Abstract
The transsynaptic retrograde transport of the pseudorabies virus Bartha (PRV-Bartha) strain has become an important neuroanatomical tract-tracing technique. Recently, dual viral transneuronal labeling has been introduced by employing recombinant strains of PRV-Bartha engineered to express different reporter proteins. Dual viral transsynaptic tracing has the potential of becoming an extremely powerful method for defining connections of single neurons to multiple neural circuits in the brain. However, the present use of recombinant strains of PRV expressing different reporters that are driven by different promoters, inserted in different regions of the viral genome, and detected by different methods limits the potential of these recombinant virus strains as useful reagents. We previously constructed and characterized PRV152, a PRV-Bartha derivative that expresses the enhanced green fluorescent protein. The development of a strain isogenic to PRV152 and differing only in the fluorescent reporter would have great utility for dual transsynaptic tracing. In this report, we describe the construction, characterization, and application of strain PRV614, a PRV-Bartha derivative expressing a novel monomeric red fluorescent protein, mRFP1. In contrast to viruses expressing DsRed and DsRed2, PRV614 displayed robust fluorescence both in cell culture and in vivo following transsynaptic transport through autonomic circuits afferent to the eye. Transneuronal retrograde dual PRV labeling has the potential to be a powerful addition to the neuroanatomical tools for investigation of neuronal circuits; the use of strain PRV614 in combination with strain PRV152 will eliminate many of the pitfalls associated with the presently used pairs of PRV recombinants.
Times of dramatic progress in neuroscience research have often been correlated with the development of new and powerful techniques that have changed the kinds of questions one can ask. The development of numerous neuroanatomical tract-tracing techniques applied singly or in combination has greatly expanded our understanding of the circuitry of the brain. With these data in hand, many functional studies have followed. The development of neurotropic alphaherpesviruses as transsynaptic tract tracers has now made it possible to examine ensembles of neurons that contribute to polysynaptic networks. The ability to describe multisynaptic neuronal circuits begins to bridge neuroanatomical and functional studies with a single technique.
Two strains of pseudorabies virus (PRV), PRV Becker and PRV-Bartha, have been used with great success to transsynaptically infect central nervous system (CNS) structures after peripheral application or direct injection into brain parenchyma (10, 19). PRV Becker is a highly virulent wild-type laboratory strain of PRV, whereas PRV-Bartha is an attenuated live vaccine strain. Pickard et al. have recently demonstrated that PRV-Bartha is transsynaptically transported in the retrograde direction only (i.e., from postsynaptic to presynaptic neuron) (15) despite earlier claims that after intravitreal injection, PRV-Bartha is transported anterogradely via the optic nerve (8). These observations are supported by recent genetic analyses indicating that due to the deletion of three genes that encode the membrane proteins gI, gE, and Us9, PRV-Bartha might be incapable of anterograde spread through chains of connected neurons (5, 9, 11, 20).
In the initial studies and often continuing to the present, PRV-Bartha-infected neurons have been identified using anti-PRV antibodies and standard immunocytochemical procedures. Although this technique yields excellent labeling with low levels of nonspecific staining and is amenable to electron microscopic analyses, PRV-infected neurons can be identified only after tissue fixation and the somewhat time-consuming and labor-intensive immunocytochemical processing of tissue sections. To eliminate the need for immunocytochemical tissue processing, Smith et al. previously constructed a PRV-Bartha recombinant, PRV152, which expresses the enhanced green fluorescent protein (EGFP) (16). EGFP diffuses throughout PRV152-infected neurons, filling the dendritic arbor completely and the axon to a great extent. The EGFP signal is very strong when viewed under standard epifluorescence light microscopy, and no additional tissue processing is required to visualize infected neurons (15). In addition to being efficient and inexpensive to use, PRV152-labeled neurons can be identified in brain slices in vitro for physiological studies (16). These studies indicate that PRV152 is a powerful tool for the transsynaptic labeling of neurons in defined CNS circuits that allows neurons to be identified in vitro by their expression of EGFP, analyzed electrophysiologically, and described in morphological detail.
