Abstract
We investigated the parameters driving nelfinavir resistance, along the D30N and L90M evolutionary pathways. The advantage of the D30N mutant was mostly due to its resistance level, while the L90M mutation allowed preservation of infectivity coupled with minimal resistance. Emergence of secondary mutations further increased the selective advantage of viruses harboring D30N.
Analysis of the Stanford database (http://hivdb.stanford.edu) on human immunodeficiency virus type 1 (HIV-1) resistance to antivirals showed that resistance to the protease inhibitor nelfinavir most often begins with the selection of the D30N mutation in the viral protease (38% of 322 isolates from 261 patients treated with nelfinavir as the first protease inhibitor). In 12% of patients, however, the alternative L90M mutation was initially selected. While D30N appears to be specific for nelfinavir resistance (7), L90M is selected in patients treated with saquinavir and nelfinavir and, to a lesser extent, other protease inhibitors (2, 10, 12). Mutants carrying both these substitutions are only rarely found in vivo and generally have additional, potentially compensatory, mutations (11). Interestingly, a major loss of replicative capacity for a mutant clone carrying both D30N and L90M substitutions was recently described (11). These observations suggest the existence of two competing alternative pathways for nelfinavir resistance. Here, we investigated the parameters that determine the emergence of resistant virus mutants following either the D30N or L90M pathway.
To this end, a series of mutant clones were compared for their replicative capacities in the absence of drug, resistance to nelfinavir, and replicative capacities as a function of drug concentration. Mutations in the protease were introduced by oligonucleotide-directed mutagenesis using a modified pBluescript II subclone that contains the entire HIV-1 protease-coding region, surrounded by unique restriction sites (4). Mutated protease alleles were then cloned into a variant of the pNL4.3 HIV-1 molecular clone (pNL-4.3XCS [4]). Three putative compensatory protease mutations are often observed in combination with D30N in nelfinavir-treated patients: L63P, a common polymorphism previously shown to compensate for the loss of infectivity resulting from several protease mutations (6); A71V, a mutation selected by several protease inhibitors; and N88D, a substitution essentially found in nelfinavir-treated patients. To compare the effects of D30N, L90M, and secondary mutations, mutants with these substitutions were cloned alone or in combination (Fig. 1). In addition, a mutant carrying both D30N and L90M was also constructed, along with clones that carry this combination in the context of additional protease mutations.
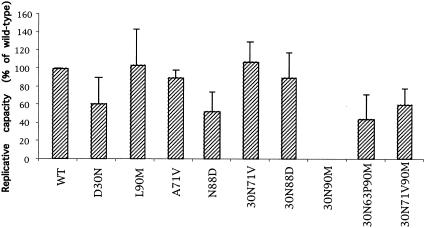
FIG. 1.
Impact of protease mutations on drug-free virus replicative capacity. Mutant virus replicative capacity was measured in a single-cycle assay in the absence of drug and expressed as a percentage of wild-type (WT) virus replicative capacity. The average values and standard deviations obtained from at least three independent experiments are shown.
Impact of nelfinavir resistance mutations on drug-free replicative capacity.
Several assays are used to compare virus replicative capacity (reviewed in reference 8). Despite differences in the magnitude of the effect of mutations measured by different techniques, these assays provide concordant results for the relative replicative capacities of mutant viruses (5, 9). We determined the impact of single and combined mutations in the protease on drug-free virus replicative capacity, as measured by a single-cycle infectivity assay. This assay is based on the measurement of β-galactosidase expression following HIV infection of CD4+ HeLa cells with a long terminal repeat (LTR) fused to the LacZ gene (HeLa-CD4-LTR-LacZ cells) (3, 4). The replicative capacity of mutant viral particles produced in drug-free cultures and normalized by p24 antigen concentration was expressed as the percentage of that of the wild type (Fig. 1). The mutation D30N was found to markedly affect drug-free replicative capacity in our single-cycle assay, in agreement with previous studies (5, 11). In contrast, the replicative capacity of the L90M mutant was not significantly different from that of wild-type virus, as previously reported (4, 5, 11). Similar to the D30N mutant, the N88D mutant was characterized by a marked loss of replicative capacity, while a clone carrying the A71V mutation alone was previously shown to display wild-type virus infectivity (4). Differences in replicative capacity were analyzed by comparison of multiple groups by one-way analysis of variance. Posttest comparisons, performed only if P was <0.05, were made by using Dunnett's multiple-comparison test and showed a statistically significant reduction of replicative capacity for both the D30N (P < 0.05) and N88D mutants (P < 0.001).
Analysis of clones that carry combinations of mutations showed that a mutant with both D30N and N88D (30N-88D mutant), the most common combination found in patients treated by nelfinavir, was more infectious than viruses carrying either of these mutations alone. This is in agreement with the idea that N88D is selected to compensate for the loss of replicative capacity resulting from D30N. The addition of the A71V mutation also efficiently compensated for the D30N defect (Fig. 1). Together these data show that D30N and N88D have stronger impact on virus replicative capacity than L90M and that the loss of replicative capacity associated with the D30N mutation can be rescued by common secondary mutations.
