Abstract
A novel Alu-long terminal repeat (LTR)-based real-time nested-PCR assay was developed to quantify integrated human immunodeficiency virus type 1 (HIV-1) DNA in infected cells with both accuracy and high sensitivity (six proviruses within 50,000 cell equivalents). Parallel assays for total HIV-1 DNA and two-LTR HIV-1 DNA circles allowed the synthesis and fate of the different HIV-1 DNA species to be monitored upon a single round of viral replication.
The RNA genome of human immunodeficiency virus type 1 (HIV-1) is, like those of other retroviruses, reverse transcribed in the cytoplasm into a double-stranded linear DNA containing a copy of the long terminal repeats (LTR) at each terminus. The resulting linear DNA molecule moves into the nucleus as a component of the preintegration complex, where it may integrate into the host cell genome (17). This process is dependent on viral integrase activity, which is essential for a productive infection (7, 8, 13, 16, 19, 21). In addition to linear DNA species, HIV-1 DNA circles with one or two LTR are detected in the nucleus (1, 11).
While the unintegrated forms of HIV-1 DNA can be readily detected by using standard PCR or Southern hybridization techniques, quantitative analysis of integrated HIV-1 DNA has been hampered by the lack of a reliable assay. Early studies have attempted to quantify integrated viral DNA by PCR using one primer annealing in the HIV-1 LTR and a second in the highly repeated chromosomal Alu element (Alu-LTR PCR) (6, 18). Alu repeated sequences are interspersed in the human genome and number at least 900,000 copies per haploid genome, giving an average distance of 4 kb between Alu elements (2). However, previous Alu-LTR PCR methods have limited sensitivity and do not take into account that the distance between an integrated viral DNA and its nearest Alu repeat is variable.
To detect and accurately quantify integrated HIV-1 DNA with high sensitivity, we have developed a new method, based on Alu-LTR real-time nested PCR. In conjunction with the study of HIV-1 DNA integration, real-time quantitative PCR was used to analyze the synthesis and fate of total HIV-1 DNA and two-LTR HIV-1 circles during a single-round of viral replication.
Generation of an integrated HIV-1 DNA standard.
To accurately determine the integrated HIV-1 DNA copy number within a sample, the standard should be representative of a natural viral infection and contain numerous integration sites with a wide distribution of distances between the provirus LTR and the nearest Alu sequence. To generate a proper integrated HIV-1 DNA standard, HeLa cells were infected with a Δenv HIV-1 R7 Neo virus pseudotyped with the G glycoprotein from vesicular stomatitis virus (VSV-G). VSV-G-pseudotyped HIV-1 R7 Neo virus stocks were prepared by cotransfection into 293T cells of the pR7 Neo Δenv vector (gift from U. Hazan) with an expression vector encoding VSV-G. The pR7 Neo Δenv vector was constructed by deleting the envelope coding sequence of the HIV-1 R7 Neo genome (9), in which a neomycin resistance gene replaces the HIV-1 nef coding sequence. Infected cells were then cultured for several weeks in the presence of G418 (500 μg/ml) to select cells that contained integrated viral DNA and to allow the loss of all unintegrated forms. Ten days after initiation of the G418 selection, neomycin-resistant cell clones were counted; they numbered approximately 5,000. For our standard cell line, designated HeLa R7 Neo, the copy number of integrated viral DNA matched the total HIV-1 DNA copy number. Thus, based on total HIV-1 DNA and β-globin quantifications (see below), we estimated that the standard cell line contained 1.24 ± 0.03 proviruses per cell (data not shown).
Alu-LTR-based real-time nested-PCR procedure.