It has been recognized for some time that a red fluorescent protein (RFP) would be highly desirable for use in multicolor labeling or fluorescence resonance energy transfer experiments. The RFP cloned from Discosoma coral, drFP583 (commercially known as DsRed), appeared to be a good candidate for a spectrally distinct companion for EGFP (14). However, slow and incomplete maturation and obligate tetramerization have hampered the evolution of DsRed as a useful tool (1, 22). Early attempts to address the rate and/or extent of maturation of DsRed (e.g., DsRed2) have provided only incremental improvements (CLONTECHniques XVI:2-3, 2001; Clontech, Palo Alto, Calif.). However, Tsien and colleagues (7) have recently described a monomeric RFP (mRFP1) derived from a rapidly maturing DsRed derivative, DsRed T1, isolated by Bevis and Glick (2). mRFP1 overcomes three critical problems associated with the wild-type tetramer of DsRed: (i) it is a monomer; (ii) it matures rapidly (about 10 times faster than DsRed); and (iii) the excitation and emission peaks are about 25 nm red shifted from DsRed, conferring greater spectral separation from autofluorescence and having minimal emission when excited at wavelengths optimal for EGFP (7). In addition, mRFP1 does not appear to generate a green intermediate. Although the fluorescence quantum yield and excitation coefficient for mRFP1 are relatively low compared to those of other DsRed variants (7), it holds great potential as a companion to EGFP for multicolor labeling.
In this report we describe the construction and characterization of recombinant PRV-Bartha strains that express a variety of RFPs. Our results indicate that one such recombinant virus, PRV614, which expresses mRFP1, promises to be an ideal companion for PRV152 for dual viral multisynaptic labeling applications.
For all recombinant viruses used in this study, the fluorescent protein expression cassette was inserted into the middle of the gG gene and was under the control of the cytomegalovirus major immediate-early promoter. A double-stranded DNA probe comprised of the PRV-Bartha SalI fragment, which encompasses the entire gG gene, was used to identify hybridizing BamHI genomic fragments from recombinant viruses. All of these recombinant viruses displayed similar hybridization profiles consistent with the expected recombination of the fluorescent protein expression cassette into the PRV-Bartha genome (data not shown).
The utility of RFP-expressing viruses as neuronal tract tracers is dependent on the capacity of viruses to replicate efficiently. All of the recombinant viruses used in the study grew to similar titers and displayed similar growth kinetics on PK15 cell monolayers, indicating that expression of EGFP, DsRed, DsRed2, and mRFP1 has no significant effect on virus replication compared to that seen with the parental PRV-Bartha strain (data not shown).
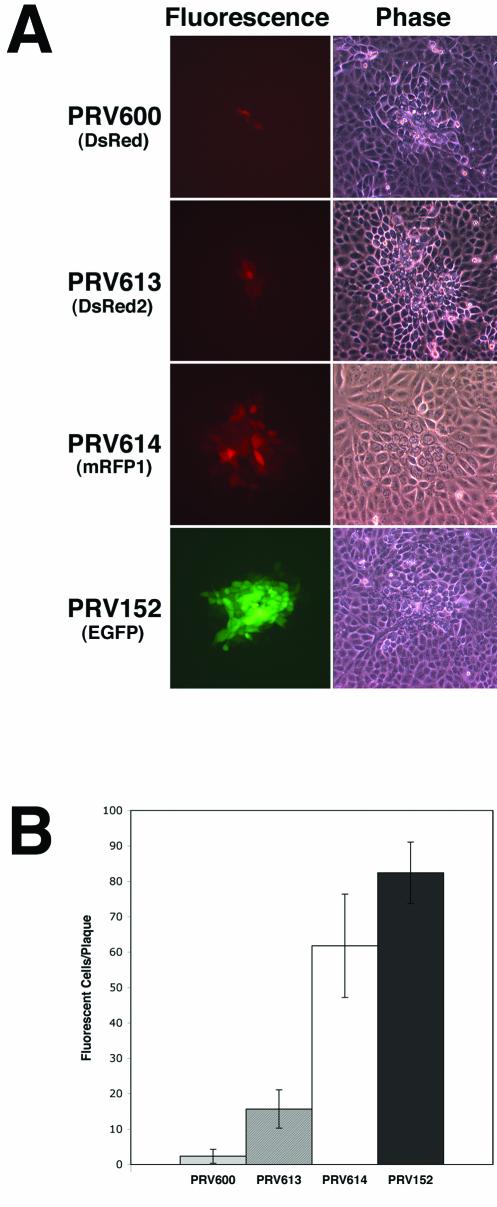
Differences in the ability to detect cells infected by PRV strains expressing different fluorescent proteins (as determined by fluorescence microscopy) were compared by examining plaques formed on PK15 cell monolayers (Fig. 1A). Consistent with the one-step growth analysis data, all viruses formed plaques of similar sizes (as determined by phase-contrast microscopy) at 17 h postinfection (Fig. 1A, right panels). By contrast, the fluorescence signals detected from cells infected with strains PRV600 (DsRed), PRV613 (DsRed2), PRV614 (mRFP1), and PRV152 (EGFP) differed considerably (Fig. 1A, left panels). At 17 h after infection, 12 random plaques formed by each virus strain were analyzed by fluorescence microscopy (Fig. 1B). An average of 2.3 infected cells expressing DsRed were detected in the center of PRV600 plaques, an average of 15.7 cells expressing DsRed2 were detected in plaques formed by PRV613, and an average of 61.8 cells were detected in plaques formed by PRV614. By comparison, an average of 82.4 cells expressing EGFP were detected at 17 h postinfection in plaques formed by PRV152. Taken together, these data indicate that the development of mRFP1 represents a substantial improvement over DsRed and DsRed2 as a red fluorescent reporter in the context of recombinant PRV infection of tissue culture cells.
