Abstract
Coronavirus genomes are the largest known autonomously replicating RNAs with a size of ca. 30 kb. They are of positive polarity and are translated to produce the viral proteins needed for the assembly of an active replicase-transcriptase complex. In addition to replicating the genomic RNA, a key feature of this complex is a unique transcription process that results in the synthesis of a nested set of six to eight subgenomic mRNAs. These subgenomic mRNAs are produced in constant but nonequimolar amounts and, in general, each is translated to produce a single protein. To take advantage of these features, we have developed a multigene expression vector based on human coronavirus 229E. We have constructed a prototype RNA vector containing the 5′ and 3′ ends of the human coronavirus genome, the entire human coronavirus replicase gene, and three reporter genes (i.e., the chloramphenicol acetyltransferase [CAT] gene, the firefly luciferase [LUC] gene, and the green fluorescent protein [GFP] gene). Each reporter gene is located downstream of a human coronavirus transcription-associated sequence, which is required for the synthesis of individual subgenomic mRNAs. The transfection of vector RNA and human coronavirus nucleocapsid protein mRNA into BHK-21 cells resulted in the expression of the CAT, LUC, and GFP reporter proteins. Sequence analysis confirmed the synthesis of coronavirus-specific mRNAs encoding CAT, LUC, and GFP. In addition, we have shown that human coronavirus-based vector RNA can be packaged into virus-like particles that, in turn, can be used to transduce immature and mature human dendritic cells. In summary, we describe a new class of eukaryotic, multigene expression vectors that are based on the human coronavirus 229E and have the ability to transduce human dendritic cells.
Human coronavirus (HCoV) 229E is a member of the Coronaviridae family within the order Nidovirales. It is an enveloped virus with a positive-strand RNA genome of 27.3 kb. About two-thirds of the genome, or 20.3 kb, constitute the replicase gene (19). The replicase gene is comprised of two overlapping open reading frames (ORFs): ORF1a and ORF1b. Translation of ORF1a results in the synthesis of a large polyprotein, pp1a, with a molecular mass of 450 kDa. Expression of ORF1b is mediated by −1 ribosomal frameshifting and results in the synthesis of a 750-kDa fusion polyprotein pp1ab (20, 21). Both polyproteins pp1a and pp1ab are extensively processed by virus-encoded proteinases to produce a functional replicase-transcriptase complex (43, 59, 60).
The coronavirus replicase-transcriptase complex is responsible for the synthesis of genomic and subgenomic mRNAs in the cytoplasm of infected cells. This process is not well understood, but some general principles have been established, either by the analysis of RNA synthesis in virus-infected cells (6, 39-41) or in cells transfected with helper virus-dependent or self-replicating RNA minigenomes (1, 13, 23, 47, 51-53). Thus, it has been shown that coronavirus subgenomic mRNAs are transcribed from subgenomic negative-strand RNA templates. Also, the process that generates subgenomic mRNAs involves the fusion of noncontiguous sequences (45). Each subgenomic mRNA contains a common 5′-leader sequence, which is only encoded at the 5′ end of the genome, and a so-called “body sequence” derived from the 3′ end of the genome. Since the subgenomic negative-strand RNA templates contain the complementary leader sequence at their 3′ end, it can be predicted that the fusion of noncontiguous sequences takes place during negative-strand synthesis (42). This model of mRNA synthesis, which has been called “discontinuous extension during negative-strand synthesis” (41), is thought to critically involve small, cis-acting elements termed transcription-associated sequences (TAS). TAS elements are located downstream of the leader sequence at the 5′ end of the genome (leader TAS) and at 3′ proximal sites (body TAS) corresponding to the 5′ end of each mRNA body (25). The exact borders of the different TAS elements have not yet been defined, but short stretches of not more than 5 to 8 nucleotides (nt) within the TAS, the so-called “core sequence,” have been shown to determine the site of leader-body fusion (14). Thus, the template “shunt” that takes place during negative-strand subgenomic RNA synthesis is guided by base pairing between the 5′ leader TAS and a complementary body TAS sequence on the nascent negative-strand RNA. Subsequently, negative-strand synthesis is completed as the anti-leader sequence is added to the 3′ end of the RNA (36, 41).
The unique transcriptional strategy of coronaviruses makes them promising candidates for the development of multigene RNA vectors (10, 14). First, they are versatile. The expression of heterologous genes in coronavirus-mediated transcription can be achieved in the context of the coronavirus genome, a helper-dependent minigenome or an autonomous replicating subgenomic RNA (replicon). In each case, it has been shown that it is possible to insert a transcriptional cassette (which is comprised of a TAS element located upstream of a gene of interest) and that transcriptional functions provided in trans are able to mediate the synthesis of a subgenomic mRNA encoding the protein of interest (2, 13, 15, 49). Second, the size of the coronavirus RNA genome implies a large cloning capacity for the insertion of heterologous genes. We have shown for HCoV vector RNA that a region of at least 5.7 kb is dispensable for discontinuous transcription (49). Thus, in theory, about one-third of the genome, or up to 9 kb, could be replaced by multiple transcriptional cassettes. Third, although we do not yet fully understand the parameters that govern TAS element activity, it is clear that different TAS elements, or modified TAS elements, could be used to regulate the levels of heterologous gene expression. Thus, it appears that coronavirus-based vector systems might be useful for the simultaneous, differential expression of multiple genes, irrespective of their size.
Another important consideration is the potential for vector RNA to transduce eukaryotic cells with specific functions. For example, the transduction of dendritic cells (DCs) is currently considered a particularly promising approach for gene-based immunotherapy (5, 22). DCs represent the most potent antigen-presenting cell population because they are specialized to induce a primary T-cell response by their unique ability to present antigens to naive T cells. It has been demonstrated in mouse models that DCs display a potent capacity to induce antiviral and antitumor activity (27). DCs can be generated from different sources, including peripheral blood monocytes, and this enables them to be loaded in vitro with polypeptide and protein antigens or pulsed with RNA that encodes antigens. It is reasonable to view DCs as an attractive target for viral expression vectors.
RNA replicon expression systems based on positive-strand RNA viruses are considered to have several advantages for DC-mediated immunotherapy. First, replication of the RNA takes place in the cytoplasm without a DNA intermediary, making integration of vector-derived nucleic acid into the target cell genome unlikely. Second, for a short period of time high levels of heterologous gene expression are achieved. Third, since RNA replicons do not encode structural proteins, they are not able to spread in the same way as replication-competent virus. However, the concept of using replicon RNA-based vectors for DC-mediated immunotherapy is dependent on the ability to efficiently transduce DCs using virus-like particles (VLPs). In this respect, it is important to note that the HCoV receptor, human aminopeptidase N (hAPN or CD13), is expressed at significant levels on human DCs (38, 56). This implies that HCoV-based vector particles could be used to efficiently transduce these cells.
