Abstract
Background
Phylogenetic comparative methods are often improved by complete phylogenies with meaningful branch lengths (e.g., divergence dates). This study presents a dated molecular supertree for all 34 world pinniped species derived from a weighted matrix representation with parsimony (MRP) supertree analysis of 50 gene trees, each determined under a maximum likelihood (ML) framework. Divergence times were determined by mapping the same sequence data (plus two additional genes) on to the supertree topology and calibrating the ML branch lengths against a range of fossil calibrations. We assessed the sensitivity of our supertree topology in two ways: 1) a second supertree with all mtDNA genes combined into a single source tree, and 2) likelihood-based supermatrix analyses. Divergence dates were also calculated using a Bayesian relaxed molecular clock with rate autocorrelation to test the sensitivity of our supertree results further.
Results
The resulting phylogenies all agreed broadly with recent molecular studies, in particular supporting the monophyly of Phocidae, Otariidae, and the two phocid subfamilies, as well as an Odobenidae + Otariidae sister relationship; areas of disagreement were limited to four more poorly supported regions. Neither the supertree nor supermatrix analyses supported the monophyly of the two traditional otariid subfamilies, supporting suggestions for the need for taxonomic revision in this group. Phocid relationships were similar to other recent studies and deeper branches were generally well-resolved. Halichoerus grypus was nested within a paraphyletic Pusa, although relationships within Phocina tend to be poorly supported. Divergence date estimates for the supertree were in good agreement with other studies and the available fossil record; however, the Bayesian relaxed molecular clock divergence date estimates were significantly older.
Conclusion
Our results join other recent studies and highlight the need for a re-evaluation of pinniped taxonomy, especially as regards the subfamilial classification of otariids and the generic nomenclature of Phocina. Even with the recent publication of new sequence data, the available genetic sequence information for several species, particularly those in Arctocephalus, remains very limited, especially for nuclear markers. However, resolution of parts of the tree will probably remain difficult, even with additional data, due to apparent rapid radiations. Our study addresses the lack of a recent pinniped phylogeny that includes all species and robust divergence dates for all nodes, and will therefore prove indispensable to comparative and macroevolutionary studies of this group of carnivores.
Background
The pinnipeds are a monophyletic group of aquatic carnivores most closely related to either mustelids or ursids. The three monophyletic families – Phocidae (earless or true seals), Otariidae (sea lions and fur seals), and Odobenidae (one extant species of walrus) last shared a common ancestor within arctoid carnivores > 25 million years ago (mya) [1,2]. Some morphological studies [3,4] and virtually all molecular studies [e.g., [5-15]] support a link between otariids and odobenids (Otarioidea). However, several morphologists prefer a phocid-odobenid clade (e.g. [2,16-18]).
There are 34 extant species of pinniped, including Monachus tropicalis (which is widely believed to have gone extinct recently) and treating Zalophus as being monotypic (Z. californianus) (Table 1). The family Phocidae contains 19 species in two subfamilies: Monachinae or "southern" hemisphere seals (nine species comprising Antarctic, elephant, and monk seals) and Phocinae or "northern" hemisphere seals (10 species that inhabit the Arctic and sub-Arctic) [19]. Some authors have questioned the monophyly of Monachinae [20-22], although recent studies have shown this subfamily to be monophyletic [4,11,14,15,23,24]. The monophyly of Phocinae has not been questioned since being established by King [25]; however, there remains considerable debate over inter-group relationships, especially within Phocina (reviewed by [11,26]). The family Otariidae contains 14 extant species that have been divided traditionally into the subfamilies Arctocephalinae (fur seals) and Otariinae (sea lions) (e.g. [27,28]). Early suggestions that this subfamilial classification might be incorrect (e.g. [29]) have received increasing support from recent molecular analyses [12,14,15,30-32]. Taken together with a number of reports of both interspecific and intergeneric hybrids within Otariidae (e.g. [19,33,34]), a reassessment of otariid taxonomy based on additional phylogenetic evidence is needed. Brunner [31] provides an extensive review of the history of otariid classification. Finally, Odobenidae today comprises only the single species of walrus, Odobenus rosmarus.