FIG. 1.
Detection of infected PK15 cells by microscopy. (A) Fluorescence and phase-contrast images of PK15 cells 17 h after inoculation with strain PRV-Bartha derivatives expressing different fluorescent proteins. Cells were infected with strains PRV600, PRV613, PRV614, and PRV152. In the plaque assay, virus is added to monolayers of susceptible cells at a concentration of roughly 100 PFU per dish. The circular plaque that forms in the monolayer is a result of the spread of virus from a single infected cell to neighboring uninfected cells. PRV600 expresses DsRed, PRV613 expresses DsRed2, PRV614 expresses mRFP1, and PRV152 expresses EGFP. (B) Quantitation of the number of fluorescent cells per plaque at 17 h postinfection. Data were obtained through the analysis of 12 plaques per virus strain tested.
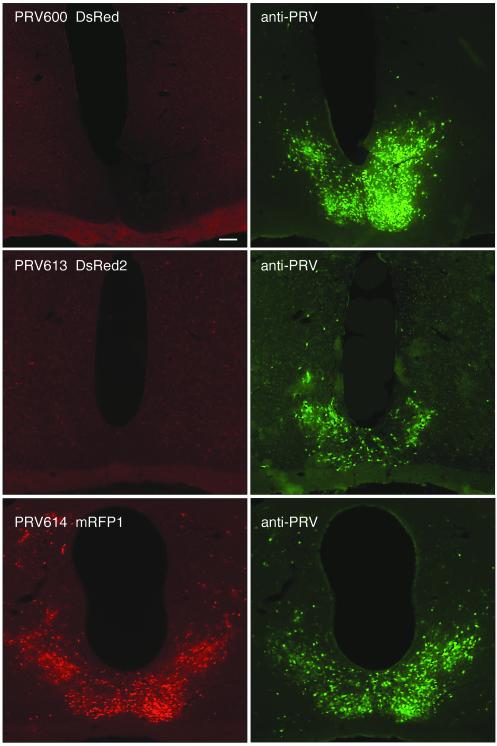
To evaluate RFP expression by the recombinant PRV strains in vivo, we examined regions in the brain (e.g., the suprachiasmatic nucleus [SCN], paraventricular nucleus [PVN], and intergeniculate leaflet [IGL]) that were infected after inoculation of the anterior chamber of the eye. To detect virus-infected neurons, a rabbit polyclonal antiserum generated against acetone-inactivated PRV (Rb133) was used followed by an Alexa-488 conjugated goat anti-rabbit second antibody (Molecular Probes). Images of coronal sections through the SCN from animals infected with viruses expressing different RFPs and killed 96 h after inoculation are provided in Fig. 2. All three viruses were able to replicate and spread to the SCN efficiently via retrograde transsynaptic transport, as evidenced by robust staining of SCN neurons with the anti-PRV antibody (Fig. 2, right panels). By contrast, direct RFP expression was only seen in the SCN of strain PRV614-infected animals. Note that no additional tissue processing (i.e., immunocytochemistry) or amplification procedures were necessary to detect PRV614 in vivo. Immunocytochemical labeling with the anti-PRV antibody confirmed that virtually all infected neurons expressed mRFP1 (Fig. 2, bottom right panel). It should also be noted that some mRFP1-expressing neurons were not immunolabeled with the PRV antibody. This was a result of the limited penetration of the antibody into the tissue sections, whereas mRFP1 diffused throughout neurons and was therefore detectable in cells located deep within the 40-μm-thick sections. Similar results have been observed in neurons infected with PRV152 and expressing EGFP (15). Consistent with our observations of infected PK15 cell cultures, these data indicate that mRFP1 expression from the Bartha genome is readily detectable in vivo whereas DsRed and DsRed2 expression is not detectable under similar conditions.