In the present study we report experiments that take advantage of the coronavirus transcriptional strategy to establish a new class of eukaryotic, multigene RNA vectors capable of expressing several heterologous proteins simultaneously. Specifically, we have constructed a prototype RNA vector, based on HCoV, that mediates the expression of three different reporter genes. Sequence analysis of the reporter gene mRNA leader-body junctions confirmed the synthesis of coronavirus-specific transcripts, and appropriate reporter gene assays confirmed the expression of chloramphenicol acetyltransferase (CAT), firefly luciferase (LUC), and green fluorescent protein (GFP) in vector RNA-transfected cells. Furthermore, we show that HCoV-based vector RNA can be packaged to transduce human DCs.
MATERIALS AND METHODS
Viruses and cells.
Recombinant vaccinia viruses and fowlpox virus strain HP1.441 (29) were propagated, titered, and purified by standard procedures (4). Human lung fibroblast (MRC-5), monkey kidney fibroblast (CV-1), and baby hamster kidney (BHK-21) cells were purchased from the European Collection of Cell Cultures and maintained in minimal essential medium supplemented with HEPES (25 mM), fetal bovine serum, and antibiotics. Human monocyte-derived DCs (MoDCs) were generated from leukapheresis blood samples. Peripheral blood monocytes were isolated from whole blood by using Ficoll-Paque Plus density gradient centrifugation (300 × g for 30 min; Amersham Biosciences, Freiburg, Germany). CD2+ cells were depleted by Ficoll-Paque Plus density gradient centrifugation after rosette formation by using 2-aminoethylisothiouronium-treated sheep red blood cells (Virion, Munich, Germany). The nonrosetted cell fraction was further purified by using Percoll density gradient centrifugation (300 × g for 30 min, 1,065 g/ml; Amersham Biosciences, Freiburg, Germany). This routinely resulted in cultures containing >90% CD14+ monocytes. Approximately 1.5 × 106 cells were plated in six-well tissue culture plates and, after 1 h of adherence, residual nonadherent cells were washed off. To generate immature MoDCs, attached cells were cultured for 6 days in RPMI 1640 supplemented with 10% fetal bovine serum, 500 U of granulocyte-macrophage colony-stimulating factor/ml, and 250 U of interleukin-4/ml (all from Strathmann Biotec, Hannover, Germany). Fresh cytokines were added every second day. To generate mature MoDCs, 100 ng of lipopolysaccharide from Escherichia coli O128:B12 (Sigma, Taufkirchen, Germany)/ml was added to the medium at day 6, and the cells were cultured for a further 24 h.
Construction of cDNAs.
Plasmids were constructed by standard methods (4) and verified by sequence analysis. The detailed cloning strategy, plasmid maps, and sequences are available from the authors upon request. The plasmid pBR-CLG is based on pBR322. The EcoRI and EagI restriction sites of pBR322 have been used to insert a DNA fragment containing an EcoRI site, a bacteriophage T7 RNA polymerase promoter, a single G nucleotide, HCoV nt 1 to 2177, HCoV nt 18227 to 20569, the gene encoding CAT, a BglII site, HCoV nt 24978 to 24994, the gene encoding LUC, HCoV nt 25654 to 25685, the gene encoding GFP, nine artificial nucleotides (AGCGGCCCG), HCoV nt 26279 to 27277, a synthetic poly(A) tail of 42 nt, a ClaI site, the hepatitis delta ribozyme, and the restriction site EagI. Consequently, in this plasmid the reporter genes encoding CAT, LUC, and GFP are preceded by a TAS of the HCoV S gene (HCoV nt 20555 to 20569), M gene (HCoV nt 24978 to 24994), and N gene (HCoV nt 25654 to 25685), respectively.
To generate a recombinant vaccinia virus containing the vecCLG cDNA, the plasmids pFE (49) and pBR-CLG and the recombinant vaccinia virus vHCoV-inf-1 DNA (48) were used. Plasmid pFE was digested with FseI, dephosphorylated, and purified. After cleavage with EcoRI, a fragment of 11.2 kb corresponding to nt 7000 to 18226 of the recombinant HCoV-inf-1 genome was isolated. Plasmid pBR-CLG was digested with EcoRI, dephosphorylated, and purified. After cleavage with XbaI, a fragment of 1.1 kb corresponding to nt 18226 to 19302 of the HCoV-inf-1 genome was isolated. Plasmid pBR-CLG was digested with XbaI and EagI and dephosphorylated, and a fragment of 5.5 kb was isolated. The 11.2-, 1.1-, and 5.5-kb DNA fragments were ligated in vitro, and the resulting products were joined by in vitro ligation to FseI- and Bsp120I-cleaved vHCoV-inf-1 DNA. Recombinant vaccinia viruses containing the vec-CLG cDNA insert were isolated after transfection of the ligation reaction into fowlpox virus-infected CV-1 cells as described previously (49).
The generation and analysis of a recombinant vaccinia virus vHCoV-PL1(−) will be described in detail elsewhere. Essentially, vHCoV-PL1(−) DNA is identical to vHCoV-inf-1 DNA with the exception of two mutated nucleotides (HCoV-inf-1 nucleotide 3452TG3453 changed to 3452GC3453). These mutations result in an amino acid change of Cys to Ala at the putative catalytic site (Cys1054) of the HCoV papain-like proteinase 1.
In vitro transcription and transfection.
HCoV nucleocapsid protein (N protein) cDNA was prepared by PCR with recombinant vHCoV-inf-1 DNA as a template and oligonucleotide primers Nup (5′-ACGTAATACGACTCACTATAGGGACGAAACCATGGCTACAGTCAAATGGGCTG-3′) and Ndown (5′-TTTTTTTTTTTTTTTTTTTTCAAACAACACAGTGGCATGTTTAG-3′). HCoV N protein cDNA was used as a template for an in vitro transcription reaction to produce synthetic HCoV N protein mRNA. In order to produce a control HCoV N protein negative-strand mRNA, PCR and in vitro transcription were carried out identically, but the oligonucleotide primer Nup was replaced by primer N(−)up (5′-ACGTAATACGACTCACTATAGGGACGAAAAGCCGGCTACAGTCAAATGGGCTG-3′). Vec-CLG DNA and HCoV-PL1(−) DNA was prepared from purified recombinant vaccinia viruses vVec-CLG and vHCoV-PL1(−), respectively, cleaved with ClaI enzyme, and used as a template for in vitro transcription reactions. Between 10 and 20 μg of in vitro-transcribed RNA was transfected into BHK-21 cells by electroporation as described previously (49).
