Abstract
Feline panleukopenia virus (FPV) and its host range variant, canine parvovirus (CPV), can bind the feline transferrin receptor (TfR), while only CPV binds to the canine TfR. Introducing two CPV-specific changes into FPV (at VP2 residues 93 and 323) endowed that virus with the canine TfR binding property and allowed canine cell infection, although neither change alone altered either property. In CPV the reciprocal changes of VP2 residue 93 or 323 to the FPV sequences individually resulted in modest reductions in infectivity for canine cells. Changing both residues in CPV to the FPV amino acids blocked the canine cell infection, but that virus was still able to bind the canine TfR at low levels. This shows that both CPV-specific changes control canine TfR binding but that binding is not always sufficient to mediate infection.
Canine parvovirus (CPV) emerged in the late 1970s as the cause of a new disease in dogs and is now prevalent in dogs worldwide. CPV was most likely derived as a host range variant of the long-known feline panleukopenia virus (FPV), which infects cats and some other carnivores, but not dogs (11, 18). FPV became adapted to dogs through a series of steps, the first being the emergence of the ancestor of the CPV viruses, which gave rise to a variant (designated CPV type 2) which spread worldwide during 1978 (30). However, by 1980 the CPV type 2 strain had been replaced worldwide by a variant strain designated CPV type 2a, which has remained prevalent in dogs with only a small number of additional changes (19, 30). Each of these evolutionary steps involved alterations in the capsid, which changed both its antigenic and host range properties (19, 22).
Parvoviruses have 25-nm-diameter T=1 icosahedral capsids which are assembled from 60 copies of a combination of the overlapping VP1 and VP2 proteins (31). The VP1 and VP2 structures contain an eight-stranded antiparallel β-barrel, with four large loops inserted between some of the β strands making up much of the viral surface (31). Features of the capsid surface include cylinders around the fivefold axes, depressions (the dimples) spanning the twofold axes of symmetry, and 22-Å-high raised regions (the threefold spikes) at the threefold axes of symmetry (1, 31, 33).
Residues in three regions of the capsid surface control the ability of CPV to infect dog cells. Two of those were seen as differences between FPV isolates and CPV type 2, where the changes in FPV of VP2 residue Lys 93 to Asn and Asp 323 to Asn allowed the virus to infect dog cells (6, 9). In a CPV background, changing either residue 93 or 323 to the FPV residue reduced the ability of the virus to infect dog cells (6). The side chain of residue 93 is exposed on the surface of the capsid on the top of loop 1 (1, 31), and in FPV Lys 93 forms two hydrogen bonds with backbone oxygen atoms of residues 225 and 227 of loop 2, while those bonds are not formed by the Asn in CPV (1, 31, and L. Govindasamy, K. Hueffer, C. R. Parrish, and M. Agbandje-McKenna, submitted for publication) (Fig. 1). VP2 residue 323 is separated from residue 93 by 20 Å, and the structure near that position is controlled in part by the binding of up to three divalent ions (25 and L. Govindasamy et al., submitted). A third region controlling infectivity for dog cells is within a ridge on the side of the threefold spike (the shoulder), and that contains three differences between CPV type 2 and FPV (VP2 residues 80, 564, and 568) which are not exposed on the surface of the capsid (13, 17). The shoulder region also acquired three additional and surface-exposed differences during the evolution to the CPV type 2a strain (19). In addition, some changes in that region can eliminate canine cell infection of CPV (13, 17).
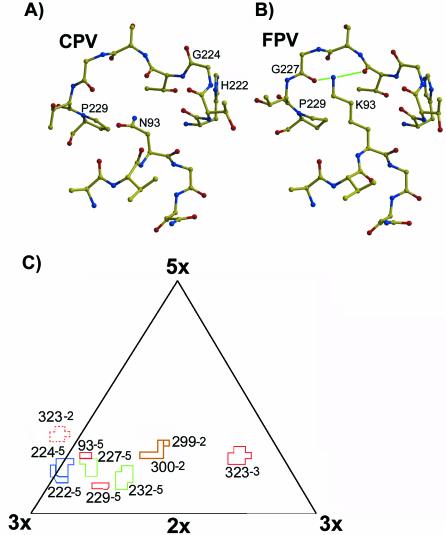
FIG. 1.