FIG. 2.
Coronal sections through the SCN, illustrating a red fluorescent signal at 96 h after inoculation of the right eye with strain PRV-Bartha recombinants. The left panels illustrate the expression of red fluorescent reporters in SCN-infected neurons. The right panels illustrate the same sections stained with an anti-PRV antibody and a secondary antibody conjugated to Alexa-488 (green). Note the absence of a red fluorescent signal in strains PRV600 (top left panel) and PRV613 (middle left panel), whereas virtually all PRV614-infected SCN cells produced a red fluorescent signal (bottom left panel).
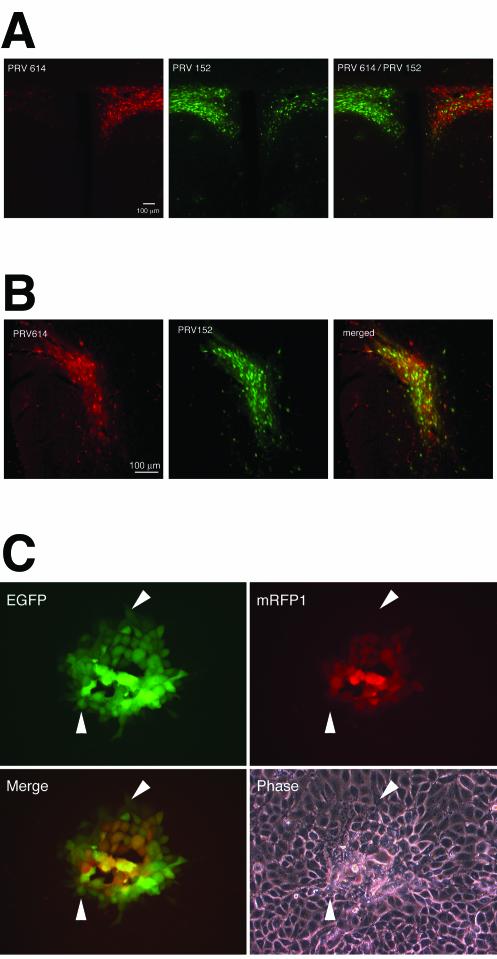
To evaluate the utility of PRV614 as a companion for the EGFP-expressing PRV152 strain in dual tract-tracing paradigms, we injected PRV614 into the right eye and PRV152 into the left eye of the same animal (n = 3). At 96 h after dual inoculation, animals were sacrificed and the brains were examined for mRFP1 and EGFP fluorescence. Figure 3A shows a coronal section through the PVN of the hypothalamus. The PVN is a major contributor to descending autonomic circuits that provide afferents to the eye and is therefore robustly labeled after PRV inoculation of the eye. Moreover, the output to the brainstem and spinal cord autonomic nuclei is primarily unilateral. As a consequence, the PVN ipsilateral to an injected eye is predominately labeled and therefore, in this case, one side is green and the other is red. The intensity and numbers of cells labeled red are comparable to the intensity and numbers of cells labeled green, suggesting two things: (i) the relative levels of detection of each of the fluorescent reporters were similar; and (ii) the PVN, which is two to three synapses away from the inoculation site, was getting infected at roughly the same time with both viruses, indicating that they were traveling throughout the nervous system at comparable rates. Moreover, we examined whether both fluorescent signals could be viewed concurrently using a dual-band filter to allow rapid identification of double-labeled neurons. The images of the PVN demonstrated that both signals were able to be isolated with presently available filter sets and also to be viewed concurrently. As seen in Fig. 3A, it appears that in the PVN, there were fewer mRFP1-expressing neurons than EGFP-expressing neurons. This was partly due to asymmetry in the plane of section but also reflects the dimmer mRFP1 signal observed under low magnification. It may also reflect a slower maturation of the mRFP1 fluorescent signal than of the EGFP signal (see below).
FIG. 3.