Table 1.
Indented taxonomy listing the 34 pinniped taxa (including the extinct Monachus tropicalis) included in the analyses.
| Pinnipedia | ||||
| Odobenidae | Walruses | |||
| Odobenus rosmarus | Walrus | |||
| Otariidae | Sea lions and fur seals | |||
| Callorhinae | ||||
| Callorhinus ursinus | Northern Fur Seal | |||
| Arctocephalinae/Otariinae | ||||
| Arctocephalus townsendi | Guadalupe Fur Seal | |||
| Arctocephalus philippii | Juan Fernandez Fur Seal | |||
| Arctocephalus galapagoensis | Galapagos Fur Seal | |||
| Arctocephalus australis | South American Fur Seal | |||
| Arctocephalus tropicalis | Subantarctic Fur Seal | |||
| Arctocephalus gazella | Antarctic Fur Seal | |||
| Arctocephalus forsteri | New Zealand Fur Seal | |||
| Arctocephalus pusillus | South African Fur Seal | |||
| Zalophus californianus | California Sea Lion | |||
| Phocarctos hookeri | Hooker's Sea Lion | |||
| Neophoca cinerea | Australian Sea Lion | |||
| Otaria byronia | Southern Sea Lion | |||
| Eumetopias jubatus | Northern Sea Lion | |||
| Phocidae | True seals | |||
| Monachinae | "Southern" true seals | |||
| Monachini | Monk seals | |||
| Monachus schauinslandi | Hawaiian Monk Seal | |||
| Monachus tropicalis† | Caribbean Monk Seal | |||
| Monachus monachus | Mediterranean Monk Seal | |||
| Miroungini | Elephant seals | |||
| Mirounga angustirostris | Northern Elephant Seal | |||
| Mirounga leonina | Southern Elephant Seal | |||
| Lobodontini | Antarctic seals | |||
| Lobodon carcinophagus | Crabeater Seal | |||
| Leptonychotes weddellii | Weddell Seal | |||
| Hydrurga leptonyx | Leopard Seal | |||
| Ommatophoca rossii | Ross Seal | |||
| Phocinae | Northern true seals | |||
| Erignathini | <no common name> | |||
| Erignathus barbatus | Bearded Seal | |||
| Cystophorini | <no common name> | |||
| Cystophora cristata | Hooded Seal | |||
| Phocini | <no common name> | |||
| Histriophocina | <no common name> | |||
| Histriophoca fasciata | Ribbon Seal | |||
| Pagophilus groenlandicus | Harp Seal | |||
| Phocina | <no common name> | |||
| Phoca largha | Largha Seal | |||
| Phoca vitulina | Harbor Seal | |||
| Pusa hispida | Ringed Seal | |||
| Pusa sibirica | Baikal Seal | |||
| Pusa caspica | Caspian Seal | |||
| Halichoerus grypus | Grey Seal |
Several recent genetic studies [11,12,14,15,24,26,32] have advanced our knowledge of relationships within Pinnipedia considerably. Unfortunately, many of these (the exceptions being [14,24,26]) did not include divergence-date estimates as required for some types of macroevolutionary studies and phylogenetic comparative analyses. In addition, none included all species. The only study to include divergence-date estimates for all extant pinnipeds was that of Bininda-Emonds et al. [23] as a part of a larger carnivore supertree. This tree has been used extensively in comparative studies of carnivores in general (e.g., [35-42]) and pinnipeds in particular (e.g., [43-47]). However, it remains that the carnivore supertree is nearly a decade old and might no longer reflect current phylogenetic opinion.
Our objective is to address the lack of a recent phylogenetic study that includes all extant pinniped species and to provide date estimates for all nodes. To accomplish this task, we used the supertree method matrix representation with parsimony (MRP, [48,49]) to derive a complete phylogeny of the group from 50 gene trees (with mtDNA markers analyzed either individually or combined as a single source tree), with corresponding maximum likelihood (ML) and Bayesian (BI) analyses of the concatenated supermatrix serving as a form of topological sensitivity analysis in a global congruence framework [50]. Divergence dates within the supertree topology were estimated using 52 genes calibrated with eight robust fossil dates using two different methods. Together, the use of a larger data set focussed exclusively on the pinnipeds should yield both a more accurate topology and divergence dates than those present in the global carnivore supertree of Bininda-Emonds et al. [23].