Structures of CPV and FPV within the areas examined in this study. (A) Atomic model of the structure of the CPV capsid in the vicinity of VP2 residue 93. Labels indicate the VP2 residues that were substituted in these studies. Loop 1 is shown between VP2 residues 91 and 95, while loop 2 is shown between residues 222 and 229. (B) Atomic model of FPV around residue 93 in the same orientation as in panel A. Labels indicate the VP2 residues that were substituted during these studies. Hydrogen bonds between residue 93 and the backbone oxygen atoms of residues 225 and 227 are indicated by green lines. (C) Residues mutated in these studies shown on a roadmap of one asymmetric unit of the CPV capsid (24), showing surface-exposed residues examined in this study. The number following the VP2 residue indicates the relationship of the VP2 molecule containing the residue relative to the reference VP2 monomer in the standard orientation (2×, twofold; 3×, threefold; 5×, fivefold) (1). Residues that were changed in CPV are marked in blue, those changed in FPV are marked in green, and those changed in both viruses are colored in red. Residues 299 and 300, which were not altered in these studies but which also affect canine host range, are shown in brown. Other surface-exposed residues are not shown here. The position of residue 323 in a neighboring asymmetric unit is indicated by the dashed lines.
CPV and FPV both bind to the feline transferrin receptor (TfR) and use that receptor to infect feline cells (16), and the host range of CPV for canine cells is determined by the specific ability of that virus to bind the canine TfR (11). Binding to the canine TfR was lost in a host range mutant of CPV which had a single substitution within the shoulder region of the threefold spike (Gly 299 to Glu), but whether the other residues controlling host range also affect canine TfR binding and its use for infection is not known (11). Here we examined the specific roles of residues 93 and 323 in controlling the binding of the canine TfR and the effects on infection of canine cells.
To determine the functions of residues 93 and 323, we replaced VP2 residue 93 and neighboring residues on an adjacent loop with alternative amino acids (Fig. 1 and 2). Residue 323 was also changed alone or in combination with residue 93 so as to explore the functional interactions between those residues (Fig. 1 and 2). Mutant viruses were prepared either by site-directed mutagenesis of M13 uracilated single-stranded DNA (12) or by using the Gene Editor protocol on the intact infectious plasmid (Promega, Madison, Wis.). Some mutants had previously been isolated as antibody neutralization escape mutants (28).
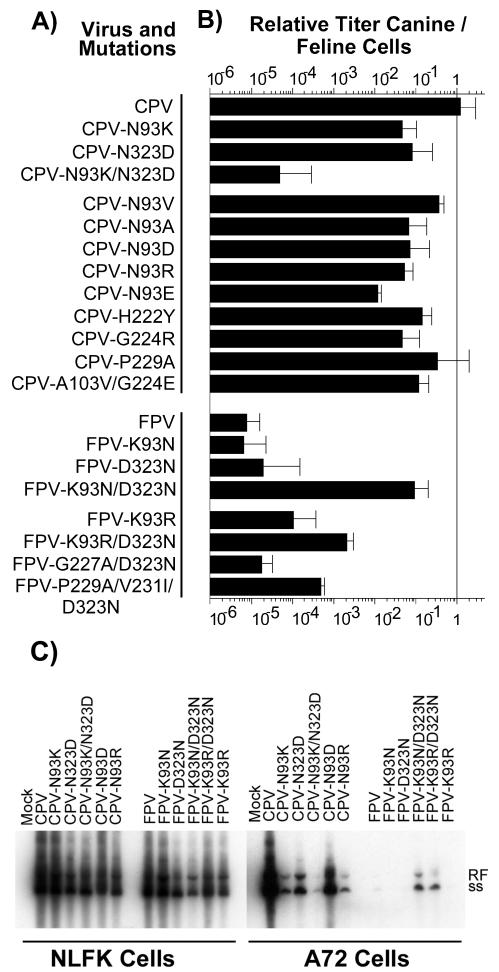
FIG. 2.