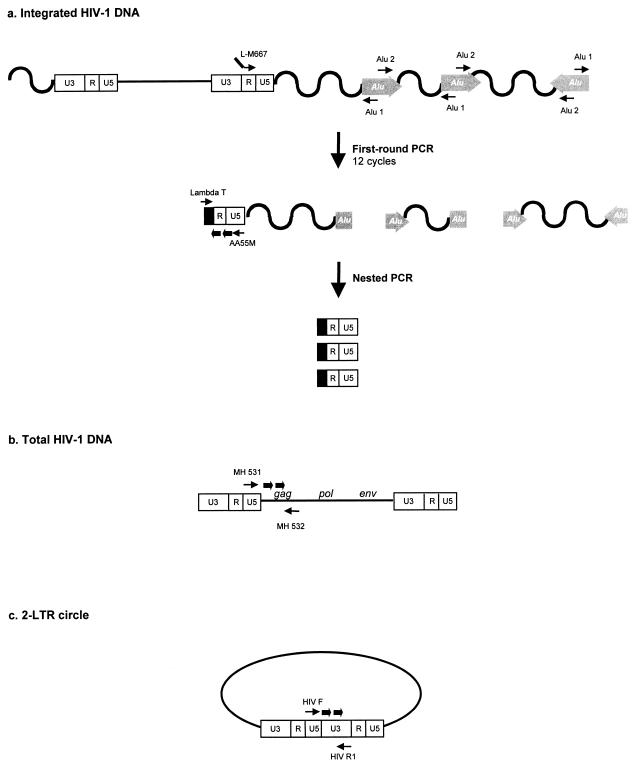
Amplification reactions were performed with the Light Cycler instrument (Roche Diagnostics, Meylan, France). In a first round of PCR, integrated HIV-1 sequences were amplified with two outward-facing Alu primers that anneal within conserved regions of the Alu repeat element together with an HIV-1 LTR-specific primer (Fig. 1a). As previously proposed (18), the use of two outward-facing Alu primers optimizes the probability of amplifying an LTR sequence, as Alu elements are present in either orientation relative to the integrated provirus (Fig. 1a). In addition, we used an LTR primer extended with a lambda phage-specific heel sequence at the 5′ end of the oligonucleotide (L-M667) in this first amplification step (Fig. 1a). Alu-LTR sequences were amplified in duplicate from 1/50 of totalcell DNA in a 20-μl reaction mixture comprising 1× Light Cycler Fast Start DNA master hybridization probes (Roche), 4 mM MgCl2, 100 nM L-M667 primer, and 300 nM (each) primers Alu 1 and Alu 2 (18) (Table 1). Given the high number of Alu elements within the human genome, abundant amplifications of inter-Alu sequences occurred simultaneously with the amplification of Alu-LTR sequences. To remain in the exponential phase, only 12 cycles of amplification were performed. Thus, the first-round PCR cycle conditions were as follows: a denaturation step of 8 min at 95°C and then 12 cycles of amplification (95°C for 10 s, 60°C for 10 s, and 72°C for 170 s).
FIG. 1.
Real-time PCR strategies and location of primers and probes for the quantification of integrated HIV-1 DNA (a), total HIV-1 DNA (b), and two-LTR circles (c). Thin arrows, primers; thick arrows, probes.
TABLE 1.
Primer and probe sequences
| Primer or probec | Sequence (5′-3′) | Target |
|---|---|---|
| MH 531 | TGTGTGCCCGTCTGTTGTGT | Total HIV-1 DNA |
| MH 532 | GAGTCCTGCGTCGAGAGAGC | |
| MH FL* | CCCTCAGACCCTTTTAGTCAGTGTGGAAa | |
| MH LC* | TCTCTAGCAGTGGCGCCCGAACAGb | |
| HIV F | GTGCCCGTCTGTTGTGTGACT | Two-LTR circle |
| HIV R1 | ACTGGTACTAGCTTGTAGCACCATCCA | |
| HIV FL* | CCACACACAAGGCTACTTCCCTGAa | |
| HIV LC* | TGGCAGAACTACACACCAGGGCb | |
| L-M667 | ATGCCACGTAAGCGAAACTCTGGCTAACTAGGGAACCCACTG | Integrated HIV-1 DNA (first-round PCR) |
| Alu 1 | TCCCAGCTACTGGGGAGGCTGAGG | |
| Alu 2 | GCCTCCCAAAGTGCTGGGATTACAG | |
| Lambda T | ATGCCACGTAAGCGAAACT | Integrated HIV-1 DNA (second-round PCR) |
| AA55M | GCTAGAGATTTTCCACACTGACTAA | |
| LTR FL* | CACAACAGACGGGCACACACTACTTGAa | |
| LTR LC* | CACTCAAGGCAAGCTTTATTGAGGCb |
Modified with fluorescein at the 3′ end.
Phosphorylated at the 3′ end and modified with LC red 640 dye at the 5′ end.
Primers and probes were purchased from TIB MOLBIOL (Berlin, Germany). *, probe sequence.