Preparation of poly(A)-containing RNA, RT-PCR, and sequencing analysis.
Poly(A)-containing RNA was isolated from BHK-21 cells by using oligo(dT)25 Dynabeads (Dynal, Oslo, Norway) and reverse transcription-PCR (RT-PCR) was done with Superscript II reverse transcriptase (Invitrogen, Karlsruhe, Germany) as described previously (50). To amplify and sequence the leader-body fusion site of the CAT mRNA, four oligonucleotide primers were used: 5′-GCCACTCATCGCAGTACTGTTGTAA-3′ (reverse transcription), 5′-TACAGATAGAAAAGTTGC-3′ (PCR, HCoV leader specific), 5′-TTAAGCATTCTGCCGACATGGAAG-3′ (PCR, CAT mRNA body specific), and 5′-CGGAATTCCGGATGAGCATTCATC-3′ (sequencing). To amplify and sequence the leader-body fusion site of the LUC mRNA, four oligonucleotide primers were used: 5′-CGGTATCCAGATCCACAACCTTCG-3′ (reverse transcription), 5′-TACAGATAGAAAAGTTGC-3′ (PCR, HCoV leader specific), 5′-ACAACACTTAAAATCGCAGTATCC-3′ (PCR, LUC mRNA body-specific), 5′-GTTCGCGGGCGCAACTGCAACTCCG-3′ (sequencing). To amplify and sequence the leader-body fusion site of the GFP mRNA, four oligonucleotide primers were used: 5′-AGAAACTTCATCACGCACTGG-3′ (reverse transcription), 5′-TACAGATAGAAAAGTTGC-3′ (PCR, HCoV leader specific), 5′-AGCAGGACCATGTGATCGCGCTTC-3′ (PCR, GFP mRNA body specific), and 5′-ACGGGCAGCTTGCCGGTGGTGCA-3′ (sequencing). Sequence analysis of plasmid DNA, RT-PCR products, and the recombinant vaccinia virus cDNA insert was done by standard cycle sequencing methods with an ABI 310 Prism Genetic Analyzer. Computer-assisted analysis of sequence data was facilitated by the Lasergene bio-computing software (DNASTAR).
Analysis of reporter protein expression.
Reporter protein expression was analyzed 3 days posttransfection in BHK-21 cells. GFP expression was analyzed by fluorescence microscopy or fluorescence-activated cell sorting (FACS). Expression of LUC and CAT were analyzed by using the luciferase reporter gene assay (Boehringer, Mannheim, Germany) or CAT-enzyme-linked immunosorbent assay (Roche, Mannheim, Germany), respectively, as described by the manufacturers.
Packaging of HCoV vector RNA.
Vec-CLG RNA (15 μg), HCoV N protein mRNA (10 μg), and synthetic HCoV genomic RNA [HCoV-PL1(−) RNA (15 μg) or HCoV-inf-1 RNA (15 μg)] were cotransfected into BHK-21 cells by electroporation (49). The cells were cultured for 3 days at 33°C until the tissue culture supernatant was harvested, and the titer [i.e., the 50% tissue culture infective dose (TCID50)] of recombinant HCoV was determined on MRC-5 cells. In addition, the MRC-5 cells were analyzed by fluorescence microscopy to determine the titer of VLPs able to mediate GFP expression. Virus stocks were concentrated by NaCl-polyethylene glycol precipitation as described by Wege et al. (54).
Transduction and analysis of cells.
MRC-5 cells or MoDCs were incubated for 2 h at 33°C with supernatant obtained from BHK-21 cells that had been transfected with Vec-CLG RNA, HCoV N protein mRNA, and synthetic HCoV genomic RNA (see above). MoDCs were then incubated at 37°C for 24 h before fluorescence microscopy or FACS analysis. MRC-5 cells were kept at 33°C for the indicated time periods before fluorescence microscopy analysis. For FACS analysis, MoDCs were incubated at 4°C for 30 min with phycoerythrin- or fluorescein isothiocyanate-conjugated monoclonal antibodies (Beckman-Coulter, Krefeld, Germany; Becton Dickinson, Heidelberg, Germany). Living cells were analyzed by using FACScan and Lysis II software with gating on morphological intact cells and by excluding 7-AAD (7-amino actinomycin D; Calbiochem, Schwalbach, Germany)-incorporating cells. Data analysis was carried out by using CellQuest software (Becton Dickinson).
RESULTS
Structure and synthesis of HCoV vector RNA.
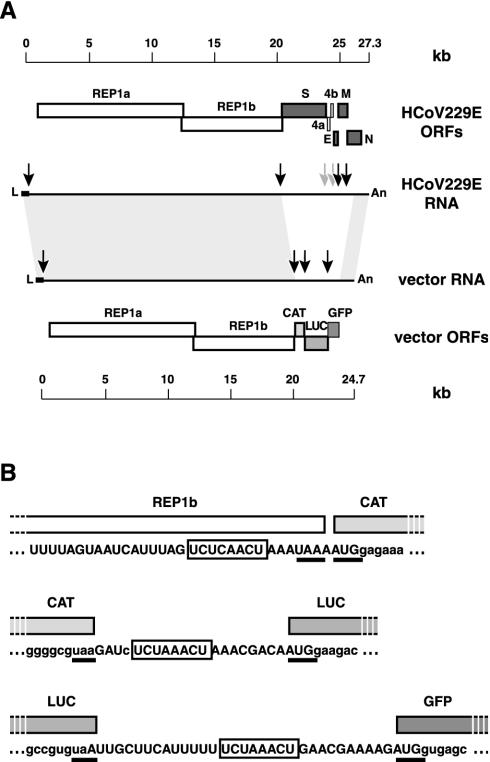
The construction of the vec-CLG vector is based upon our recently established reverse genetic system for HCoV (48). We used a vaccinia virus vector to clone a 24.7-kb cDNA containing the 5′ and 3′ ends of the HCoV genome, the entire HCoV replicase gene, and three reporter genes. The 24.7-kb cDNA was assembled and introduced into the vaccinia virus genome by in vitro ligation as described in Materials and Methods. After transfection of the in vitro-ligated DNA into fowlpox virus-infected CV-1 cells, recombinant vaccinia viruses containing the 24.7-kb cDNA insert were identified by Southern blotting (data not shown). We chose one clone, designated vVec-CLG, and confirmed its identity by sequencing the reporter gene region of the cDNA insert (nt 20000 to 24700 [data not shown]). In order to produce vec-CLG vector RNA, recombinant vaccinia virus vVec-CLG was grown to high titers and purified, and vVec-CLG DNA was isolated. The vec-CLG DNA was cleaved with restriction endonuclease ClaI and used as a template for bacteriophage T7 RNA polymerase-driven in vitro transcription in the presence of m7G(5′)ppp(5′)G cap analogue. The structure of the resulting synthetic vec-CLG RNA is illustrated in Fig. 1. Essentially, a sequence of 5,708 nt, corresponding to HCoV 229E genome nt 20570 to 26278 and containing mainly the HCoV 229E structural protein genes, was replaced in the HCoV genome by three transcriptional cassettes encoding CAT, LUC, and GFP. As illustrated in Fig. 1B, each reporter gene is located downstream of an HCoV TAS element, derived from sequences upstream of the HCoV S gene, M gene and N gene, respectively.