Mutants prepared in these studies and their relative titers in feline and canine cells. (A) The genetic background (CPV or FPV) of each virus is indicated, as well as the changes(s) introduced into the VP2 sequence. The single-letter code for amino acids was used, with the letter before the number indicating the amino acid of the wild-type virus and that following indicating the change(s) introduced. (B) Relative infectivity of each virus for canine cells shown as the ratio of the TCID50 titer of the virus stock in A72 cells to that in NLFK cells. Error bars show one standard deviation of the mean for at least three independent experiments. For the CPV-P229A mutant, data from one (ratio, 2.8 log10) of seven independent experiments was excluded from the analysis. (C) Infection and replication in NLFK and A72 cells of selected viruses examined by the production of viral RF and single-stranded DNA (ss). Cells seeded in six-well plates were inoculated with 0.5 TCID50 per cell and were incubated for 2 days, and then the low-molecular-weight DNA was recovered, electrophoresed in an agarose gel, transferred to a membrane, and detected with a 32P-labeled DNA probe. The samples from NLFK cells were exposed for half the exposure of the A72 cell-derived samples.
When mutant genomes were tested for viability and host range, it was apparent that considerable variation was allowed in the structure of the CPV capsid near residue 93 without affecting feline cell infection. All the viruses retained feline TfR binding and feline cell infection (Fig. 2). Most CPV mutants with single substitutions also retained the ability to infect dog cells, although many showed lower titers in those cells than in feline cells (Fig. 2B). In that assay, NLFK or A72 cells in 96-well plates were inoculated with 10-fold dilutions of virus, were incubated for 1 h, and then were incubated for a further 2 days in growth medium, after which the cells were fixed and stained for viral antigen (17). The ratio between the titers obtained for each virus in dog and cat cells measured the efficiency of canine cell infection (Fig. 2B). Wild-type CPV infected both NLFK and A72 cells to similar titers, but all CPV mutants with single changes of residue 93 infected dog cells at 10- to 100-fold lower titers than were seen with the feline cells (Fig. 2B). CPV with a single change of residue 323 to Asp (CPV-N323D) also showed a 10-fold reduction in canine cell infection, but the double mutant CPV-N93K/N323D was restricted by 10−4, similar to the case of FPV (Fig. 2B).
FPV was highly restricted in dog cells, infecting only to 10−5 the titer seen in cat cells. However, FPV-K93N/D323N infected canine cells to levels only 10-fold lower than levels for CPV, but changing either residue 93 or 323 alone to Asn resulted in no increase in FPV infectivity for canine cells (Fig. 2B). Replacing residue 93 in FPV with Arg instead of Lys along with the 323 change to Asn (FPV-K93R/D323N) resulted in a 250-fold increase in canine cell infectivity compared to that for wild-type FPV (Fig. 2B).
Infectivities for NLFK and A72 cells were also examined by testing for replicative-form (RF) DNA production after inoculation with CPV and FPV mutants containing reciprocal substitutions of residues 93 and 323 or with alternative changes of residue 93 (CPV-N93D, CPV-N93R, FPV K93R, and FPV-K93R/D323N). A72 or NLFK cells seeded at 104 cells per cm2 in 9.6-cm2 cultures were inoculated with a 50% tissue culture infective dose (TCID50) of each virus of 0.5 per cell (as determined in NLFK cells), and then low-molecular-weight DNA was recovered 2 days later according to a modification of the method of Hirt (8, 17). After electrophoresis in a 1% agarose gel containing 1 μg of ethidium bromide/ml, the DNA was blotted to a membrane and was probed with a 32P-labeled DNA sequence from the CPV genome. All mutants were infected at similar levels in NLFK cells, while in A72 cells CPV-N93K, CPV-N93D, CPV-N93R, and CPV-N323D showed reduced levels of both infection and RF DNA production compared to those of wild-type CPV, but CPV-N93K/N323D did not infect to a detectable level (Fig. 2C). FPV-K93N/D323N infected more efficiently than FPV-K93R/D323N but did so at lower levels than CPV, and little or no infection was observed for wild-type FPV, FPV-K93N, FPV-D323N, or FPV-K93R (Fig. 2C). Although the results of the TCID50 and Southern blots were closely correlated, differences in the assays occurred for CPV-N93D, which showed more DNA in the canine cell cultures than might be predicted from the TCID50 assay. This is likely explained by the fact that the TCID50 detects only the initial cell infection, while the RF DNA would measure multiple rounds of replication for the viruses that can infect the cells.