Kinetics of mRFP1 and EGFP expression directed by PRV strains. (A) A coronal section of the PVN 96 h after injection of strain PRV614 into the right eye and strain PRV152 into the left eye as viewed using a Texas Red filter set to identify PRV614-infected neurons (left panel) and an EGFP filter set to identify PRV152-infected neurons (middle panel). The right panel image was captured using a dual-band EGFP-Texas Red filter set to identify PRV614- and PRV152-infected neurons simultaneously. (B) A coronal section of the IGL 96 h after a PRV152/PRV614 cocktail was injected into one eye. Images were captured with a Texas Red filter set (left panel) to identify PRV614-infected neurons and an EGFP filter set (right panel) to identify PRV152-infected neurons. A merged image is shown in the right panel. Approximately 75% of infected IGL neurons were labeled with both viruses. (C) Kinetics of fluorescent protein expression in PK15 cells infected with PRV615. PRV615 was engineered to express both mRFP1 and EGFP. Four images of the same PRV615 plaque are shown: top left panel, EGFP signal; top right panel, mRFP1 signal; bottom left panel, merge of EGFP and mRFP1 signals; bottom right panel, phase-contrast image. Arrowheads point to cells that are EGFP positive and mRFP1 negative.
To directly compare the rates of infection and spread of EGFP- versus mRFP1-expressing strains, the anterior chamber of one eye was injected with a 2.0-μl cocktail containing equal titers of strains PRV152 and PRV614 (2 × 108 PFU/ml) and the brain was examined at 96 h postinfection (Fig. 3B). All regions examined contained double-labeled neurons, and no region contained neurons infected with only a single virus. The IGL was examined under high magnification in three tissue sections that contained a moderate number of labeled cells to facilitate counting; 146 PRV-labeled neurons were identified. Most neurons in each of the three sections were double labeled (77, 76, and 73%), whereas neurons that were green labeled only (i.e., in strain PRV152) (17, 22, and 24%) outnumbered neurons that were red labeled only (PRV614) (6, 2, and 3%). IGL neurons are at least fourth-order retrogradely labeled neurons (eye, ciliary ganglion, Edinger-Westphal nucleus, olivary pretectal nucleus, and IGL) and thus, the potential slower maturation of the mRFP1 signal may have resulted in the greater number of neurons that were green labeled only. These data suggest that PRV614 can be used in combination with PRV152 as dual viral transsynaptic retrograde tracers.
An important issue when considering dual synaptic labeling is the relative rates at which reporters can be detected. To test our ability to detect the fluorescent reporters in cells expressing both EGFP and mRFP1, a new recombinant PRV strain, PRV615, was constructed. PRV615 expresses EGFP from the Us3 locus and mRFP1 from the gG locus located immediately downstream. In PRV615, both fluorescent reporters are under the control of the cytomegalovirus major immediate-early promoter. PRV615 infection of PK15 cells resulted in the formation of plaques that were indistinguishable in size from those formed by strains PRV152, PRV600, PRV613, and PRV614. Figure 3C shows a representative plaque formed by PRV615 at 17 h postinfection. More EGFP-expressing cells than mRFP1-expressing cells were detected per plaque; EGFP expression was detected in the outer ring of cells in the plaque (Fig. 3C), whereas no mRFP1 expression was observed in these cells. The lag between detectable EGFP expression and mRFP1 expression in these cells was typically less than 1.5 h (data not shown).
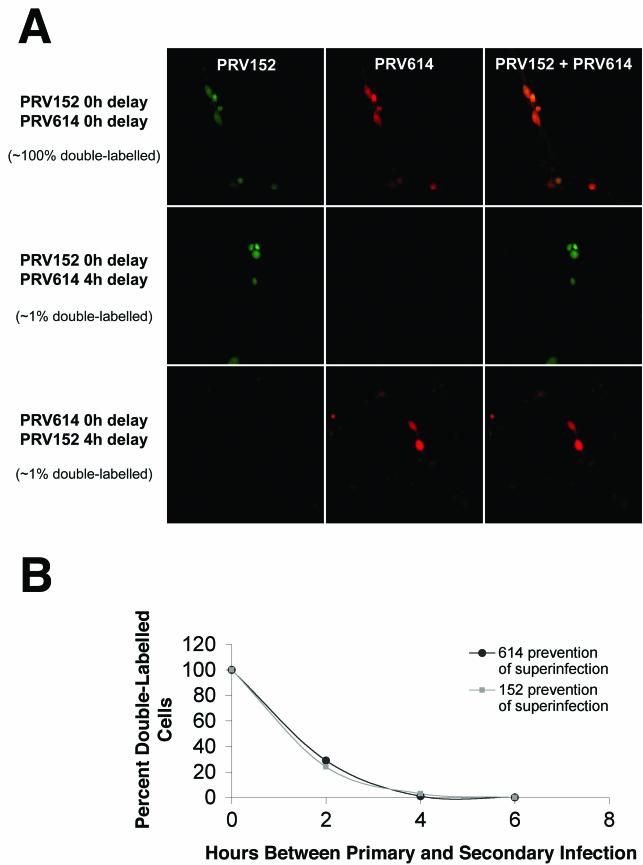