FIG. 1.
Structure of HCoV vector RNA. (A) The structural relationship and sizes of the RNAs and ORFs of HCoV 229E and vec-CLG vector is shown. TAS elements encoded by HCoV 229E or vec-CLG vector are shown as arrows. TAS elements that mediate the synthesis of HCoV 229E subgenomic mRNAs encoding S, M, and N proteins have been inserted into vec-CLG RNA and are indicated by black arrows. (B) TAS element-containing RNA sequences of vec-CLG RNA are shown in relation to ORFs encoding the replicase gene, CAT gene, LUC gene, and GFP gene, respectively. Core sequences are depicted in boxes; stop and start codons of the respective ORFs are underlined. Nucleotides that match the HCoV 229E sequence are shown as capital letters.
Multigene expression in vec-CLG-RNA-transfected cells: influence of the N protein.
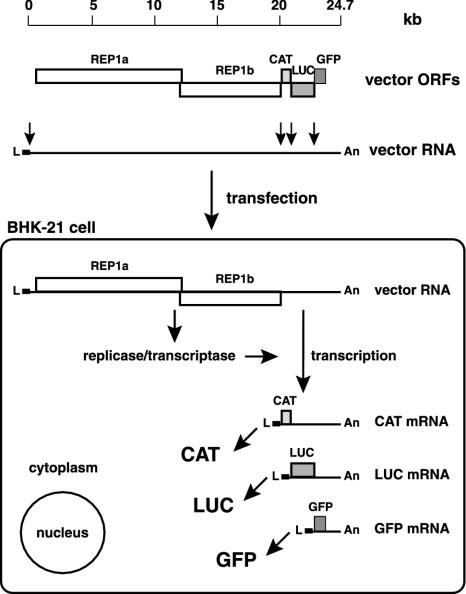
We predicted that a functional replicase-transcriptase complex should be formed after translation of vec-CLG RNA in eukaryotic cells. This complex should then mediate coronavirus-specific discontinuous transcription of CAT, LUC, and GFP mRNAs and result, after translation, in reporter protein expression (Fig. 2). To test this prediction, we introduced vec-CLG RNA into BHK-21 cells and monitored the cells for green fluorescence. At 2 days posttransfection we could detect a small number of green fluorescent BHK-21 cells (data not shown). This result confirms that the replicase gene products suffice for coronavirus discontinuous transcription. However, as described previously for HCoV-vec-1 RNA (49), only ca. 0.1% of the transfected BHK-21 cell culture displayed GFP expression. With this transfection frequency, we were unable to detect LUC or CAT reporter gene expression (data not shown).
FIG. 2.
Heterologous gene expression with vec-CLG vector RNA. The structural relationship of vec-CLG ORFs, TAS elements, and the intracellular mRNAs produced by coronavirus-mediated transcription is illustrated, together with the predicted intracellular translation products (i.e., the HCoV replicase/transcriptase, CAT, LUC, and GFP).
In our studies with the coronavirus reverse genetic system, we have observed that expression of the N protein facilitates the recovery of recombinant coronaviruses (12) (V. Thiel, unpublished data). Although, the reason for this effect is not known, it prompted us to include the N protein in our vector RNA expression system. Therefore, we coelectroporated vec-CLG RNA with a synthetic mRNA encoding the HCoV N protein. As determined by this approach, the percentage of green fluorescent BHK-21 cells increased to 0.8 to 3% at 3 days posttransfection and GFP expression was detectable by as early as 30 h posttransfection compared to 48 h posttranfection when vec-CLG RNA was electroporated alone. Moreover, when we coelectroporated a synthetic mRNA in which the N protein start codon had been changed from AUG to CCG, the percentage of green fluorescent cells decreased to 0.1%. This finding indicates that the N protein itself, rather than its mRNA, provides an important function in the HCoV-based vector system.
In addition, the increased number of transduced cells after coelectroporation of vec-CLG RNA with the N protein mRNA enabled us to detect LUC and CAT expression. A total of 107 BHK-21 cells were electroporated with 15 μg of vec-CLG RNA and 10 μg of N protein mRNA and, after 3 days, the cells were analyzed (Table 1). As is shown, the percentage of green fluorescent cells, as determined by FACS analysis, was found to be 0.8 to 3% in this experiment. Second, CAT and LUC activity were easily detectable by using the CAT-enzyme-linked immunosorbent assay and the luciferase reporter gene assay (see Materials and Methods). Finally, when we analyzed poly(A)-containing RNA from transfected cells, we were able to detect CAT, LUC, and GFP reporter gene mRNAs by RT-PCR. As expected, these mRNAs show leader-body junctions characteristic for coronavirus transcriptase-mediated discontinous transcription (Fig. 3). Taken together, these data demonstrate the ability of vec-CLG RNA to facilitate the simultaneous expression of three heterologous genes.
TABLE 1.
Analysis of reporter gene expression
| Transfection | % Green fluorescent cells | Expression of:
|
|
|---|---|---|---|
| CAT (ng/106 cells) | LUC (RLUa/106 cells) | ||
| 1 | 0.8 | 0.067 | 14,163 |
| 2 | 1.4 | 0.207 | 36,981 |
| 3 | 3.0 | 0.463 | 57,008 |
| Mock | 0.0 | 0.000 | 36 |
RLU, relative light units.
FIG. 3.
Sequencing analysis of subgenomic mRNAs isolated from vec-CLG-transfected cells. Poly(A)-containing RNA was isolated from vec-CLG RNA and N protein mRNA transfected cells. The TAS regions of vec-CLG subgenomic mRNAs encoding CAT (A), LUC (B), and GFP (C) were analyzed by RT-PCR amplification and cycle sequencing. The sequences corresponding to HCoV leader (L), core sequences of the TAS elements (in boxes), and the reporter gene start codons (underlined) are shown.
Packaging of HCoV vector RNA.