In earlier studies using plaque titrations, a greater decrease in canine cell infection was observed for single-point mutants in a CPV background, although the results of DNA analysis were similar to those observed here (6). The TCID50 assay used here is considerably more sensitive than the plaque assay for detecting infections by viruses in cells that are relatively resistant to the virus, as multiple rounds of infection of cells would be required to form a detectable plaque (6).
The antigenic properties of the mutant viruses were tested using monoclonal antibodies (MAb) in a hemagglutination inhibition (HI) assay (20, 21, 28). Most mutated viruses showed reduced binding to several MAb that recognize antigenic site A, which includes the regions around residue 93 (28) (Fig. 3). An Asn at residue 93 creates an epitope recognized by all the CPV-specific MAb tested (MAb 14, 7, 13, and 2A9), and substitution of residue 93 with any of several other amino acids reduced or eliminated that epitope (Fig. 3). Antibodies recognizing antigenic site A were clearly subdivided into two groups: the CPV-specific antibodies that were affected by changes of residue 93 from Asn to most other amino acids and those that were not affected by changes of residue 93 but which were affected by changes of residues 222 and 224 (Fig. 3). These mutants show that the wild-type CPV structure was not required for the virus to infect canine cells, as many mutant viruses showing reduced or no reactivity with CPV-specific MAb still infected canine cells, albeit with reduced efficiency compared to that of the wild type (Fig. 2 and 3). Changing residue 323 alone did not alter the reactivity with any of the MAb tested, and none of the mutants showed altered reactivity with MAb 8, which recognizes antigenic site B (Fig. 3).
FIG. 3.
Antigenic analysis of the viruses examined by using MAb in an HI assay. Eight hemagglutinin units of virus were incubated for 1 h with twofold dilutions of monoclonal antibodies in the assay, and then erythrocytes were added. The results are shown as the titer of the antibodies for each virus relative to that of wild-type CPV. Filled boxes, mutant HI at 10 to 100% wild-type titer; cross-hatched boxes, less than 10% but greater than 1% of the wild-type titer; open boxes, less than 1% of the wild-type titer.
Infection of canine cells partially correlated with the degree of virus binding and uptake into those cells (Fig. 4). For the binding assays, cells seeded at 5 × 104 cells per cm2 in 10-cm2 dishes were incubated for 1 h at 37°C with a 10-μg/ml concentration of purified empty capsids of the indicated viruses (Fig. 4) (11). After being washed twice in cold Hanks buffered saline solution without Mg2+ or Ca2+ (HBSS), cells were detached with 1 mM EDTA in HBSS for 10 min at 4°C and then were fixed for 10 min at 22°C with 4% paraformaldehyde. Cell-associated virus was detected with MAb 8 conjugated to Cy2 (Amersham Biosciences, Piscataway, N.J.) in phosphate-buffered saline with 0.5% (wt/vol) bovine serum albumin and 0.5% (vol/vol) Triton X-100. Cells were analyzed by using a FACScalibur flow cytometer and Cell Quest software (Becton Dickinson, San Jose, Calif.). All viruses tested bound efficiently to the feline NLFK cells, although FPV, FPV mutants, and CPV-N93D and CPV-N93R all showed two- to threefold reduced binding levels compared to those for CPV (Fig. 4A).
FIG. 4.