In a second round of PCR using the lambda-specific primer (Lambda T) and an LTR primer (AA55M), only products from the first-round PCR could be amplified (Fig. 1a). In contrast to the first-round PCR, which generated fragments of various lengths, the nested amplification resulted in discrete DNA fragments. Nested PCR was performed on 1/10 of the first-round PCR product in a mixture comprising 1× Light Cycler Fast Start DNA master hybridization probes, 4 mM MgCl2, 300 nM Lambda T primer, 300 nM AA55M primer, and 200 nM (each) hybridization probes LTR FL and LTR LC (Table 1). The nested-PCR cycling profile began with a denaturation step (95°C for 8 min), followed by 50 cycles of amplification (95°C for 10 s, 60°C for 10 s, and 72°C for 9 s).
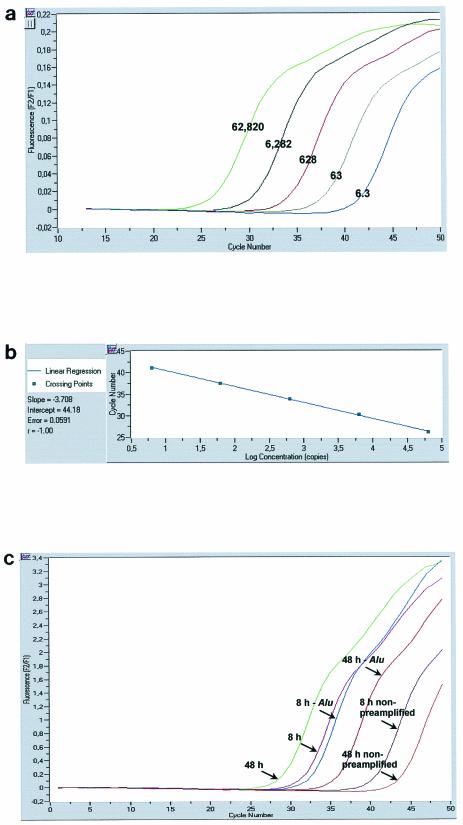
The copy number of integrated HIV-1 DNA was determined in reference to a standard curve generated by concomitant two-stage PCR amplification of a serial dilution of the standard HeLa R7 Neo cell DNA (Fig. 2a) mixed with uninfected-cell DNA to yield 50,000 cell equivalents. Quantification was achieved by the second-derivative maximum method provided by the Light Cycler quantification software, version 3.5 (Roche Diagnostics). In our system, the linear regression obtained from amplification of serial dilutions of the integrated HIV-1 DNA standard was linear over a 5-log10-unit range, and our Alu-LTR nested-PCR procedure allowed the detection of approximately six proviruses within 50,000 cell equivalents (Fig. 2b).
FIG. 2.
Characteristics of Alu-LTR nested PCR. (a) Fluorescence curves generated by two-step amplification of serial dilutions of HeLa R7 Neo cell DNA. The copy numbers for each standard dilution are shown over the corresponding fluorescence curve. (b) Linear regression for quantifying integrated HIV-1 DNA. (c) Specificity of the Alu-LTR nested PCR. Fluorescence curves generated by amplification of viral DNA from CEM cells infected with the VSV-G-pseudotyped HIV-1 R7 Neo virus and collected in the presence of Alu primers (pink, 8 h p.i.; green, 48 h p.i.), in the absence of Alu primers (blue, 8 h p.i.; red, 48 h p.i.), or without a preamplification step (purple, 8 h p.i.; orange, 48 h p.i.) are shown.
Specificity of the Alu-LTR nested-PCR assay.
During the first-round PCR, the L-M667 oligonucleotide can prime the formation of a single-stranded DNA from all LTR containing HIV-1 DNA, leading to an overestimation of the actual integrated HIV-1 DNA copy number. To control these linear amplifications, we performed a nested-PCR procedure omitting or not Alu primers in the first-round PCR. Figure 2c displays fluorescence curves obtained during amplification of cell DNA from CEM lymphoid cells infected with the VSV-G-pseudotyped HIV-1 R7 Neo virus and collected 8 or 48 h postinfection (p.i.). In the presence of Alu primers, the fluorescence curves shifted, moderately but reproductively, to the left, compared to an amplification performed in the absence of Alu primers (Fig. 2c). The shifts were about 1 cycle and 7 cycles, corresponding to 2-fold and 50-fold increases in copy numbers, for the 8- and 48-h time points, respectively. Furthermore, to determine the extent of second-round amplification of nonpreamplified viral DNA, we performed an additional control where the first-round PCR was replaced by a 4°C incubation of the PCR mixture. As expected, the amplification curves shifted dramatically to the left when a preamplification step was performed. The shift corresponded to approximately 2.3- (9 cycles) and 3.5-log10-unit (14 cycles) increases in copy numbers for the 8- and 48-h time points, respectively. Taken together, these findings indicate that the PCR signal obtained in the absence of Alu primers must be subtracted from the total signal, especially at early times following infection, and that the second-round amplification of nonpreamplified viral DNA is efficiently prevented.