Next, we addressed the question of whether vec-CLG RNA can be packaged into VLPs. The obvious experiment would be to transfect vec-CLG RNA into HCoV-infected MRC-5 cells. However, although MRC-5 cells can be infected with HCoV, transfection of RNA into MRC-5 cells is very inefficient (data not shown). Therefore, we again used BHK-21 cells into which we transfected by electroporation vec-CLG RNA, the synthetic N protein mRNA and the synthetic genomic HCoV-inf-1 RNA (48) (Fig. 4A). The cells were cultured for 3 days at 33°C before the supernatant, referred as SN-CLG/inf1, was harvested. To test whether vec-CLG-RNA-containing VLPs have been produced and whether these particles can be used to transduce MRC-5 cells, we then transferred serial dilutions of the supernatant SN-CLG/inf1 on to MRC-5 cells. After 24 h, fluorescent MRC-5 cells could be detected, in wells that have been incubated with undiluted and a 10-fold dilution of the SN-CLG/inf1 supernatant, indicating that vec-CLG particles were indeed present in the supernatant and that the cells could be transduced with these particles. After 6 days, the tissue culture wells were analyzed for characteristic cytopathic effects of HCoV infection, and the virus titer of supernatant SN-CLG/inf1 was calculated to be 2.9 × 105 TCID50/ml.
FIG. 4.
Packaging of vec-CLG RNA and transduction of MRC-5 cells. (A) Tissue culture supernatants containing HCoV and presumably vector-derived VLPs were obtained by coelectroporation of synthetic HCoV genomic RNA, vec-CLG RNA, and N protein mRNA into BHK cells. (B) GFP expression analyzed by fluorescence microscopy 1 day or 4 days posttransduction of MRC-5 cells with tissue culture supernatant obtained from HCoV-PL1(−) RNA, vec-CLG RNA, and N protein mRNA-transfected BHK-21 cells.
In order to achieve a higher titer of vec-CLG particles and a lower titer of recombinant HCoV virus, we repeated this experiment but replaced the recombinant HCoV full-length RNA with a recombinant RNA derived from vHCoV-PL1(−). This cDNA copy of the HCoV genome contains a mutation that results in a Cys-to-Ala change at the predicted catalytic residue of the HCoV papain-like proteinase 1. The detailed phenotypic analysis of recombinant HCoV-PL1(−) coronavirus will be described elsewhere but, in this context, it is important to note that this phenotype is viable, despite the lack of proteolytic active papain-like proteinase 1. However, HCoV-PL1(−) grows to reduced titers and reversion to wild-type virus (Ala to Cys) occurs within two passages. Three days after coelectroporation of vec-CLG RNA, N protein mRNA, and genomic HCoV-PL1(−) RNA, the tissue culture supernatant, SN-CLG/PL1(−), was harvested and concentrated by polyethylene glycol precipitation. Titration of the concentrated SN-CLG/PL1(−) supernatant revealed a titer of 3.0 × 104/ml for infectious HCoV-PL1(−) virus and 3.5 × 103/ml for transducing vec-CLG particles, as determined by TCID50 assay and fluorescent microscopy, respectively. We then transferred 100 μl of SN-CLG/PL1(−) on to 2 × 105 MRC-5 cells. Single green fluorescent MRC-5 cells could be observed after 24 h and, after 3 to 4 days, additional foci of green fluorescent plaques became apparent (Fig. 4B). Although we cannot exclude the possibility that recombination events have led to the generation of an infectious coronavirus encoding GFP, we interpret this result to indicate spread of the vec-CLG RNA, along with infectious virus, at foci of HCoV-PL1(−) and vec-CLG VLP coinfection. It remains to be determined whether this spread is intra- or intercellular.
Transduction of human DCs with HCoV vector particles.
We have shown that HCoV-based vector particles can be used to transduce MRC-5 cells. This result was not surprising, since MRC-5 cells express the HCoV 229E receptor, human aminopeptidase N (hAPN or CD13). In order to test whether we can transduce human DCs, which also express hAPN, we prepared immature and mature human DCs from peripheral blood monocytes and exposed them to the concentrated tissue culture supernatant SN-CLG/PL1(−) described above.
First, we generated immature DCs derived from CD14+ purified monocytes by using medium supplemented with granulocyte-macrophage colony-stimulating factor and interleukin-4. The cells were analyzed by FACS and displayed the characteristic CD83− CD86low CD80low HLA-DRlow phenotype (data not shown). Within 24 h after we had transferred the concentrated supernatant SN-CLG/PL1(−) to immature DCs, we could detect green fluorescent cells, indicative of GFP expression (Fig. 5A). Second, we generated mature DCs by adding LPS to immature DCs for 24 h. FACS analysis revealed the characteristic CD83+ CD86high CD80high HLA-DRhigh phenotype for mature DCs (data not shown). We then transferred the concentrated supernatant SN-CLG/PL1(−) to mature DC and 24 h later we could, again, detect green fluorescent cells (Fig. 5A).
FIG. 5.
vec-CLG-mediated GFP expression in human DCs. (A) GFP expression was analyzed by fluorescence microscopy of immature and mature DCs transduced with supernatant SN-CLG/PL1(−). (B) Flow cytometry analysis of CD83, CD86, and GFP expression of mature DCs that have been either mock infected, infected with HCoV 229E, or transduced with supernatant SN-CLG/PL1(−). Indicated values represent the percentage of green fluorescent cells.
In order to further confirm this result by FACS analysis, we incubated 2 × 105 mature DCs with 1.5 ml of concentrated SN-CLG/PL1(−) for 24 h at 33°C. As controls, we used mock-infected and HCoV-infected mature DCs. FACS analysis revealed that ca. 1.4% of the SN-CLG/PL1(−)-treated cells displayed green fluorescence, whereas mock-infected and HCoV-infected DCs did not show any green fluorescence. In addition, green fluorescent cells were found to express CD83 and CD86, a characteristic indicative for mature human DCs (Fig. 5B). It is noteworthy that the observed percentage of green fluorescent cells (1.4%) correlates with the titer of vec-CLG particles determined on MRC-5 cells. Moreover, we could increase the percentage green fluorescent cells up to 50% when we incubated fewer than 103 mature DCs with 1 ml of SN-CLG/PL1(−), indicating that the percentage of green fluorescent cells is dependent on the titer of vec-CLG transducing particles. In summary, these data indicate that HCoV-based vectors can be used to transduce both immature and mature human DCs.
DISCUSSION
In this report we describe a new class of eukaryotic, multigene expression vectors based on the unique transcriptional strategy of coronaviruses. We have constructed an HCoV vector RNA that simultaneously mediates the expression of three reporter proteins. To our knowledge, this is the first example demonstrating the ability of coronavirus vectors to mediate the expression of multiple heterologous genes. We further show evidence that HCoV vector RNA can be packaged to VLPs, which can be used to transduce human DCs. In the long term this system may provide an attractive strategy to manipulate DCs by genetically transferring multiple antigens and stimulatory molecules for immunotherapy applications.