Binding of CPV, FPV, or mutant virus capsids to feline or canine cells. Cells were incubated with 10 μg of virus/ml for 1 h at 37°C, and then after cell detachment, fixation, and permeabilization the capsid antigen was detected by antibody staining and flow cytometry. (A) Binding to feline cells of CPV capsids and mutants in a CPV background (upper panel) or FPV capsids and mutants in an FPV background (lower panel). (B) Binding to canine cells of CPV capsid and mutants in a CPV background (upper panel) or FPV capsids and mutants in an FPV background (lower panel). (C) Effect of temperature on binding of CPV or FPV capsids to feline or canine cells. Cells were incubated with 10 μg of virus/ml for 1 h at 4°C or at 37°C, and then after detachment, fixation, and permeabilization viral antigen was detected by antibody staining and flow cytometry.
Levels of virus binding to canine Cf2Th cells were more variable than those for binding to feline cells (Fig. 4B), and similar results were obtained for A72 cell binding of the same capsids (data not shown). CPV showed the strongest canine cell binding, while mutants of CPV with residue 93 or 323 replaced bound at lower levels, some at close to background levels (Fig. 4). The correlation between binding and infectivity of dog cells was most clearly seen for FPV and its mutants, where FPV-K93N/D323N bound and infected while wild-type FPV and the mutants FPV-K93N and FPV-K93R/D323N did not bind or infect those cells (Fig. 2 and 4B).
In previous studies viruses labeled with 35S or 3H were incubated with cells at 4°C, and no significant differences in binding to canine cells were seen between the closely related mink enteritis virus and CPV or between a host range mutant of CPV and wild-type CPV (10, 17). We examined the binding of the viruses to feline and canine cells at 37 and 4°C and found that at 4°C the CPV capsids bound to canine cells at levels close to background, similar to the binding seen for FPV capsids or for no capsids (Fig. 4C). At 4°C the binding to NLFK cells showed moderate decreases of three- to fourfold for CPV and 1.5- to 3.5-fold for FPV (Fig. 4C). This shows that binding is temperature dependent and that temperature directly influences the association between the capsids and the cell surface or the receptor(s).
We have shown that the specificity of capsid binding to the feline and canine TfRs correlates with the host range of the viruses and the susceptibility of the host cells (11). The binding of mutant capsids to the canine TfR was therefore tested on TRVb cells expressing the canine TfR from a plasmid after transfection, and the cells were simultaneously incubated with iron-loaded canine transferrin conjugated to Cy5 (11). All viruses tested efficiently bound the feline TfR expressed on TRVb cells (results not shown) (11, 16). All of the viruses that infected canine cells showed efficient binding to the expressed canine TfR at 37°C (Fig. 5). Mutants CPV-N93K, CPV-N93D, and CPV-N93R all bound the canine TfR, but they did so at levels lower than were seen for the wild-type virus, as higher expression levels of the TfR on the cells (as seen by transferrin binding) were necessary before virus binding was detected (Fig. 5A). Although CPV-N93K/N323D was not able to infect the canine cells, it still bound the canine TfR, although at levels that were lower than those for the single mutants (Fig. 5A). Of the FPV mutants, FPV-K93N and FPV-D323N did not show detectable binding to the canine TfR, while FPV-K93N/D323N, which was able to infect canine cells, bound at levels that were similar to those of CPV-N93K/N323D (Fig. 5B). It therefore appears that canine TfR binding is required for infection of canine cells, that additional host-specific steps are required for successful infection, and that infection is influenced by residues that act in combination with VP2 residues 93 and 323. In previous studies of CPV binding to the canine TfR, binding to the canine TfR expressed on TRVb cells led to only low levels of infection by the CPV type 2 strain, while those cells were more efficiently infected by the CPV type 2b strain (11).
FIG. 5.