A recombinant virus clone carrying the combination of D30N and L90M mutations displayed an infectivity that was at the detection limit of our system, confirming that the association of these mutations is deleterious for virus infectivity. Interestingly, however, the addition of the common polymorphism L63P or the mutation A71V largely rescued virus replicative capacity (Fig. 1), indicating that the defect associated with the D30N-L90M combination is highly context dependent. Interestingly, both L63P and A71V mutations are frequently observed in the few patients in the Stanford database whose viruses carry D30N together with L90M.
Effect of mutations in the protease on resistance to nelfinavir.
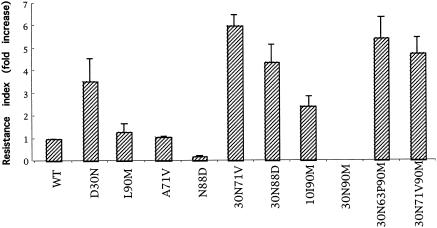
To compare the effects of the above-described mutations on the level of resistance to nelfinavir, the concentrations inhibiting 50 and 90% of the infectious events (IC50 and IC90, respectively) were determined. HeLa cells were transfected with wild-type or mutant full-length molecular clones. At 12 h posttransfection, cells were trypsinized and plated in 96-well plates. Producer cells in triplicate wells were immediately treated with 0, 0.8, 4, 20, 100, 500, or 2,500 nM nelfinavir for 24 h, and virions produced in the supernatant of these cultures were used to infect HeLa-CD4-LTR-LacZ target cells. Target cells were lysed 40 h after infection, a β-galactosidase chromogenic substrate (CPRG) was added, and optical densities were measured as previously described (4). IC50 and IC90 values were calculated by comparing the optical densities obtained with virions produced in the absence and in the presence of increasing concentrations of nelfinavir in at least three independent experiments. Resistance was expressed as the change in susceptibility to nelfinavir compared to that of wild-type virus (resistance index). Mean changes in IC90 values for the different mutants are reported in Fig. 2.
FIG. 2.
Effect of protease mutations on nelfinavir resistance. Resistance was expressed as the change in susceptibility to nelfinavir compared to that of wild-type (WT) virus (resistance index). Mean changes in IC90 values for the different mutants, obtained from at least three independent experiments, are shown.
In our assay system, mean IC50 and IC90 values for wild-type virus were 21 and 83 nM, respectively. Among the single mutants, only the D30N mutant was significantly more resistant to nelfinavir than the wild type (3.6-fold increase). The susceptibilities of the L90M and A71V mutants were similar to that of the wild type, while the N88D mutant was found to be highly susceptible to nelfinavir. The N88S (an alternative to N88D) mutant has also been reported to be highly susceptible to amprenavir (9). The addition of mutation A71V to D30N conferred high-level resistance to nelfinavir (sixfold). Relatively strong resistance was also observed for the 30N-88D mutant. The 10I-90M double mutant was more resistant to nelfinavir than the L90M clone, but it was still more susceptible than the mutant containing D30N alone. These data show that the genetic barrier to nelfinavir resistance is lower for mutants that engage in the D30N pathway than for those in the L90M series.
Given the extremely low infectivity of the 30N-90M mutant, we could not determine the level of resistance to nelfinavir for this mutant. Mutants with an additional change (L63P or A71V) that compensated for the replicative defect of the 30N-90M mutant (30N-63P-90M and 30N-71V-90M mutants; Fig. 2) appeared to be relatively resistant to nelfinavir. Similar results were obtained when resistance levels based on IC50 values were compared (not shown).
Selective advantage as a function of drug concentration.
The selective advantage conferred by a combination of protease mutations depends both on the impact of the mutations on virus replicative capacity and on the gain in resistance (reviewed in references 1 and 8). Previous work conducted with some of the mutants described here was limited to the independent assessment of these parameters (7) or to the exclusive measurement of drug-free infectivity (11). In contrast, we have previously shown that, by comparing the ratio of mutant to wild-type virus replicative capacities at increasing drug concentrations, the range of concentrations for which a given combination of mutations conferred a selective advantage and the extent of this advantage can be directly measured (4). We applied this approach to the series of mutants described above (Fig. 3). The optical density obtained after infection with viruses produced in the presence of increasing nelfinavir concentrations was measured, and the ratio of mutant to wild-type virus was calculated and plotted against drug concentration in Fig. 3. The curves represent the averages of at least three independent experiments. Comparison of mutants carrying single amino acid changes (Fig. 3A) shows that both D30N and L90M conferred a selective advantage to the virus in the presence of nelfinavir. The ranges of concentrations for which the two mutant viruses displayed higher replicative capacity than wild-type virus largely overlapped, but the extent of the advantage (heights of the peaks) was consistently greater for the D30N mutant. The increased susceptibility of the N88D mutant to nelfinavir (Fig. 2), together with its reduced replicative capacity in the absence of the drug (Fig. 1), resulted in a peculiar shape of the curve for this virus, which was found to display a replicative capacity lower than that of the wild-type virus for all drug concentrations used in our study (Fig. 3A). The A71V mutant displayed a wild-type replicative capacity over the range of nelfinavir concentrations tested (Fig. 3A). Statistical analysis was performed by comparison of multiple groups using analysis of variance. A posttest comparison, performed only if P was <0.05, was made using Dunnett's multiple-comparison test. The replicative capacities of D30N and L90M mutants were confirmed to be significantly different from that of wild-type virus at 100 nM nelfinavir (P < 0.001 for the D30N mutant, and P < 0.05 for the L90M mutant).