To construct the HCoV-based vector RNA, we made use of our reverse genetic system for HCoV. We have cloned a prototype vector cDNA of 24.7 kb into the vaccinia virus genome in order to produce a DNA template for the generation of synthetic vector RNA by in vitro transcription. HCoV vector RNA, containing the 5′ and 3′ ends of the HCoV genome and the entire HCoV replicase gene, has previously been shown to mediate the synthesis of coronavirus-specific transcripts (49). In order to demonstrate the expression of multiple heterologous genes, we replaced the HCoV structural gene region by three transcriptional cassettes, encoding CAT, LUC, and GFP. The HCoV vector RNA described here is similar to replicon RNA constructs known from other positive-strand RNA virus systems (18, 24, 26). Since this type of vector RNA does not encode any coronaviral structural genes, it is not capable of spreading independently. In the particular construct we describe, we have deleted ca. 5.7 kb of the HCoV structural gene region and this has been replaced by 3.1 kb of heterologous gene sequence. Theoretically, there should be room for, at least, another 2.6 kb of heterologous sequence, which would then correspond to the authentic size of the HCoV genome. It seems quite likely that we can still increase the number of heterologous genes that can be expressed by HCoV replicon-based vector RNAs.
The expression of reporter proteins by using the HCoV-based vector system was achieved via coronavirus-specific synthesis of subgenomic mRNAs. A prerequisite for coronavirus discontinuous transcription is the TAS element. In the vec-CLG RNA construct, we have inserted sequences of naturally occurring TAS elements, derived from the HCoV S gene, the M gene, and the N gene. Since the borders of these TAS elements are not known exactly, we took care to position the inserted TAS elements relative to the AUG start codon of each heterologous gene. For example, the position of the AUG start codon of the CAT gene relative to the S gene TAS in vec-CLG RNA parallels the position of the HCoV S gene TAS and start codon within the HCoV genome. Also, in order to express LUC and GFP, we had to position TAS elements of the HCoV M gene and N gene between the ORFs of CAT and LUC and between the ORFs of LUC and GFP, respectively. As a consequence, these TAS elements are true intergenic sequences and not part of an upstream ORF. Nevertheless, coronavirus-specific mRNA encoding LUC and GFP could be detected in the present study, indicating that it is possible to separate TAS elements from upstream ORFs. This finding has an important impact on future HCoV vector constructs, since it appears to be most likely that HCoV transcriptional cassettes, comprised of a upstream TAS element and a downstream gene of interest, will be sufficient to direct the synthesis of a coronavirus-specific subgenomic mRNA.
During the course of the present study we have observed that the HCoV N protein or mRNA provides an important function in the efficacy of our system. Although we could detect GFP expression in a small fraction of vector RNA-transfected cells in the absence of N protein expression, cotransfection of a functional N protein mRNA, together with vector RNA, significantly increased the percentage of green fluorescent cells. This observation is in agreement with data obtained from rescue experiments with full-length recombinant coronavirus genomes of transmissible gastroenteritis virus (57), infectious bronchitis virus (12), mouse hepatitis virus (MHV) (58), and HCoV (V. Thiel, unpublished data). In these studies, the rescue of recombinant coronaviruses either failed in the absence of a cotransfected N gene construct (12, 57), or it appeared to be enhanced by cotransfection of N protein mRNA (58) (V. Thiel, unpublished data). However, from these experiments, it was not clear whether the N protein or the N protein mRNA was responsible for this effect. Based on the observation that the percentage of green fluorescent cells was greatly enhanced by the coelectroporation of a synthetic N protein mRNA but not by the coelectroporation of a mutagenized version of this RNA (where the AUG start codon has been changed to CCG), we propose that the N protein itself, rather than the N protein mRNA is the important molecule in our system. Nevertheless, the function(s) provided by the N protein in our system has not been addressed systematically and additional studies are under way to determine the role of N protein in coronavirus gene expression.
Coronavirus particles are comprised of viral RNA and four structural proteins, namely, the surface glycoprotein S, the integral membrane protein M, the envelope protein E, and the N protein. Some coronaviruses have an additional virion protein, the hemagglutinin-esterase protein HE. In general, only the genomic RNA is efficiently packaged into coronavirus particles, indicating that there is a specific mechanism for packaging (31). In the case of MHV, a packaging signal located in the replicase-coding region, has been identified (8, 55). Although the position of the HCoV packaging signal has not been determined, the HCoV vectors described here contain the unique region of HCoV genomic RNA, and we expected packaging of vector RNA when the HCoV structural proteins were coexpressed. In our experiments, we essentially used infectious virus particles to provide the structural proteins (albeit via an infectious RNA), and the results indicate that HCoV vector RNA was packaged into VLPs. In the longer term, however, we want to study the interaction of HCoV vectors with specific cells (e.g., DCs) in the absence of helper virus and, therefore, we need to establish an efficient packaging system devoid of infectious virus. It has been shown that recombinant MHV defective-interfering particles can be produced in the absence of helper virus (9) and that replication-competent but propagation-deficient transmissible gastroenteritis virus vector RNA that lacks the E gene can be packaged when the E protein is expressed under the control of the cytomegalovirus promoter or by using alphavirus-based expression systems (13, 34). Thus, the efficacy of using versatile packaging systems to generate biosafe coronavirus vectors has clearly been demonstrated.
We have shown that we can use HCoV-based vectors to transduce eukaryotic cells expressing the HCoV receptor hAPN. Specifically, we demonstrate that human lung fibroblasts (MRC-5 cells) and human DCs are susceptible to HCoV-based vector transduction. Human DCs are of particular interest, since they play an important role as professional antigen-presenting cells. They are potent immunostimulatory cells facilitating antigen transport to lymphoid tissues and efficient stimulation of T cells. For example, a series of preclinical experimental studies in mice have demonstrated that antitumor immunity can be induced by using DCs exogenously pulsed with tumor-derived peptides (35, 61) or crude tumor cell preparations (3, 16). Alternatively, similar results can be achieved with DCs that have been transfected with tumor cell-derived mRNA (7, 30) or infected with recombinant viruses (37, 44, 46). This preclinical experience has been reproduced in a variety of clinical trials, which have shown that the application of DCs is safe and clinically effective (17, 33). However, important limitations of these approaches have been identified: the suboptimal delivery of antigens to the DCs, the ability of tumors to escape from specific immune responses if too few antigens are presented by the DCs, and inappropriate concomitant immunostimulation (11, 28, 32). The ability to manipulate DCs by genetically transferring simultaneously several antigens and immunostimulatory molecules is an attractive strategy to overcome these limitations. The HCoV-based vector system represents a promising system to genetically deliver multiple heterologous genes to cells expressing the HCoV 229E receptor molecule and may provide a valuable tool for future DC-based immunotherapy.