Binding of transferrin and capsids of CPV, FPV, or mutant virus to TRVb cells expressing the canine TfR from plasmids. Transiently transfected TRVb cells expressing the canine TfR were incubated with both capsids and Cy5-labeled canine transferrin for 1 h at 37°C, and then after detachment, fixation, and permeabilization the viral antigen was detected by antibody staining and flow cytometry. Virus signal is shown on the X axis, while transferrin is shown on the Y axis. The numbers in the upper quadrants of the graphs represent the percentage of events that fell within those quadrants, and the numbers in parentheses show the percentage of positive cells (as shown by transferrin binding in the upper two quadrants) which bound virus in each case. (A) Binding to the canine TfR expressed on TRVb cells of CPV or mutants derived from CPV. (B) Binding to the canine TfR expressed on TRVb cells as well as on mock-inoculated cells of FPV or mutants derived from FPV.
VP2 residues 93 and 323 are separated by 20 Å in the capsid structure and there is no direct association between them, but both sites are required for successful capsid interactions with the canine TfR. Other sites in the CPV capsid are also involved in controlling canine TfR binding and canine cell infection. This was seen for CPV-N93K/N323D, which bound the canine TfR despite the FPV-derived Lys and Asp at residues 93 and 323, indicating that other structural differences in that virus must allow binding to the canine TfR in the presence of those changes. However, that virus-receptor interaction was not sufficient for infection. Mutant CPV-G299E does not infect canine cells or bind the canine TfR (11, 17), and residue 299 is >30 Å from both residues 93 and 323 (Fig. 1C), indicating that there is a third capsid region that also influences the canine TfR-virus interaction (Fig. 1C). From the structures of CPV and FPV, and also from new structures of additional mutant viruses CPV-N93D and CPV-N93R (L. Govindasamy et al., submitted), it is clear that the changes within each host-range-controlling region do not affect the structure at the other site. This suggests that the TfR apical domain, which mediates interaction with the capsid (15), engages all three host-range-determining regions to give a binding that can lead to infection.
These results have parallels with those for other parvoviruses. In the minute virus of mice (MVM), two strains can be distinguished on the basis of their tropisms for lymphoid and fibroblastic cells. MVM(p) can infect fibroblasts, while MVM(i) infects erythropoetic progenitor cells and lymphoid cells, including T cells (3, 7, 29). The fibrotropic phenotype of MVM(p) was controlled by a combination of residues 317 and 321 in VP2, which determine the host cell specificity in a coordinated manner (4). Those residues are located on the surface of the capsid on the side of the threefold spike and correspond to residues 318 and 323 of CPV (2). When MVM(i) containing single-point mutations of either residue were grown on fibroblasts to select for variants, viruses were isolated with mutations of residues 317 or 321 or of residues in nearby regions of the capsids. In MVM VP2 residue 95, which corresponds to CPV residue 93, is a Lys and forms a hydrogen bond to the backbone oxygen of residue 230, similar to FPV (2). In contrast to our data for FPV, the restricted infection of MVM strains was reported to be blocked after cell binding and uptake of the capsids (23, 26). In Aleutian mink disease virus (ADV) the ADV Utah strain does not replicate in cell culture but infects mink, while the ADV-G strain replicates in cell culture but not in mink (5). The differences controlling these tropisms were mapped to a region of the capsid gene corresponding to the tip of loop 1 in CPV (14, 27). One of these changes (Gln 94 to Lys) aligns with residue 93 in CPV. These similarities of changes controlling host range and tropism between CPV, MVM, and ADV suggest that the regions examined in this study play a central role in the general virus-cell interactions of parvoviruses.
Most viruses are well-adapted parasites and are restricted to a defined range of hosts or cell types. Rare transfers of mutant viruses with new host range properties into new hosts can lead to widespread distribution. From CPV and from other examples of host range adaptation, such as that of the influenza viruses (32), it is clear that emerging viruses of this type can become very successful in their new host. The finding that in CPV specific combinations of residues act in concert to control the new receptor-virus interactions gives a clearer understanding of the molecular mechanisms that permitted CPV emergence and evolution to occur. Several changes were required for FPV to become a new successful pathogen of dogs, and those most likely needed to arise in a particular order (11, 30). This suggests that the probability of this type of host shift is extremely low and likely occurs through multiple adaptive events. Early identification of new viruses may therefore allow control of the new virus before it becomes fully adapted to its new host.