Results and Discussion
General structure of the supertree
Our preferred hypothesis of pinniped evolution is that derived from the molecular supertree with all genes analyzed individually (Fig. 1; see Methods). It agrees broadly with other recent studies (e.g., [10-15,23,24,26,32]). In particular, the monophyly of each of Pinnipedia, Otarioidea, Phocidae, Otariidae, and the two phocid subfamilies was supported. Many of these nodes are among the most strongly supported in the supertree. The high level of congruence across numerous studies using different data sources and methodologies would suggest that higher-level pinniped relationships are well resolved. However, many relationships closer to the tips of the tree, particularly those within each of Arctocephalus and Phocina, remain contentious.
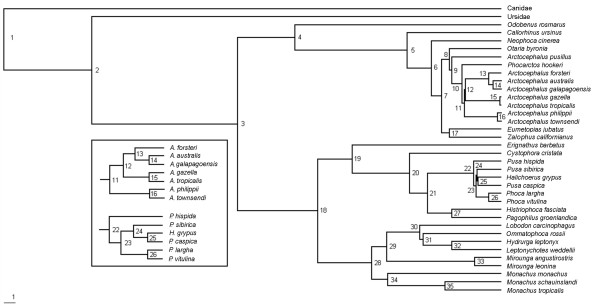
Figure 1.
Molecular supertree of the world's extant pinnipeds (plus one recently extinct Monachus species) based on a weighted matrix representation with parsimony analysis of 50 maximum likelihood gene trees. Node numbers correspond to divergence dates in Table 2. Branch lengths correspond to time with the scale bar indicating one million years. Boxed subset provides additional detail on branching order for two parts of the supertree where divergences occurred over a short timeframe.
Support values within the supertree (Table 2) were generally much higher than values typically reported for the supertree-specific support measure rQS (see [51,52]), with an average rQS value (± SD) across the tree of 0.234 ± 0.214. As such, most nodes are directly supported by a majority of the 50 source trees containing all the relevant taxa. The only exception is the node comprising Halichoerus grypus, Pusa caspica and Pusa sibirica, which has a slightly negative rQS value (-0.040). Even so, all more inclusive nodes possess positive rQS values, indicating that the conflict has more to do with the exact placement of Halichoerus within Pusa rather than the placement of it within this genus per se.
Table 2.
Divergence dates for the world's pinnipeds based on the median of up to 52 relative molecular and/or one fossil date analyzed using the relDate method.
| Confidence Interval | Number of date estimates | |||||||
| Node | rQS | Input date | Corrected date (SE) | Lower | Upper | Total | Molecular | Fossil |
| 1 | n/a | 43.35 | 43.4 | 1 | 0 | 1 | ||
