray bars interact with HRV16 when mutICAM-1 is fitted into the cryo-EM electron density map of the virus–receptor complex. ICAM-1 residues identified by mutational studies as being involved in binding HRVs are marked with ∗ (11), ▵ (12), and ▿ (13). Residues marked with ♦ have been identified by single amino acid mutations as being important in binding LFA-1 (24). Residues probably involved in binding to PFIE are marked with × (11). The wavy line (∼) indicates residues that interact with LFA-1 for the docking shown in Fig. 5. Sequence identities are shown in boxes.
Loops identified as making contact between HRV16 and human ICAM-1 correspond with the mutational studies of Register et al. (13), who showed that mutations of residues in the BC and FG loops had the greatest effect on the binding of six different HRV serotypes (Fig. 4). Mutational and immunological studies of McClelland et al. (12) also pointed to interaction between ICAM-1 and major-group HRVs in the FG loop and in β-strand D. Staunton et al. (11) reported mutational changes in the BC, CD, DE, and FG loops, but also elsewhere on the molecule, that affect HRV binding.
Adhesion of ICAM-1 to LFA-1.
LFA-1 binds to domain D1 of ICAM-1, ICAM-2, and ICAM-3 (9, 11, 24, 34). Hence, its site of binding on ICAM-1 should have little conformational or sequence differences when compared with the corresponding site on ICAM-2. It follows that the LFA-1-binding site on ICAMs should be distinct from that of the HRV-binding site on ICAM-1. The latter corresponds to the largest structural differences with ICAM-2. The separation of the HRV- and LFA-1-binding sites on ICAM-1 is consistent with mutational and immunological data (4, 5). There is a consensus, based on mutational studies, that Glu-34 (on β-strand C) and Gln-73 (on β-strand F) strongly influence LFA-1 binding to ICAM-1 (4, 11). The equivalent residues have also been identified for binding of LFA-1 to ICAM-2 (24) and ICAM-3 (34). Other residues that have been found to affect binding of LFA-1 to ICAM molecules are mostly on the side of the Ig β-barrel formed by the antiparallel β-strands C, F, and G (4).
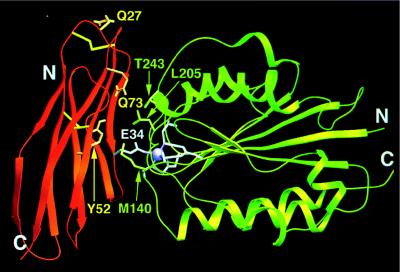
It was suggested by Lee et al. (17) that a divalent cation, associated with the I-domain of LFA-1, may coordinate with Glu-34 of ICAM-1. The I-domain has a “nucleotide binding fold” (35) with the metal-binding site in the usual substrate-binding site at the carboxyl-terminal end of the six-stranded parallel β-sheet. Docking of the I-domain of human LFA-1 (36), with the metal ion liganding to Glu-34 of ICAM-1, is highly restrictive if steric conflicts are to be avoided. A model of D1 mutICAM-1 docked with the LFA-1 I-domain was built by rotating D1 around an axis about 45° to the CFG sheet, while keeping Glu-34 coordinated with the metal ion and minimizing steric conflicts. In the best model (Fig. 5), ICAM-1 residue Gln-73 is associated with residue Thr-243 on the surface of the LFA-1 I-domain; hydrophobic residues Leu-30, Pro-36, Leu-44, and Met-64 on ICAM-1 and Met-140 and Leu-205 of LFA-1 become involved in the docking interface, consistent with the mutational results on ICAM-1 (11) and the suggestions of Qu and Leahy (36) for LFA-1. The entry and exit of the integrin α chain into the I-domain are conveniently on the opposite side of the interaction with ICAM-1.
Figure 5.
Ribbon diagram showing docking of the I-domain of LFA-1 (green) with domain D1 of mutICAM-1 (orange). Coordination of the metal ion (purple) on the I-domain is completed by Glu-34 (white) on the β-strand C of mutICAM-1. Additional residues of the I-domain (36) and of ICAM-1 (24) considered important for binding are shown in green and yellow, respectively.
There have been several reports that ICAM-1 forms dimers and that this dimerization results in an enhanced affinity for LFA-1 (4, 18). The stoichiometry of this union is, however, unknown and could imply an interaction between a dimer of ICAM-1 with either one or two LFA-1 molecules. In the former case, the second ICAM-1/LFA-1 contact could involve one of the additional cation-binding sites identified on LFA-1 integrin (37). In the crystal structure described here, there is an extensive antiparallel β-sheet created by the β-strand G of molecules related by a crystallographic twofold axis (Fig. 2). This may be of biological significance, although such dimerization of Ig domains also occurs in some crystal structures and may be merely a lattice effect. Nevertheless, the dimer extends the three-stranded antiparallel sheet (CFG) to one of six strands, (CFGGFC) with four main-chain hydrogen bonds between the two monomers. The critical residue, Glu-34, is in both monomers pointing out and away from the surface of this extended β-sheet. The docking of the I-domain of LFA-1 to ICAM-1 as described above places the two I-domains so as to make extensive contacts between them when associated with the ICAM-1 dimer. Such a structure would increase the area of contact in an (ICAM-1)2:(LFA-1)2 complex.