Synthesis and fate of HIV-1 DNA species in a single-round infection assay.
To validate our integration assay, we quantified integrated HIV-1 DNA, together with total HIV-1 DNA and two-LTR circles in a single round of viral replication. Eighteen million CEM cells were infected by VSV-G-pseudotyped HIV-1 R7 Neo virus by using 35 ng of p24gag antigen (measured by enzyme-linked immunosorbent assay; Perkin-Elmer Life Sciences, Paris, France) per 106 cells. One hour p.i., cells were washed in phosphate-buffered saline (PBS), exposed to trypsin (25 μg/ml) for 1 min at 37°C, and washed once with medium and twice with PBS. The cells were then cultured in RPMI 1640 supplemented with 10% fetal calf serum. At each time point, 1 × 106 to 3 × 106 infected cells were collected. To eliminate residual pR7 Neo Δenv vector DNA, the samples were washed in PBS and incubated with 750 to 1,500 U of DNase I (Invitrogen, Cergy Pontoise, France) in a buffer comprising 20 mM Tris-Cl, pH 8.3, 50 mM KCl, and 2 mM MgCl2 for 1 h at room temperature. Cells were washed in PBS, and dry cell pellets were frozen at −80°C until use. Total cell DNA was extracted with a QIAamp blood DNA minikit (Qiagen, Courtaboeuf, France).
The total HIV-1 DNA copy number was determined with previously described primers (4) that annealed in the U5 region of the LTR (MH 531) and in the 5′ end of the gag gene (MH 532) (Fig. 1b). The two-LTR circles were amplified with primers spanning the LTR-LTR junction (HIV F and HIV R1), as described elsewhere (3) (Fig. 1c). U5-gag sequences and two-LTR junctions were amplified in duplicate from 1/50 of total cell DNA. Reaction mixtures contained 1× Light Cycler Fast Start DNA master hybridization probes (Roche Diagnostics), 4 mM MgCl2, 300 nM (each) forward and reverse primers, and 200 nM (each) fluorogenic hybridization probes in a final volume of 20 μl. After an initial denaturation step (95°C for 8 min), the cycling profile for total HIV-1 DNA was 50 cycles consisting of 95°C for 10 s, 60°C for 10 s, and 72°C for 6 s and that for two-LTR circles was 15 cycles consisting of 95°C for 10 s, 66°C for 10 s, and 72°C for 10 s, followed by 35 cycles at the beginning of which the annealing temperature was decreased by 0.5°C per cycle to the secondary target temperature (59°C). The copy numbers of total HIV-1 DNA and two-LTR circles were determined in reference to a standard curve prepared by amplification of quantities ranging from 10 to 105 copies of cloned DNA with matching sequences. The cell equivalents in sample DNA were calculated based on the amplification of the β-globin gene (two copies per diploid cell) with the Light Cycler instrument and commercially available materials (Control kit DNA; Roche Diagnostics). The quantification results for two-LTR circle, total HIV-1 DNA, and integrated HIV-1 DNA were expressed as copy numbers per 106 cells.