| 2 | n/a | 35.7 | 35.7 (2.63) | 30.56 | 40.85 | 14 | 13 | 1 |
| 3 | 0.60 | 23 | 23 (1.36) | 20.33 | 25.67 | 27 | 26 | 1 |
| 4 | 0.12 | 18 | 18 (1.40) | 15.25 | 20.75 | 16 | 15 | 1 |
| 5 | 0.42 | 8.22 | 8.2 (2.09) | 4.12 | 12.32 | 5 | 4 | 1 |
| 6 | 0.36 | 6.11 | 6.1 | 1 | 1 | 0 | ||
| 7 | 0.36 | 5.15 | 5.2 (1.09) | 3.01 | 7.30 | 16 | 16 | 0 |
| 8 | 0.12 | 4.36 | 4.5 (0.21) | 3.95 | 4.77 | 12 | 12 | 0 |
| 9 | 0.20 | 4.36 | 4.3 | 1 | 1 | 0 | ||
| 10 | 0.20 | 3.21 | 3.4 (0.34) | 2.55 | 3.88 | 2 | 2 | 0 |
| 11 | 0.20 | 2.46 | 3.2 | 1 | 1 | 0 | ||
| 12 | 0.20 | 3.96 | 3.1 (3.43) | -2.76 | 10.68 | 3 | 3 | 0 |
| 13 | 0.20 | 1.05 | 1.1 (0.25) | 0.55 | 1.55 | 12 | 12 | 0 |
| 14 | 0.02 | 0.74 | 0.7 | 1 | 1 | 0 | ||
| 15 | 0.02 | 0.13 | 0.1 | 1 | 1 | 0 | ||
| 16 | 0.02 | 0.32 | 0.3 | 1 | 1 | 0 | ||
| 17 | 0.06 | 4.52 | 4.5 (0.37) | 3.79 | 5.24 | 5 | 5 | 0 |
| 18 | 0.50 | 16 | 16 (0.93) | 14.18 | 17.82 | 23 | 22 | 1 |
| 19 | 0.36 | 12.96 | 13 (0.90) | 11.20 | 14.72 | 12 | 12 | 0 |
| 20 | 0.42 | 7.97 | 8 (0.42) | 7.15 | 8.78 | 12 | 12 | 0 |
| 21 | 0.26 | 6.4 | 6.4 (0.40) | 5.62 | 7.18 | 13 | 13 | 0 |
| 22 | 0.38 | 2.29 | 2.4 (0.23) | 1.84 | 2.73 | 12 | 12 | 0 |
| 23 | 0.10 | 2.2 | 2.2 (0.62) | 0.99 | 3.41 | 18 | 18 | 0 |
| 24 | -0.04 | 2.2 | 2.1 (0.21) | 1.79 | 2.61 | 3 | 3 | 0 |
| 25 | 0.00 | 1.99 | 2 (0.14) | 1.71 | 2.27 | 3 | 3 | 0 |
| 26 | 0.12 | 1.07 | 1.1 (0.18) | 0.71 | 1.43 | 12 | 12 | 0 |
| 27 | 0.02 | 4.34 | 4.3 (0.51) | 3.35 | 5.33 | 5 | 5 | 0 |
| 28 | 0.22 | 11.33 | 11.3 (0.60) | 10.16 | 12.51 | 15 | 14 | 1 |
| 29 | 0.18 | 9.97 | 10 (0.65) | 8.69 | 11.25 | 21 | 20 | 1 |
| 30 | 0.30 | 7.07 | 7.1 (0.34) | 6.41 | 7.73 | 16 | 16 | 0 |
| 31 | 0.06 | 6.81 | 6.8 (0.26) | 6.29 | 7.32 | 17 | 17 | 0 |
| 32 | 0.34 | 4.32 | 4.3 (0.55) | 3.24 | 5.39 | 21 | 21 | 0 |
| 33 | 0.32 | 2.28 | 2.3 (0.85) | 0.61 | 3.96 | 21 | 21 | 0 |
| 34 | 0.08 | 9.95 | 9.9 (0.28) | 9.40 | 10.49 | 12 | 12 | 0 |
| 35 | n/a | 4.9 | 4.9 | 0 | 0 | 0 | ||
Node numbers correspond to Figure 1. Dates and 95% confidence intervals are in millions of years ago, with the latter applying to the input dates only. Fossil dates correspond to those listed in Table 5 and act as constraints on the minimum divergence time for the node in question. The date for node 35 was interpolated according to a constant birth model (see Methods). Support values for each node, as measured by rQS [51, 52] are also provided.