Acknowledgments
Wendy Weichert and Gail Sullivan provided expert technical assistance. Hollis Erb provided help with data analysis.
This work was supported by grants AI 28385 and AI 33468 from the National Institutes of Health to C.R.P.
REFERENCES
- 1.Agbandje, M., R. McKenna, M. G. Rossmann, M. L. Strassheim, and C. R. Parrish. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16:155-171. [DOI] [PubMed] [Google Scholar]
- 2.Agbandje-McKenna, M., A. L. Llamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 6:1369-1381. [DOI] [PubMed] [Google Scholar]
- 3.Ball-Goodrich, L. J., R. D. Moir, and P. Tattersall. 1991. Parvoviral target cell specificity: acquisition of fibrotropism by a mutant of the lymphotropic strain of minute virus of mice involves multiple amino acid substitutions within the capsid. Virology 184:175-186. [DOI] [PubMed] [Google Scholar]
- 4.Ball-Goodrich, L. J., and P. Tattersall. 1992. Two amino acid substitutions within the capsid are coordinately required for acquisition of fibrotropism by the lymphotropic strain of minute virus of mice. J. Virol. 66:3415-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom, M. E., B. D. Berry, W. Wei, S. Perryman, and J. B. Wolfinbarger. 1993. Characterization of chimeric full-length molecular clones of Aleutian mink disease parvovirus (ADV): identification of a determinant governing replication of ADV in cell culture. J. Virol. 67:5976-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, S. F., J. Y. Sgro, and C. R. Parrish. 1992. Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J. Virol. 66:6858-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colomar, M. C., B. Hirt, and P. Beard. 1998. Two segments in the genome of the immunosuppressive minute virus of mice determine the host-cell specificity, control viral DNA replication and affect viral RNA metabolism. J. Gen. Virol. 79:581-586. [DOI] [PubMed] [Google Scholar]
- 8.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 9.Horiuchi, M., H. Goto, N. Ishiguro, and M. Shinagawa. 1994. Mapping of determinants of the host range for canine cells in the genome of canine parvovirus using canine parvovirus/mink enteritis virus chimeric viruses. J. Gen. Virol. 75:1319-1328. [DOI] [PubMed] [Google Scholar]
- 10.Horiuchi, M., N. Ishiguro, H. Goto, and M. Shinagawa. 1992. Characterization of the stage(s) in the virus replication cycle at which the host-cell specificity of the feline parvovirus subgroup is regulated in canine cells. Virology 189:600-608. [DOI] [PubMed] [Google Scholar]
- 11.Hueffer, K., J. S. Parker, W. S. Weichert, R. E. Geisel, J. Y. Sgro, and C. R. Parrish. 2003. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J. Virol. 77:1718-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llamas-Saiz, A. L., M. Agbandje-McKenna, J. S. L. Parker, A. T. M. Wahid, C. R. Parrish, and M. G. Rossmann. 1996. Structural analysis of a mutation in canine parvovirus which controls antigenicity and host range. Virology 225:65-71. [DOI] [PubMed] [Google Scholar]
- 14.McKenna, R., N. H. Olson, P. R. Chipman, T. S. Baker, T. F. Booth, J. Christensen, B. Aasted, J. M. Fox, M. E. Bloom, J. B. Wolfinbarger, and M. Agbandje-McKenna. 1999. Three-dimensional structure of Aleutian mink disease parvovirus: implications for disease pathogenicity. J. Virol. 73:6882-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palermo, L. M., K. Hueffer, and C. R. Parrish. 2003. Residues in the apical domain of the feline and canine transferrin receptor control the host specific binding and cell infection of canine and feline parvoviruses. J. Virol. 77:8915-8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker, J. S. L., W. J. Murphy, D. Wang, S. J. O'Brien, and C. R. Parrish. 2001. Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J. Virol. 75:3896-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker, J. S. L., and C. R. Parrish. 1997. Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J. Virol. 71:9214-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parrish, C. R. 1990. Emergence, natural history, and variation of canine, mink, and feline parvoviruses. Adv. Virus Res. 38:403-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parrish, C. R., C. Aquadro, M. L. Strassheim, J. F. Evermann, J.-Y. Sgro, and H. Mohammed. 1991. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J. Virol. 65:6544-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parrish, C. R., and L. E. Carmichael. 1983. Antigenic structure and variation of canine parvovirus type 2, feline panleukopenia virus, and mink enteritis virus. Virology 129:401-414. [DOI] [PubMed] [Google Scholar]
- 21.Parrish, C. R., L. E. Carmichael, and D. F. Antczak. 1982. Antigenic relationships between canine parvovirus type 2, feline panleukopenia virus and mink enteritis virus using conventional antisera and monoclonal antibodies. Arch. Virol. 72:267-278. [DOI] [PubMed] [Google Scholar]
- 22.Parrish, C. R., P. H. O'Connell, J. F. Evermann, and L. E. Carmichael. 1985. Natural variation of canine parvovirus. Science 230:1046-1048. [DOI] [PubMed] [Google Scholar]
- 23.Previsani, N., S. Fontana, B. Hirt, and P. Beard. 1997. Growth of the parvovirus minute virus of mice MVMp3 in EL4 lymphocytes is restricted after cell entry and before viral DNA amplification: cell-specific differences in virus uncoating in vitro. J. Virol. 71:7769-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossmann, M. G., and A. C. Palmenberg. 1988. Conservation of the putative receptor attachment site in picornaviruses. Virology 164:373-382. [DOI] [PubMed] [Google Scholar]
- 25.Simpson, A. A., V. Chandrasekar, B. Hebert, G. M. Sullivan, M. G. Rossmann, and C. R. Parrish. 2000. Host range and variability of calcium binding by surface loops in the capsids of canine and feline parvoviruses. J. Mol. Biol. 300:597-610. [DOI] [PubMed] [Google Scholar]
- 26.Spalholz, B. A., and P. Tattersall. 1983. Interaction of minute virus of mice with differentiated cells: strain-dependent target cell specificity is mediated by intracellular factors. J. Virol. 46:937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevenson, M. A., J. M. Fox, J. B. Wolfinbarger, and M. E. Bloom. 2001. Effect of a valine residue at codon 352 of the VP2 capsid protein on in vivo replication and pathogenesis of Aleutian disease parvovirus in mink. Am. J. Vet. Res. 62:1658-1663. [DOI] [PubMed] [Google Scholar]
- 28.Strassheim, L. S., A. Gruenberg, P. Veijalainen, J.-Y. Sgro, and C. R. Parrish. 1994. Two dominant neutralizing antigenic determinants of canine parvovirus are found on the threefold spike of the virus capsid. Virology 198:175-184. [DOI] [PubMed] [Google Scholar]
- 29.Tattersall, P., and J. Bratton. 1983. Reciprocal productive and restrictive virus-cell interactions of immunosuppressive and prototype strains of minute virus of mice. J. Virol. 46:944-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Truyen, U., A. Gruenberg, S. F. Chang, B. Obermaier, P. Veijalainen, and C. R. Parrish. 1995. Evolution of the feline-subgroup parvoviruses and the control of canine host range in vivo. J. Virol. 69:4702-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsao, J., M. S. Chapman, M. Agbandje, W. Keller, K. Smith, H. Wu, M. Luo, T. J. Smith, M. G. Rossmann, R. W. Compans, and C. R. Parrish. 1991. The three-dimensional structure of canine parvovirus and its functional implications. Science 251:1456-1464. [DOI] [PubMed] [Google Scholar]
- 32.Webby, R. J., and R. G. Webster. 2001. Emergence of influenza A viruses. Philos. Trans. R. Soc. Lond. (B) Biol. Sci. 356:1817-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie, Q., and M. S. Chapman. 1996. Canine parvovirus capsid structure, analyzed at 2.9 Å resolution. J. Mol. Biol. 264:497-520. [DOI] [PubMed] [Google Scholar]