Acknowledgments
The work was supported by a grant (SFB 479/B4) from the German Research Council (DFG) to S.G.S.
REFERENCES
- 1.Alonso, S., A. Izeta, I. Sola, and L. Enjuanes. 2002. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J. Virol. 76:1293-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso, S., I. Sola, J. P. Teifke, I. Reimann, A. Izeta, M. Balasch, J. Plana-Duran, R. J. Moormann, and L. Enjuanes. 2002. In vitro and in vivo expression of foreign genes by transmissible gastroenteritis coronavirus-derived minigenomes. J. Gen. Virol. 83:567-579. [DOI] [PubMed] [Google Scholar]
- 3.Ashley, D. M., B. Faiola, S. Nair, L. P. Hale, D. D. Bigner, and E. Gilboa. 1997. Bone marrow-generated dendritic cells pulsed with tumor extracts or tumor RNA induce antitumor immunity against central nervous system tumors. J. Exp. Med. 186:1177-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. D. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Banchereau, J., B. Schuler-Thurner, A. K. Palucka, and G. Schuler. 2001. Dendritic cells as vectors for therapy. Cell 106:271-274. [DOI] [PubMed] [Google Scholar]
- 6.Baric, R. S., and B. Yount. 2000. Subgenomic negative-strand RNA function during mouse hepatitis virus infection. J. Virol. 74:4039-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boczkowski, D., S. K. Nair, D. Snyder, and E. Gilboa. 1996. Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. J. Exp. Med. 184:465-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bos, E. C., J. C. Dobbe, W. Luytjes, and W. J. Spaan. 1997. A subgenomic mRNA transcript of the coronavirus mouse hepatitis virus strain A59 defective interfering (DI) RNA is packaged when it contains the DI packaging signal. J. Virol. 71:5684-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bos, E. C., W. Luytjes, H. V. van der Meulen, H. K. Koerten, and W. J. Spaan. 1996. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 218:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bredenbeek, P. J., and C. M. Rice. 1992. Animal RNA virus expression systems. Semin. Virol. 3:297-310. [Google Scholar]
- 11.Brossart, P., A. Zobywalski, F. Grunebach, L. Behnke, G. Stuhler, V. L. Reichardt, L. Kanz, and W. Brugger. 2000. Tumor necrosis factor alpha and CD40 ligand antagonize the inhibitory effects of interleukin 10 on T-cell stimulatory capacity of dendritic cells. Cancer Res. 60:4485-4492. [PubMed] [Google Scholar]
- 12.Casais, R., V. Thiel, S. G. Siddell, D. Cavanagh, and P. Britton. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 75:12359-12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curtis, K. M., B. Yount, and R. S. Baric. 2002. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 76:1422-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enjuanes, L., I. Sola, F. Almazan, J. Ortego, A. Izeta, J. M. Gonzalez, S. Alonso, J. M. Sanchez, D. Escors, E. Calvo, C. Riquelme, and C. Sanchez. 2001. Coronavirus derived expression systems. J. Biotechnol. 88:183-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer, F., C. F. Stegen, C. A. Koetzner, and P. S. Masters. 1997. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J. Virol. 71:5148-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flamand, V., T. Sornasse, K. Thielemans, C. Demanet, M. Bakkus, H. Bazin, F. Tielemans, O. Leo, J. Urbain, and M. Moser. 1994. Murine dendritic cells pulsed in vitro with tumor antigen induce tumor resistance in vivo. Eur. J. Immunol. 24:605-610. [DOI] [PubMed] [Google Scholar]
- 17.Fong, L., and E. G. Engleman. 2000. Dendritic cells in cancer immunotherapy. Annu. Rev. Immunol. 18:245-273. [DOI] [PubMed] [Google Scholar]
- 18.Frolov, I., T. A. Hoffman, B. M. Pragai, S. A. Dryga, H. V. Huang, S. Schlesinger, and C. M. Rice. 1996. Alphavirus-based expression vectors: strategies and applications. Proc. Natl. Acad. Sci. USA 93:11371-11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herold, J., T. Raabe, B. Schelle-Prinz, and S. G. Siddell. 1993. Nucleotide sequence of the human coronavirus 229E RNA polymerase locus. Virology 195:680-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herold, J., T. Raabe, and S. Siddell. 1993. Molecular analysis of the human coronavirus (strain 229E) genome. Arch. Virol. Suppl. 7:63-74. [DOI] [PubMed]
- 21.Herold, J., and S. G. Siddell. 1993. An “elaborated” pseudoknot is required for high-frequency frameshifting during translation of HCV 229E polymerase mRNA. Nucleic Acids Res. 21:5838-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenne, L., G. Schuler, and A. Steinkasserer. 2001. Viral vectors for dendritic cell-based immunotherapy. Trends Immunol. 22:102-107. [DOI] [PubMed] [Google Scholar]
- 23.Jeong, Y. S., J. F. Repass, Y. N. Kim, S. M. Hwang, and S. Makino. 1996. Coronavirus transcription mediated by sequences flanking the transcription consensus sequence. Virology 217:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai, M. M., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 27.Ludewig, B., F. Barchiesi, M. Pericin, R. M. Zinkernagel, H. Hengartner, and R. A. Schwendener. 2001. In vivo antigen loading and activation of dendritic cells via a liposomal peptide vaccine mediates protective antiviral and anti-tumor immunity. Vaccine 19:23-32. [DOI] [PubMed] [Google Scholar]
- 28.Ludewig, B., K. McCoy, M. Pericin, A. F. Ochsenbein, T. Dumrese, B. Odermatt, R. E. Toes, C. J. Melief, H. Hengartner, and R. M. Zinkernagel. 2001. Rapid peptide turnover and inefficient presentation of exogenous antigen critically limit the activation of self-reactive CTL by dendritic cells. J. Immunol. 166:3678-3687. [DOI] [PubMed] [Google Scholar]
- 29.Mayr, A., and K. Malicki. 1966. Attenuation of virulent fowl pox virus in tissue culture and characteristics of the attenuated virus. Zentbl. Veterinärmed. 13:1-13. (In German.) [PubMed]
- 30.Nair, S. K., D. Boczkowski, M. Morse, R. I. Cumming, H. K. Lyerly, and E. Gilboa. 1998. Induction of primary carcinoembryonic antigen (CEA)-specific cytotoxic T lymphocytes in vitro using human dendritic cells transfected with RNA. Nat. Biotechnol. 16:364-369. [DOI] [PubMed] [Google Scholar]