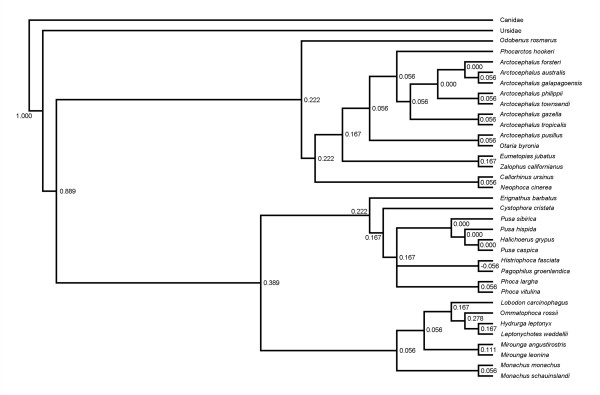
Alternative analyses of the molecular data set (supertree analysis with all mtDNA forming a single source tree or ML or BI analyses of the combined supermatrix; Figures 2 and 3, respectively) yield topologies that agree broadly with that in Figure 1. The rQs support measure across the supertree (0.18 ± 0.11) again showed that most nodes are directly supported by a majority of the 12 source trees containing all the relevant taxa. In all cases, the changes occur in parts of the tree with noticeably weaker support and/or branch lengths, indicating general regions of uncertainty: 1) Neophoca cinerea nests deeper within otariids, either as the sister taxon to Phocarctos hookeri (ML) or to the clade comprising the genera Arctocephalus, Otaria, and Phocarctos (BI), or forms the sister taxon to Callorhinus ursinus (supertree); 2) the formation of a sister-group relationship between Otaria byronia and Arctocephalus pusillus, which were previously adjacent to one another (all analyses); 3) the clades (Arctocephalus townsendii + A. phillippi) and (A. gazella + A. tropicalis) trade places (all analyses); and 4) changes to the internal relationships of Phocina, either with Halichoerus grypus and Pusa caspica being pulled basally with respect to the remainder of the group, with Halichoerus forming the sister group to the remaining species (ML), or with Pusa hispida and the clade of Histriophoca fasciata and Pagophilus groenlandicus nesting deeper within the group (BI), or with Pusa hispida moving inside P. sibirica and with a polytomy at the base of Phocini (supertree).
Figure 2.
Molecular supertree of the world's extant pinnipeds (excluding the recently extinct Monachus tropicalis) based on a weighted matrix representation with parsimony analysis of 12 maximum likelihood gene trees, where all mtDNA genes were combined to form a single source tree. Support values for each node, as measured by rQS [51, 52] are also provided.
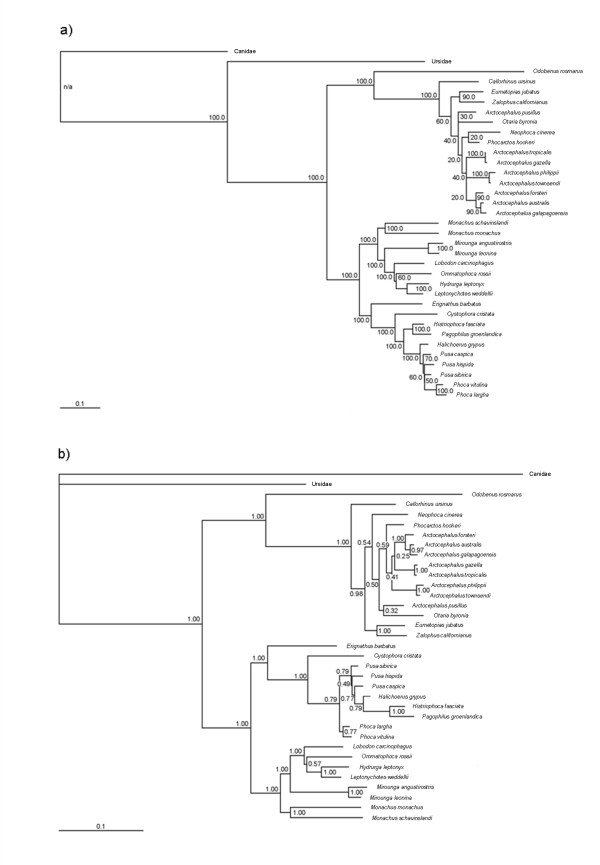
Figure 3.
Likelihood-based analyses of the molecular supermatrix of 50 gene trees: a) ML tree with bootstrap proportions and b) BI tree with posterior probabilities. Scale bars indicate average number of substitutions per site per unit time.