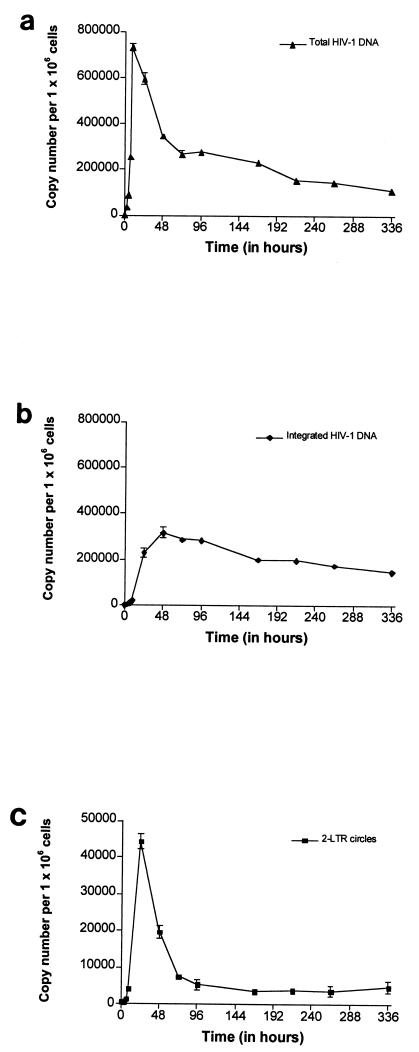
Viral DNA was detected by 3 h p.i. The level of total HIV-1 DNA increased rapidly until 9 h p.i., reaching a maximum of 734,996 copies per 106 cells (Fig. 3a). Then the level of total HIV-1 DNA underwent a steep decrease until 72 h p.i., followed by a slow decay phase. In previous single-round infection assays, the initial steep decrease in the level of total HIV-1 DNA was reported to represent the proteasome-mediated degradation of unintegrated linear HIV-1 DNA (5). We detected integrated HIV-1 DNA by 3 h p.i., attesting to the rapid nuclear import of reverse transcription products and emphasizing the sensitivity of our integration assay. The level of integrated HIV-1 DNA increased until 48 h p.i to reach a maximum of 316,578 copies per 106 cells (Fig. 3b). By 72 h p.i., the level of integrated HIV-1 DNA matched that of total HIV-1 DNA, as expected in a single-round of viral replication. This confirmed the accuracy of our quantification method. The slow decay in the level of integrated HIV-1 DNA may reflect the death of infected cells and/or the slow division rate of infected cells carrying an integrated provirus compared to that of uninfected ones. Two-LTR circles, as detected by the presence of LTR-LTR junctions, were observed as early as 3 h p.i. The level of two-LTR circles reached 44,253 copies per 106 cells by 24 h p.i. and declined thereafter, reaching a quasisteady level from 96 h p.i. until the end of the culture (Fig. 3c). Two-LTR circles were shown to be stable DNA forms incapable of self-replication, and, consequently, they are diluted as a function of cell division (5, 15). Accordingly, a dilution effect resulting from cell division may account for the decrease in the level of two-LTR circles observed before 96 h p.i. In contrast, steady LTR-LTR junction levels from 96 h p.i. until the end of the culture were unexpected. Nevertheless, the nonspecific integration of some two-LTR circles (less than 10% in this experiment) into the host cell genome might explain the stability of LTR-LTR junction levels. Integrated LTR-LTR junctions may have been transmitted from mother to daughter cells at each division, leading to steady LTR-LTR junction levels in spite of cell proliferation. Likewise, it was previously shown that some linear HIV-1 DNA could be integrated into the cell genome by an integrase-independent process (10). This finding indicates that detection of LTR-LTR junctions does not necessarily indicate the presence of unintegrated forms of HIV-1 DNA.
FIG. 3.
Quantification of total HIV-1 DNA (a), integrated HIV-1 DNA (b), and two-LTR circles (c) during a single round of replication of the VSV-G-pseudotyped HIV-1 R7 Neo virus in CEM cells. Values are means ± standard errors of the means. The depicted results were obtained from a representative experiment.
Using a new sensitive assay to quantify integrated viral DNA, we analyzed the integration kinetics of viral DNA following a single cycle of replication. Our integration kinetics were similar to those obtained by Butler et al., who employed a one-step real-time Alu-LTR PCR-based amplification method (4). However, we were able to detect integrated HIV-1 DNA as early as 3 h p.i., while the one-step quantification method allowed the detection of proviral DNA no earlier than 12 (5) or 24 h p.i. (4). The limited sensitivity of the one-step Alu-LTR PCR method (1,000 copies in 100,000 cell equivalents) should account for the delayed detection of integrated HIV-1 DNA. Recently, a second method for the quantification of integrated HIV-1 DNA was devised. This method is based on a linker-primer PCR (LP-PCR) amplification (12, 20). The LP-PCR method was shown to detect four copies of proviruses in 200,000 cell equivalents (12), allowing the detection of integrated HIV-1 DNA as early as 4 h p.i. Unfortunately, the LP-PCR protocol involves many experimental steps. More recently, a real-time nested-PCR method (14) which used one primer annealing in the HIV-1 gag sequence and a second annealing in the Alu element was described. This assay has a detection limit of one provirus per 10,000 cells, i.e., comparable to ours. However, this Alu-LTR PCR method did not prevent the second-round amplification of nonpreamplified HIV-1 DNA, and its range of linearity was less extensive than ours (2.5 versus 5 log10 units).