FIG. 3.
Virus replicative capacity as a function of drug concentration. The ratio of mutant to wild-type virus replicative capacities was measured for virions produced in the presence of increasing nelfinavir concentrations. The curves represent average values obtained from at least three independent experiments. (A) Single mutants; (B) mutants carrying multiple protease substitutions (note the different scale).
The advantage of mutants carrying multiple amino acid changes in the protease is shown in Fig. 3B (note that different scales are used for panels A and B). The selective advantage of the 30N-71V mutant was significant at both 100 and 500 nM nelfinavir (P < 0.001 for both concentrations), and the extent of the advantage for this clone was far greater than those for all the other clones analyzed here. For the other clones carrying multiple protease mutations, the selective advantage was statistically significant at 100 nM nelfinavir (P < 0.001 for all mutants). The 30N-88D clone was reproducibly the second-best double mutant evaluated in our study, and the advantage conferred by this combination was greater than that conferred by the 10I-90M combination. These results are consistent with the more frequent detection of mutants carrying the D30N mutation in nelfinavir-treated patients. These findings may also explain why mutants carrying L90M are not observed in in vitro selection experiments, because relatively high drug concentrations are commonly used (7).
Our study, comparing the selective advantage for viral replication in the presence of nelfinavir conferred by different combinations of resistance mutations, reveals several findings. Both D30N and L90M confer a selective advantage for replication in the presence of low nelfinavir concentrations. The advantage for the D30N mutant mostly comes from resistance, while the advantage for the L90M mutant reflects preservation of infectivity coupled with a minimal reduction in susceptibility. At higher nelfinavir concentrations, viruses engaged in the D30N pathway can reach higher levels of resistance and display a higher selective advantage than mutants of the L90M series. Thus, the initial selection of D30N results in engagement in an evolutionary pathway with a lower genetic barrier for nelfinavir resistance. In the absence of compensatory mutations, the combination of D30N and L90M mutations dramatically reduces virus replicative capacity. Nevertheless, a relatively frequent polymorphism (L63P) or the presence of the common resistance mutation A71V relieved this barrier, reinforcing the notion that the genetic context of the virus may substantially influence the impact of resistance mutations.
Acknowledgments
This work was supported in part by a grant from the Agence Nationale de Recherche sur le SIDA and by European Union grant QLK2-CT-2001-02360.
We thank Mark Becker (Agouron Pharmaceutical, Inc.) for kindly providing nelfinavir.
REFERENCES
- 1.Clavel, F., E. Race, and F. Mammano. 2000. HIV drug resistance and viral fitness. Adv. Pharmacol. 49:41-66. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 3.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mammano, F., V. Trouplin, V. Zennou, and F. Clavel. 2000. Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: virus fitness in the absence and in the presence of drug. J. Virol. 74:8524-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez-Picado, J., A. V. Savara, L. Sutton, and R. T. D'Aquila. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 7.Patick, A. K., M. Duran, Y. Cao, D. Shugarts, M. R. Keller, E. Mazabel, M. Knowles, S. Chapman, D. R. Kuritzkes, and M. Markowitz. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob. Agents Chemother. 42:2637-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quinones-Mateu, M. E., and E. J. Arts. 2001. HIV-1 fitness: implications for drug resistance, disease progression, and global epidemic evolution, p134-170. In C. Kuiken (ed.), HIV sequence compendium. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 9.Resch, W., R. Ziermann, N. Parkin, A. Gamarnik, and R. Swanstrom. 2002. Nelfinavir-resistant, amprenavir-hypersusceptible strains of human immunodeficiency virus type 1 carrying an N88S mutation in protease have reduced infectivity, reduced replication capacity, and reduced fitness and process the Gag polyprotein precursor aberrantly. J. Virol. 76:8659-8666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts, N. A. 1995. Drug-resistance patterns of saquinavir and other HIV proteinase inhibitors. AIDS 9(Suppl. 2):S27-S32. [PubMed] [Google Scholar]
- 11.Sugiura, W., Z. Matsuda, Y. Yokomaku, K. Hertogs, B. Larder, T. Oishi, A. Okano, T. Shiino, M. Tatsumi, M. Matsuda, H. Abumi, N. Takata, S. Shirahata, K. Yamada, H. Yoshikura, and Y. Nagai. 2002. Interference between D30N and L90M in selection and development of protease inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 46:708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaillancourt, M., D. Irlbeck, T. Smith, R. W. Coombs, and R. Swanstrom. 1999. The HIV type 1 protease inhibitor saquinavir can select for multiple mutations that confer increasing resistance. AIDS Res. Hum. Retrovir. 15:355-363. [DOI] [PubMed] [Google Scholar]