In the supertree, nodes 1 and 2 (see Fig. 1) represent the divergences of the canid and ursid lineages, respectively, and nodes 3 to 35 represent the various pinniped divergences. The total sample size (molecular and fossil date estimates) underlying the divergence times for each node ranged from 0 (node 35 – the split between Monachus schauinslandi and M. tropicalis, where the date was interpolated using a constant birth model) to 27 (Table 2). Over half (19) of the pinniped nodes were dated using at least 12 separate estimates. The remaining 14 nodes were dated by five or fewer estimates. Ten of these 14 nodes relate to otariid relationships, and seven concern Arctocephalus species. Divergences within the Pusa + Halichoerus clade were also dated by a comparatively small number of estimates. However, no obvious relationship existed between the variability in a date estimate (given by the coefficient of variation, CV) and the number of estimates it was derived from (R2 = 0.02, P = 0.4849, df = 26).
Our inferred relDate dates for the supertree topology (see Methods) are also significantly correlated with those for comparable nodes (which are restricted largely to Phocidae) in the two major studies to estimate divergence times within pinnipeds, those of Bininda-Emonds et al. [23] (R2 = 0.52, P = 0.004) and Arnason et al. [14] (R2 = 0.958, P < 0.0001) (df = 12 in both cases using ln-transformed values). However, whereas our dates did not differ significantly from those of Bininda-Emonds et al. [23] (paired-t of ln-transformed values = -1.36, P = 0.197; df = 13), they were significantly more recent than those of Arnason et al. [14] (paired-t of ln-transformed values = -9.82, P < 0.0001; df = 13), probably reflecting their use of a only single and more distant calibration point (the caniform-feliform split at 52 mya) as well as topological differences between the trees and different methodologies used to derive the dates.
Both sets of multidivtime divergence dates (Table 3) are significantly different from the relDate divergence dates (paired-t of ln-transformed values = -11.39, P < 0.0001; df = 32, for relDate versus multidivtime all genes; paired-t of ln-transformed values = -4.53, P < 0.0001; df = 32, for relDate versus multidivtime mtDNA only). The supertree (relDate) divergence dates underestimate the multidivtime dates from all genes and mtDNA genes by 88% and 51% on average, respectively. With respect to confidence intervals (CIs), only 9 and 7 (of 33) of the relDate dates fall into the range provided by the multidivtime CIs for mtDNA or all genes, respectively. Conversely, only 3 and 4 (of 33) dates for all genes and mtDNA only, respectively, fall within the CIs of the relDate dates. However, it is important to note that the two sets of multidivtime dates themselves are also significantly different from one another (paired-t of ln-transformed values = 2.36, P = 0.02; df = 32). In the following sections, we compare both sets of divergence dates (i.e., the relDate and multidivtime dates) with those from the fossil record and other studies.
Table 3.
Divergence dates calculated using Bayesian relaxed molecular clock method implemented by multidivtime [122, 123] for all genes combined and for mtDNA genes only, each fitted to the preferred supertree topology (Fig. 1).
| MultiDivTime dates (rttm = 1.95; bigtime = 4.335) | ||||||||