Studies were also conducted on primary cultures of rat dorsal root ganglion (DRG) neurons to evaluate the ability of a second virus to infect a cell in which an ongoing infection exists. DRG cells were infected with PRV152 and PRV614 simultaneously and examined 24 and 48 h later. Additionally, DRG cultures were infected first with PRV614 and 4, 6, 12, 18, and 24 h later with PRV152 and examined 24 h later. A similar series of experiments was conducted with the order of the virus infections reversed. When both PRV152 and PRV614 were applied simultaneously to DRG cultures, virtually all of the infected DRG cells were found to be double labeled when examined 24 or 48 h after inoculation. When the second virus was applied with a delay of ≥4 h after the first virus, almost no (∼1%) double-labeled DRG cells were observed (Fig. 4A). These data suggest that the late-arriving PRV must arrive at DRG cells less than 4 h after the initial infection to achieve double infection of cells. When the experiment was repeated and cultures were infected simultaneously with both viruses, with PRV614 first followed by PRV152 at 2, 4, and 6 h afterwards or with PRV152 first and then PRV614 at 2, 4, and 6 h afterwards, the results were indistinguishable (Fig. 4B). When the delay between the first and second infections was 6 h, no double-labeled cells were detected. As described above, when the delay between the first and second infections was 4 h, only 1 to 3% of the cells were double labeled. When the delay between the first and second infections was 2 h, 24 to 29% of the cells were double labeled. These data suggest that a significant amount of superinfection inhibition occurs prior to 2 h postinfection of primary rat DRG neurons.
FIG.4.
Analysis of dual viral infection of cultured DRG cells. (A) DRG cells were infected with strains PRV152 and PRV614 simultaneously (top panels), PRV152 4 h prior to PRV614 infection (middle panels), or PRV614 4 h prior to PRV152 infection (bottom panels). The PRV152 signal (left panels), PRV614 signal (center panels), and merged signals (right panels) are shown. (B) Kinetics of superinfection inhibition. DRG cells were infected with strains PRV152 and PRV614 simultaneously, infected with PRV614 2, 4, or 6 h prior to PRV152 infection (closed circles), or infected with PRV152 2, 4, or 6 h prior to PRV614 infection (closed squares). The percentage of double-labeled cells was determined under each set of conditions and plotted as a function of time. A minimum of 196 cells was scored for each data point. Simultaneous infections were performed in duplicate, 2-h-delay experiments were performed in quadruplicate, 4-h-delay experiments were performed in duplicate (with PRV614 infection performed first) or triplicate (PRV152 first), and 6-h-delay experiments were performed in duplicate.
It has been recognized for several years that a PRV-Bartha strain expressing an RFP would be a valuable companion to the PRV152 EGFP-expressing strain for dual transneuronal tract-tracing studies. The result of our first attempt to engineer such a virus was strain PRV600, constructed to express DsRed. After 17 h of infection in cell culture, PRV600 produced an RFP with a very weak signal that was just detectable in PK15 cells whereas 96 h after anterior chamber injection and retrograde transsynaptic transport to several regions in the brain, the slow maturation of the reporter yielded no detectable signal. Immunocytochemical staining with an anti-PRV antibody confirmed that neurons were infected with PRV600 but that they did not produce a detectable red fluorescent signal. With the evolution of DsRed to DsRed2, PRV613 was constructed. Although the DsRed2 protein generated by PRV613 produced a brighter signal in PK15 cells in vitro, the reporter again failed to generate a suitable fluorescent signal for in vivo use. These initial experiments also emphasized the differences in the kinetics of PRV infection and the expression of reporters in PK15 cells in vitro compared to those of CNS neurons in vivo. The development of mRFP1 by Tsien and colleagues (7) led to our efforts to construct strain PRV614, which expresses this new monomeric RFP. The red fluorescent signal generated by PRV614 in PK15 cells was much stronger than that produced in either of the previous attempts, and PRV614 generated a strong signal in vivo nearly equivalent to the EGFP signal produced by PRV152.