The rapidity and sensitivity of our Alu-LTR-based real-time nested PCR should allow the routine analysis of HIV-1 DNA integration and may be a valuable tool to test future integrase inhibitors or to evaluate the integration efficiency of retroviral vectors designed for gene therapy.
Acknowledgments
A. Brussel was supported by a fellowship from the Agence Nationale de Recherche sur le SIDA.
We thank U. Hazan for constant advice and help in this work. We acknowledge Olfert Landt (TIB MOLBIOL) for excellent technical assistance with the design of primers and hybridization probes. We also thank C. Petit, O. Delelis, S. Pierre, M. Alizon, and especially B. Canque for testing and optimizing the assay and C. Sonnenschein for reviewing the manuscript.
REFERENCES
- 1.Barbosa, P., P. Charneau, N. Dumey, and F. Clavel. 1994. Kinetic analysis of HIV-1 early replicative steps in a coculture system. AIDS Res. Hum. Retrovir. 10:53-59. [DOI] [PubMed] [Google Scholar]
- 2.Britten, R. J., W. F. Baron, D. B. Stout, and E. H. Davidson. 1988. Sources and evolution of human Alu repeated sequences. Proc. Natl. Acad. Sci. USA 85:4770-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brussel, A., D. Mathez, S. Broche-Pierre, R. Lancar, T. Calvez, P. Sonigo, and J. Leibowitch. 2003. Longitudinal monitoring of 2-long terminal repeat circles in peripheral blood mononuclear cells from patients with chronic HIV-1 infection. AIDS 17:645-652. [DOI] [PubMed] [Google Scholar]
- 4.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 5.Butler, S. L., E. P. Johnson, and F. D. Bushman. 2002. Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. J. Virol. 76:3739-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelman, A., G. Englund, J. M. Orenstein, M. A. Martin, and R. Craigie. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Englund, G., T. S. Theodore, E. O. Freed, A. Engleman, and M. A. Martin. 1995. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J. Virol. 69:3216-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feinberg, M. B., D. Baltimore, and A. D. Frankel. 1991. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl. Acad. Sci. USA 88:4045-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaur, M., and A. D. Leavitt. 1998. Mutations in the human immunodeficiency virus type 1 integrase D, D(35)E motif do not eliminate provirus formation. J. Virol. 72:4678-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, S. Y., R. Byrn, J. Groopman, and D. Baltimore. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 63:3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar, R., N. Vandegraaff, L. Mundy, C. J. Burrell, and P. Li. 2002. Evaluation of PCR-based methods for the quantitation of integrated HIV-1 DNA. J. Virol. Methods 105:233-246. [DOI] [PubMed] [Google Scholar]
- 13.LaFemina, R. L., C. L. Schneider, H. L. Robbins, P. L. Callahan, K. LeGrow, E. Roth, W. A. Schleif, and E. A. Emini. 1992. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 66:7414-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Doherty, U., W. J. Swiggard, D. Jeyakumar, D. McGain, and M. H. Malim. 2002. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J. Virol. 76:10942-10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pierson, T. C., T. L. Kieffer, C. T. Ruff, C. Buck, S. J. Gange, and R. F. Siliciano. 2002. Intrinsic stability of episomal circles formed during human immunodeficiency virus type 1 replication. J. Virol. 76:4138-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakai, H., M. Kawamura, J. Sakuragi, S. Sakuragi, R. Shibata, A. Ishimoto, N. Ono, S. Ueda, and A. Adachi. 1993. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J. Virol. 67:1169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherman, M. P., and W. C. Greene. 2002. Slipping through the door: HIV entry into the nucleus. Microbes Infect. 4:67-73. [DOI] [PubMed] [Google Scholar]
- 18.Sonza, S., A. Maerz, N. Deacon, J. Meanger, J. Mills, and S. Crowe. 1996. Human immunodeficiency virus type 1 replication is blocked prior to reverse transcription and integration in freshly isolated peripheral blood monocytes. J. Virol. 70:3863-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevenson, M., T. L. Stanwick, M. P. Dempsey, and C. A. Lamonica. 1990. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 9:1551-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vandegraaff, N., R. Kumar, C. J. Burrell, and P. Li. 2001. Kinetics of human immunodeficiency virus type 1 (HIV) DNA integration in acutely infected cells as determined using a novel assay for detection of integrated HIV DNA. J. Virol. 75:11253-11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiskerchen, M., and M. A. Muesing. 1995. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 69:376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]