| All gennes | mtDNA genes only | |||||||
| Node | Point estimate | 1 SD | 95% CI (lower) | 95% CI (upper) | Point estimate | 1 SD | 95% CI (lower) | 95% CI (upper) |
| 1 | ||||||||
| 2 | 35.27 | 3.53 | 29.91 | 42.52 | 36.34 | 3.39 | 30.61 | 42.74 |
| 3 | 26.67 | 2.64 | 23.15 | 32.44 | 26.73 | 2.36 | 23.28 | 31.86 |
| 4 | 21.67 | 2.27 | 18.47 | 26.77 | 21.16 | 2.01 | 18.29 | 25.77 |
| 5 | 11.91 | 1.98 | 8.18 | 16.05 | 10.72 | 1.84 | 7.45 | 14.66 |
| 6 | 9.98 | 1.86 | 6.58 | 13.94 | 9.27 | 1.71 | 6.29 | 12.98 |
| 7 | 9.16 | 1.76 | 6.00 | 12.89 | 8.72 | 1.64 | 5.87 | 12.25 |
| 8 | 7.35 | 1.54 | 4.67 | 10.68 | 6.86 | 1.39 | 4.50 | 9.96 |
| 9 | 7.07 | 1.50 | 4.46 | 10.32 | 6.58 | 1.36 | 4.29 | 9.59 |
| 10 | 5.98 | 1.43 | 3.54 | 9.11 | 5.56 | 1.29 | 3.41 | 8.43 |
| 11 | 4.87 | 1.21 | 2.86 | 7.58 | 4.58 | 1.08 | 2.81 | 6.99 |
| 12 | 4.63 | 1.17 | 2.69 | 7.26 | 4.34 | 1.05 | 2.64 | 6.68 |
| 13 | 2.02 | 0.63 | 1.07 | 3.51 | 1.91 | 0.54 | 1.09 | 3.21 |
| 14 | 0.95 | 0.55 | 0.11 | 2.24 | 0.90 | 0.51 | 0.11 | 2.07 |
| 15 | 0.50 | 0.40 | 0.03 | 1.51 | 0.02 | 0.02 | 0.00 | 0.07 |
| 16 | 0.79 | 0.59 | 0.07 | 2.32 | 0.70 | 0.49 | 0.06 | 1.96 |
| 17 | 6.57 | 1.50 | 3.98 | 9.83 | 6.10 | 1.37 | 3.81 | 9.14 |
| 18 | 22.22 | 2.33 | 18.95 | 27.40 | 21.37 | 2.00 | 18.56 | 26.01 |
| 19 | 19.89 | 2.21 | 16.57 | 24.84 | 18.63 | 1.85 | 16.15 | 22.98 |
| 20 | 14.45 | 1.93 | 11.21 | 18.72 | 12.53 | 1.62 | 9.87 | 16.21 |
| 21 | 12.68 | 1.85 | 9.51 | 16.75 | 10.93 | 1.56 | 8.33 | 14.44 |
| 22 | 6.86 | 1.48 | 4.37 | 10.15 | 4.48 | 1.04 | 2.85 | 6.90 |
| 23 | 6.47 | 1.41 | 4.10 | 9.62 | 4.05 | 0.95 | 2.56 | 6.26 |
| 24 | 6.06 | 1.36 | 3.78 | 9.12 | 3.77 | 0.91 | 2.36 | 5.89 |
| 25 | 5.46 | 1.29 | 3.33 | 8.35 | 3.31 | 0.83 | 2.03 | 5.26 |
| 26 | 2.11 | 0.56 | 1.23 | 3.40 | 1.75 | 0.46 | 1.04 | 2.82 |
| 27 | 8.34 | 1.66 | 5.45 | 11.93 | 7.40 | 1.41 | 5.01 | 10.53 |
| 28 | 18.16 | 2.23 | 14.54 | 23.15 | 16.80 | 1.92 | 13.76 | 21.21 |
| 29 | 16.54 | 2.19 | 12.82 | 21.45 | 15.05 | 1.87 | 11.97 | 19.30 |
| 30 | 13.43 | 2.09 | 9.78 | 18.05 | 11.91 | 1.75 | 8.96 | 15.82 |
| 31 | 12.92 | 2.07 | 9.30 | 17.51 | 11.41 | 1.72 | 8.52 | 15.29 |
| 32 | 8.93 | 1.77 | 5.84 | 12.86 | 7.47 | 1.40 | 5.14 | 10.63 |
| 33 | 4.64 | 1.49 | 2.47 | 8.24 | 3.45 | 0.97 | 2.07 | 5.82 |
| 34 | 16.25 | 2.22 | 12.44 | 21.17 | 14.98 | 1.91 | 11.81 | 19.27 |
| 35 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Origins of major pinniped groups
The split between ursids and pinnipeds is estimated to be 35.7 ± 2.63 (= mean ± SE) mya (relDate, Table 2; the multidivtime dates for this node were similar (Table 3)), although this should not be taken to imply that ursids are the closest living relatives of pinnipeds among arctoid carnivores. Early pinnipeds (pinnipedimorphs) are held to have originated in the North Pacific during the late Oligocene (34-24 mya) ([2,22,45,53], but see [14], who speculate on an origin on the southern shores of North America), which is consistent with our estimate. Thereafter, a substantial lag is apparent, with the basal pinniped split between Phocidae and Otarioidea occurring some 12 million years later at 23.0 ± 1.36 mya (Table 2) (ca. 26 mya with multidivtime, Table 3). Both values are more recent than the 28.1 mya and 33.0 mya estimates obtained by Bininda-Emonds et al. [23] and Arnason et al. [14], respectively.