Dual viral tracing methods were introduced using variants of PRV-Bartha genetically engineered to express different viral membrane proteins and the reporter β-galactosidase (12, 13, 18). Using two different PRV-Bartha recombinants, Loewy and colleagues demonstrated the power of this technique by showing that individual hypothalamic neurons provide a dual input to sympathetic preganglionic neurons regulating cardiac and adrenal medullary functions (12). PRV-Bartha recombinants expressing EGFP have also been used in dual viral tracing experiments, in combination with other PRV-Bartha recombinants that required immunocytochemical processing, to differentiate the two antigenically distinct recombinants (3, 21). Although potentially a very valuable technique, the use of the presently available recombinant viruses for dual viral transneuronal tracing is not without pitfalls that must be carefully addressed (19).
For the several different PRV-Bartha recombinants presently in use, different promoters have been used to drive reporter gene expression and, in some cases, the promoters have been inserted into different loci of the viral genome. These differences may affect both virulence and reporter gene expression (13, 17). There are a number of factors that can influence the dual labeling of a single neuron by two different reporter viruses. Depending on the reporter, the promoter used to drive reporter expression, and the site at which the reporter is inserted into the viral genome, the viruses constructed may not be equally fit. When one reporter virus has reduced transport or replication capabilities compared to the second reporter virus, these deficiencies are amplified as the number of neurons in the multisynaptic circuit increases. This may result in the arrival of the more fit virus to the population of neurons under study before the less fit virus arrives. To complicate things further, the rate at which the reporter matures or becomes detectable is also an important consideration. The delayed arrival or detection of the second reporter virus has important ramifications for dual viral tract-tracing experiments.
Prior infection of a cell with one virus has the potential to inhibit subsequent infection by another, a process referred to as superinfection inhibition. Indeed, investigators have shown that BHK cells constitutively expressing the alphaherpesvirus envelope glycoprotein, gD, are resistant to infection by virus (4, 6). This inhibition of infection is thought to occur through gD sequestration of critical herpesvirus receptors. It is likely that other mechanisms for superinfection inhibition downstream of the virus entry pathway also exist. Although the window for double infections is probably small (i.e., on the order of a few hours), there are no data in the literature from studies addressing this issue in an established experimental transsynaptic tracing paradigm in vivo. Data from sensory neurons maintained in vitro as presented in the present report affirm the small temporal window available for dual PRV infections. An additional potential for false-negative data may also result from the differences in sensitivity of the different methods used to detect different reporter proteins (e.g., EGFP fluorescence versus immunocytochemical detection). Fixation parameters, penetration of antibodies into tissue sections, etc., may differentially affect the visualization of different reporter proteins. The ability to differentiate two isogenic PRV-Bartha recombinants solely on the basis of the use of different fluorescent reporters represents a major advance in the development of the potential of dual viral tract tracing.
Acknowledgments
We thank R.Y. Tsien (HHMI, UCSD) for providing plasmid mRFP1 in pRSETB. Antibodies against pseudorabies virus were a kind gift from L. W. Enquist, Princeton University. We thank Matt Lyman and Renée Finnen for critical reviews of the manuscript.
This work was supported in part by Basil O'Connor Starter Scholar Research Award grant 5-FY00-631 from the March of Dimes Birth Defects Foundation and NIH grant AI48626 to B.W.B. and NIH grants MH62296 and NS35615 to G.E.P.