Other ICAM-1 Functions.
The binding site on ICAM-1 for malaria-infected erythrocytes is located in domain D1 but is distinct from the binding sites for LFA-1 and HRVs (4, 5). Immunological peptide scans and mutagenesis have shown that ICAM-1 residues 15–19 (5), 20–22 (4), and 40–49 (4), and possibly also 36–38 (4), can bind to antibodies that block binding to infected erythrocytes. These residues are primarily on the β-strand B and on the CD and the DE loops, and are, therefore, away from the LFA-1-binding site, but partially overlap the HRV-binding site (Figs. 1 and 2). This region is fairly variable in amino acid sequence among ICAM molecules and might account for the specificity of infected erythrocytes binding to only human ICAM-1.
No specific function has yet been associated with domain D2 of ICAM-1, other than its role as a spacer to extend the functional domain D1 away from the cell surface. Thus, the evolution of this domain may be less constrained by a need to maintain multiple functions. This is evident in the bigger structural differences between the D2 domains of mutICAM-1 and ICAM-2 compared with those for domain D1. The difference of 15° in angular relationship of the two domains in mutICAM-1 compared with those in ICAM-2 is caused by the conformational changes in domain D2. The contact surface between domains D1 and D2 is complementary in shape. In addition, there is a salt bridge between Arg-13 on D1 and Glu-87 on D2 that to some extent “freezes” the elbow angle and supports the fit of the D1D2 structure into the cryo-EM density without alteration of the elbow angle between the two domains.
The structure of mutICAM-1 offers a molecular interpretation of the interactions between HRVs and their cellular receptor. This may further guide the development of antiviral compounds that alter the ability of HRVs to bind to ICAM-1 (38) or by blocking the receptor-binding site on the virus by ICAM-1 mimics. The structure also suggests modes of interaction between ICAM-1 and LFA-1 required for cell adhesion and utilized to capture leukocytes at the site of an injury or infection.
Acknowledgments
We are grateful for outstanding help with data collection by Robert Sweet at the X12C beam line and Craig Ogata at the X4A beam line of the Brookhaven National Light Source, and the staff of the Cornell High Energy Synchrotron Source. We greatly appreciate the use of the cryo-EM maps provided by Norman Olson and Timothy Baker of Purdue University. We thank Karl Wood and R. Graham Cooks of Purdue University for their carbohydrate analysis of mutICAM-1, as well as Carla Forte, Susan Barkowski-Clark, Robert Kuhn, and Art Robbins at Bayer Biotechnology. We acknowledge the assistance of Cheryl Towell and Sharon Wilder in the preparation of this manuscript. The work was supported by a National Institutes of Health grant to M.G.R., a Jane Coffin Childs postdoctoral fellowship to P.R.K., a Lucille P. Markey Foundation grant for the advancement of structural biology at Purdue University, and a Purdue University reinvestment grant.
ABBREVIATIONS
- ICAM-1
human intercellular adhesion molecule-1
- LFA-1
leukocyte function-associated antigen
- HRV
human rhinovirus
- cryo-EM
cryoelectron microscopy
- Mac-1
macrophage-1 antigen
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (reference 1iam).
References
- 1.van de Stolpe A, van der Saag P T. J Mol Med. 1996;74:13–33. doi: 10.1007/BF00202069. [DOI] [PubMed] [Google Scholar]
- 2.Greve J M, Davis G, Meyer A M, Forte C P, Yost S C, Marlor C W, Kamarck M E, McClelland A. Cell. 1989;56:839–847. doi: 10.1016/0092-8674(89)90688-0. [DOI] [PubMed] [Google Scholar]
- 3.Staunton D E, Merluzzi V J, Rothlein R, Barton R, Marlin S D, Springer T A. Cell. 1989;56:849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- 4.Berendt A R, McDowall A, Craig A G, Bates P A, Sternberg M J E, Marsh K, Newbold C I, Hogg N. Cell. 1992;68:71–81. doi: 10.1016/0092-8674(92)90207-s. [DOI] [PubMed] [Google Scholar]
- 5.Ockenhouse C F, Betageri R, Springer T A, Staunton D E. Cell. 1992;68:63–69. doi: 10.1016/0092-8674(92)90206-r. [DOI] [PubMed] [Google Scholar]
- 6.Dustin M L, Rothlein R, Bhan A K, Dinarello C A, Springer T A. J Immunol. 1986;137:245–254. [PubMed] [Google Scholar]
- 7.Simmons D, Makgoba M W, Seed B. Nature (London) 1988;331:624–627. doi: 10.1038/331624a0. [DOI] [PubMed] [Google Scholar]
- 8.Marlin S D, Springer T A. Cell. 1987;51:813–819. doi: 10.1016/0092-8674(87)90104-8. [DOI] [PubMed] [Google Scholar]