- 31.Narayanan, K., C. J. Chen, J. Maeda, and S. Makino. 2003. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 77:2922-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nestle, F. O. 2000. Dendritic cell vaccination for cancer therapy. Oncogene 19:6673-6679. [DOI] [PubMed] [Google Scholar]
- 33.Nestle, F. O., S. Alijagic, M. Gilliet, Y. Sun, S. Grabbe, R. Dummer, G. Burg, and D. Schadendorf. 1998. Vaccination of melanoma patients with peptide- or tumor lysate-pulsed dendritic cells. Nat. Med. 4:328-332. [DOI] [PubMed] [Google Scholar]
- 34.Ortego, J., D. Escors, H. Laude, and L. Enjuanes. 2002. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 76:11518-11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ossevoort, M. A., M. C. Feltkamp, K. J. van Veen, C. J. Melief, and W. M. Kast. 1995. Dendritic cells as carriers for a cytotoxic T-lymphocyte epitope-based peptide vaccine in protection against a human papillomavirus type 16-induced tumor. J. Immunother. Emphasis Tumor Immunol. 18:86-94. [DOI] [PubMed] [Google Scholar]
- 36.Pasternak, A. O., E. van den Born, W. J. Spaan, and E. J. Snijder. 2001. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 20:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rea, D., M. J. Havenga, M. van Den Assem, R. P. Sutmuller, A. Lemckert, R. C. Hoeben, A. Bout, C. J. Melief, and R. Offringa. 2001. Highly efficient transduction of human monocyte-derived dendritic cells with subgroup B fiber-modified adenovirus vectors enhances transgene-encoded antigen presentation to cytotoxic T cells. J. Immunol. 166:5236-5244. [DOI] [PubMed] [Google Scholar]
- 38.Rosenzwajg, M., L. Tailleux, and J. C. Gluckman. 2000. CD13/N-aminopeptidase is involved in the development of dendritic cells and macrophages from cord blood CD34+ cells. Blood 95:453-460. [PubMed] [Google Scholar]
- 39.Sawicki, D., T. Wang, and S. Sawicki. 2001. The RNA structures engaged in replication and transcription of the A59 strain of mouse hepatitis virus. J. Gen. Virol. 82:385-396. [DOI] [PubMed] [Google Scholar]
- 40.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215-219. [DOI] [PubMed] [Google Scholar]
- 42.Sethna, P. B., M. A. Hofmann, and D. A. Brian. 1991. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J. Virol. 65:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siddell, S. G., and E. J. Snijder. 1998. Coronaviruses, toroviruses and arteriviruses, p. 463-484. In B. W. J. Mahy and L. Collier (ed.), Topley & Wilson's microbiology and microbial infections, 9th ed. Arnold, London, United Kingdom.
- 44.Song, W., H. L. Kong, H. Carpenter, H. Torii, R. Granstein, S. Rafii, M. A. Moore, and R. G. Crystal. 1997. Dendritic cells genetically modified with an adenovirus vector encoding the cDNA for a model antigen induce protective and therapeutic antitumor immunity. J. Exp. Med. 186:1247-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spaan, W., H. Delius, M. Skinner, J. Armstrong, P. Rottier, S. Smeekens, B. A. van der Zeijst, and S. G. Siddell. 1983. Coronavirus mRNA synthesis involves fusion of noncontiguous sequences. EMBO J. 2:1839-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Specht, J. M., G. Wang, M. T. Do, J. S. Lam, R. E. Royal, M. E. Reeves, S. A. Rosenberg, and P. Hwu. 1997. Dendritic cells retrovirally transduced with a model antigen gene are therapeutically effective against established pulmonary metastases. J. Exp. Med. 186:1213-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stirrups, K., K. Shaw, S. Evans, K. Dalton, R. Casais, D. Cavanagh, and P. Britton. 2000. Expression of reporter genes from the defective RNA CD-61 of the coronavirus infectious bronchitis virus. J. Gen. Virol. 81:1687-1698. [DOI] [PubMed] [Google Scholar]
- 48.Thiel, V., J. Herold, B. Schelle, and S. G. Siddell. 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 82:1273-1281. [DOI] [PubMed] [Google Scholar]
- 49.Thiel, V., J. Herold, B. Schelle, and S. G. Siddell. 2001. Viral replicase gene products suffice for coronavirus discontinuous transcription. J. Virol. 75:6676-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thiel, V., A. Rashtchian, J. Herold, D. M. Schuster, N. Guan, and S. G. Siddell. 1997. Effective amplification of 20-kb DNA by reverse transcription PCR. Anal. Biochem. 252:62-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thiel, V., S. G. Siddell, and J. Herold. 1998. Replication and transcription of HCV 229E replicons. Adv. Exp. Med. Biol. 440:109-113. [DOI] [PubMed] [Google Scholar]
- 52.van der Most, R. G., R. J. de Groot, and W. J. Spaan. 1994. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J. Virol. 68:3656-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Marle, G., W. Luytjes, R. G. van der Most, T. van der Straaten, and W. J. Spaan. 1995. Regulation of coronavirus mRNA transcription. J. Virol. 69:7851-7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wege, H., K. Nagashima, and V. ter Meulen. 1979. Structural polypeptides of the murine coronavirus JHM. J. Gen. Virol. 42:37-47. [DOI] [PubMed] [Google Scholar]
- 55.Woo, K., M. Joo, K. Narayanan, K. H. Kim, and S. Makino. 1997. Murine coronavirus packaging signal confers packaging to nonviral RNA. J. Virol. 71:824-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeager, C. L., R. A. Ashmun, R. K. Williams, C. B. Cardellichio, L. H. Shapiro, A. T. Look, and K. V. Holmes. 1992. Hum. aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yount, B., K. M. Curtis, and R. S. Baric. 2000. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 74:10600-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ziebuhr, J., and S. G. Siddell. 1999. Processing of the human coronavirus 229E replicase polyproteins by the virus-encoded 3C-like proteinase: identification of proteolytic products and cleavage sites common to pp1a and pp1ab. J. Virol. 73:177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]
- 61.Zitvogel, L., J. I. Mayordomo, T. Tjandrawan, A. B. DeLeo, M. R. Clarke, M. T. Lotze, and W. J. Storkus. 1996. Therapy of murine tumors with tumor peptide-pulsed dendritic cells: dependence on T cells, B7 costimulation, and T helper cell 1-associated cytokines. J. Exp. Med. 183:87-97. [DOI] [PMC free article] [PubMed] [Google Scholar]