REFERENCES
- 1.Baird, G. S., D. A. Zacharias, and R. Y. Tsien. 2000. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA 97:11984-11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bevis, B. J., and B. S. Glick. 2002. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 20:83-87. [DOI] [PubMed] [Google Scholar]
- 3.Billig, I., J. M. Foris, L. W. Enquist, J. P. Card, and B. J. Yates. 2000. Definition of neuronal circuitry controlling the activity of phrenic and abdominal motoneurons in the ferret using recombinant strains of pseudorabies virus. J. Neurosci. 20:7446-7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandimarti, R., T. Huang, B. Roizman, and G. Campadelli-Fiume. 1994. Mapping of herpes simplex virus 1 genes with mutations which overcome host restrictions to infection. Proc. Natl. Acad. Sci. USA 91:5406-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brideau, A. D., J. P. Card, and L. W. Enquist. 2000. Role of pseudorabies virus Us9, a type II membrane protein, in infection of tissue culture cells and the rat nervous system. J. Virol. 74:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campadelli-Fiume, G., M. Arsenakis, F. Farabegoli, and B. Roizman. 1988. Entry of herpes simplex virus 1 in BJ cells that constitutively express viral glycoprotein D is by endocytosis and results in degradation of the virus. J. Virol. 62:159-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Card, J. P., M. E. Whealy, A. K. Robbins, R. Y. Moore, and L. W. Enquist. 1991. Two alpha-herpesvirus strains are transported differentially in the rodent visual system. Neuron 6:957-969. [DOI] [PubMed] [Google Scholar]
- 9.Enquist, L. W. 2002. Exploiting circuit-specific spread of pseudorabies virus in the central nervous system: insights to pathogenesis and circuit tracers. J. Infect. Dis. 186(Suppl. 2):S209-S214. [DOI] [PubMed] [Google Scholar]
- 10.Enquist, L. W., P. J. Husak, B. W. Banfield, and G. A. Smith. 1999. Infection and spread of alphaherpesviruses in the nervous system. Adv. Virus Res. 51:237-347. [DOI] [PubMed] [Google Scholar]
- 11.Husak, P. J., T. Kuo, and L. W. Enquist. 2000. Pseudorabies virus membrane proteins gI and gE facilitate anterograde spread of infection in projection-specific neurons in the rat. J. Virol. 74:10975-10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansen, A. S. P., X. V. Nguyen, V. Karpitskiy, T. C. Mettenleiter, and A. D. Loewy. 1995. Central command neurons of the sympathetic nervous system: basis of the fight-or-flight response. Science 270:644-646. [DOI] [PubMed] [Google Scholar]
- 13.Kim, J. S., L. W. Enquist, and J. P. Card. 1999. Circuit-specific coinfection of neurons in the rat central nervous system with two pseudorabies virus recombinants. J. Virol. 73:9521-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matz, M. V., A. F. Fradkov, Y. A. Labas, A. P. Savitsky, A. G. Zaraisky, M. L. Markelov, and S. A. Lukyanov. 1999. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 17:969-973. [DOI] [PubMed] [Google Scholar]
- 15.Pickard, G. E., C. A. Smeraski, C. C. Tomlinson, B. W. Banfield, J. Kaufman, C. L. Wilcox, L. W. Enquist, and P. J. Sollars. 2002. Intravitreal injection of the attenuated pseudorabies virus PRV Bartha results in infection of the hamster suprachiasmatic nucleus only by retrograde transsynaptic transport via autonomic circuits. J. Neurosci. 22:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith, B. N., B. W. Banfield, C. A. Smeraski, C. L. Wilcox, F. E. Dudek, L. W. Enquist, and G. E. Pickard. 2000. Pseudorabies virus expressing enhanced green fluorescent protein: a tool for in vitro electrophysiological analysis of transsynaptically labeled neurons in identified central nervous system circuits. Proc. Natl. Acad. Sci. USA 97:9264-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith, R. L., D. L. Traul, J. Schaack, G. H. Clayton, K. J. Staley, and C. L. Wilcox. 2000. Characterization of promoter function and cell-type-specific expression from viral vectors in the nervous system. J. Virol. 74:11254-11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Standish, A., L. W. Enquist, R. R. Miselis, and J. S. Schwaber. 1995. Dendritic morphology of cardiac related medullary neurons defined by circuit-specific infection by a recombinant pseudorabies virus expressing β-galactosidase. J. Neurovirol. 1:359-368. [DOI] [PubMed] [Google Scholar]
- 19.Ter Horst, G. J. 2000. Transneuronal retrograde dual viral labelling of central autonomic circuitry: possibilities and pitfalls. Autonomic Neurosci. 83:134-139. [DOI] [PubMed] [Google Scholar]
- 20.Tomishima, M. J., and L. W. Enquist. 2001. A conserved alpha-herpesvirus protein necessary for axonal localization of viral membrane proteins. J. Cell Biol. 154:741-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ueyama, T., K. E. Krout, X. V. Nguyen, V. Karpitskiy, A. Kollert, T. C. Mettenleiter, and A. D. Loewy. 1999. Suprachiasmatic nucleus: a central autonomic clock. Nat. Neurosci. 2:1051-1053. [DOI] [PubMed] [Google Scholar]
- 22.Yarbrough, D., R. M. Wachter, K. Kallio, M. V. Matz, and S. J. Remington. 2001. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution. Proc. Natl. Acad. Sci. USA 98:462-467. [DOI] [PMC free article] [PubMed] [Google Scholar]