- 9.Landis R C, McDowall A, Holness C L L, Littler A J, Simmons D L, Hogg N. J Cell Biol. 1994;126:529–537. doi: 10.1083/jcb.126.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diamond M S, Garcia-Aguilar J, Bickford J K, Corbi A L, Springer T A. J Cell Biol. 1993;120:1031–1043. doi: 10.1083/jcb.120.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staunton D E, Dustin M L, Erickson H P, Springer T A. Cell. 1990;61:243–254. doi: 10.1016/0092-8674(90)90805-o. [DOI] [PubMed] [Google Scholar]
- 12.McClelland A, deBear J, Yost S C, Meyer A M, Marlor C W, Greve J M. Proc Natl Acad Sci USA. 1991;88:7993–7997. doi: 10.1073/pnas.88.18.7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Register R B, Uncapher C R, Naylor A M, Lineberger D W, Colonno R J. J Virol. 1991;65:6589–6596. doi: 10.1128/jvi.65.12.6589-6596.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olson N H, Kolatkar P R, Oliveira M A, Cheng R H, Greve J M, McClelland A, Baker T S, Rossmann M G. Proc Natl Acad Sci USA. 1993;90:507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoover-Litty H, Greve J M. J Virol. 1993;67:390–397. doi: 10.1128/jvi.67.1.390-397.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossmann M G. Protein Sci. 1994;3:1712–1725. doi: 10.1002/pro.5560031010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J, Rieu P, Arnaout M A, Liddington R. Cell. 1995;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- 18.Reilly P L, Woska J R, Jr, Jeanfavre D D, McNally E, Rothlein R, Bormann B-J. J Immunol. 1995;155:529–532. [PubMed] [Google Scholar]
- 19.Ryu S E, Kwong P D, Truneh A, Porter T G, Arthos J, Rosenberg M, Dai X, Xuong N, Axel R, Sweet R W, Hendrickson W A. Nature (London) 1990;348:419–426. doi: 10.1038/348419a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Yan Y, Garrett T P J, Liu J, Rodgers D W, Garlick R L, Tarr G E, Husain Y, Reinherz E L, Harrison S C. Nature (London) 1990;348:411–418. doi: 10.1038/348411a0. [DOI] [PubMed] [Google Scholar]
- 21.Jones E Y, Davis S J, Williams A F, Harlos K, Stuart D I. Nature (London) 1992;360:232–239. doi: 10.1038/360232a0. [DOI] [PubMed] [Google Scholar]
- 22.Jones E Y, Harlos K, Bottomley M J, Robinson R C, Driscoll P C, Edwards R M, Clements J M, Dudgeon T J, Stuart D I. Nature (London) 1995;373:539–544. doi: 10.1038/373539a0. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Pepinsky R B, Stehle T, Liu J, Karpusas M, Browning B, Osborn L. Proc Natl Acad Sci USA. 1995;92:5714–5718. doi: 10.1073/pnas.92.12.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casasnovas J M, Springer T A, Liu J, Harrison S C, Wang J. Nature (London) 1997;387:312–315. doi: 10.1038/387312a0. [DOI] [PubMed] [Google Scholar]
- 25.Harpaz Y, Chothia C. J Mol Biol. 1994;238:528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- 26.Kolatkar P R, Oliveira M A, Rossmann M G, Robbins A H, Katti S K, Hoover-Litty H, Forte C, Greve J M, McClelland A, Olson N H. J Mol Biol. 1992;225:1127–1130. doi: 10.1016/0022-2836(92)90110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossmann M G. Acta Crystallogr. 1961;14:383–388. [Google Scholar]
- 28.Hendrickson W A. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 29.Bella J, Rossmann M G. Acta Crystallogr. 1998;D54:159–174. doi: 10.1107/s0907444997010469. [DOI] [PubMed] [Google Scholar]
- 30.Navaza J. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- 31.Brünger A T. x-plor, Version 3.1 Manual: A System for X-ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 32.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. San Diego: Academic; 1996. [Google Scholar]
- 33.Colonno R J, Condra J H, Mizutani S, Callahan P L, Davies M E, Murcko M A. Proc Natl Acad Sci USA. 1988;85:5449–5453. doi: 10.1073/pnas.85.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holness C L, Bates P A, Little A J, Buckley C D, McDowall A, Bossy D, Hogg N, Simmons D L. J Biol Chem. 1995;270:877–884. doi: 10.1074/jbc.270.2.877. [DOI] [PubMed] [Google Scholar]
- 35.Rossmann M G, Moras D, Olsen K W. Nature (London) 1974;250:194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- 36.Qu A, Leahy D J. Proc Natl Acad Sci USA. 1995;92:10277–10281. doi: 10.1073/pnas.92.22.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanley P, Bates P A, Harvey J, Bennett R I, Hogg N. EMBO J. 1994;13:1790–1798. doi: 10.1002/j.1460-2075.1994.tb06447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hadfield A T, Oliveira M A, Kim K H, Minor I, Kremer M J, Heinz B A, Shepard D, Pevear D C, Rueckert R R, Rossmann M G. J Mol Biol. 1995;253:61–73. doi: 10.1006/jmbi.1995.0536. [DOI] [PubMed] [Google Scholar]