Odobenidae includes a single extant species and at least 20 fossil species in 14 genera [2], with the most basal taxa known from the late early Miocene (ca. 21-16 mya). Deméré et al. [2] suggest that odobenoids first evolved in the North Pacific region sometime before 18 mya (late early Miocene), and our data indicate the upper bound to be 20.8 mya. The multidivtime dates were similar at ca. 21 mya. Both values are substantially older than the 14.2 mya estimate obtained by Bininda-Emonds et al. [23], but younger than the 26.0 mya estimate of Arnason et al. [14].
Modern fur seals and sea lions are thought to have evolved from the ancestral family Enaliarctidae ca. 11 mya [54-56], with our data showing that the diversification of the crown group occurred shortly thereafter at 8.2 ± 2.09 mya (the dates estimated using multidivtime were again older, ca. 11 mya). Arnason et al. [14] consider the late Oligocene Enaliarctinae [57] to be the oldest otarioid lineage so far described (25–27 mya; [58]). However, Deméré et al. [2] consider this group to be early pinnipedimorphs that originated before the evolution of the modern crown-group pinnipeds.
The first phocid fossils date from the middle Miocene (ca. 16-14 mya) (but see [59,60]) in the North Atlantic [61], although some authors (e.g., [2,4,62]) have speculated over a North Pacific origin. Koretsky and Sanders [59,60] recently described the "Oligocene seal" from the late Oligocene (ca. 28 mya) in South Carolina as the oldest known true seal, a fossil that predates our estimate for the basal-most split in all pinnipeds. However, because this new description was based on a very small sample (two partial femora), and because Deméré et al. [2] noted that its stratigraphic provenience may be in question, we instead used 23 mya as a conservative fossil calibration point for the split between Phocidae and Otarioidea. Obviously, acceptance of the "Oligocene seal" as the oldest known phocid (and therefore crown-group pinniped) would cause all divergence times within the pinnipeds to be older than the ones that we report.
Otariidae
Phylogeny
The supertree resolved Callorhinus ursinus as sister to all remaining otariids (as is now generally accepted [12-14,23,32]), with neither the sea lions nor Arctocephalus forming clades. The five sea lion genera were generally positioned basally to the various Arctocephalus species. The exception was Phocarctos (and possibly Otaria in the supermatrix analyses), which nested within Arctocephalus. Wynen et al. [32] also reconstructed Neophoca as being the next otariid species to diverge (contra the supermatrix results) and found Zalophus + Eumetopias to form the sister clade to the remaining forms (Arctocephalus, Otaria and Phocarctos). These results add to the already large body of evidence, both molecular and morphological, that subfamilial descriptions in Otariidae, traditionally based on the single character of presence or absence of underfur, are incorrect [7,12,14,15,30-32,53,63]. However, resolution of most of the more inclusive otariid clades remains problematic [14,15,32], especially the relationships among the various Arctocephalus species, and the placements of the A. australis + A. forsteri + A. galapagoensis and A. philippii + A. townsendi clades in particular. The likelihood-based supermatrix analyses reinforce the generally weak or conflicting phylogenetic signal in the data set for otariids, with both suggesting what is to our knowledge a novel, more nested position for Neophoca (although the inferred location differs between the analyses).
The supertree resolved A. forsteri as the sister to A. australis + A. galapagoensis, with all three as sister to an A. gazella + A. tropicalis clade, an arrangement with relatively moderate support (Table 2). Wynen et al. [32] found a similar result, placing A. gazella as sister to the A. australis + A. forsteri + A. galapagoensis clade, but placed A. tropicalis as sister to A. pusillus on a more basal branch separate from other arctocephaline species. Our results also support a polyphyletic Arctocephalus, but with A. pusillus as the separate lineage. The separation of A. pusillus from other Arctocephalus species (and possible pairing with Otaria as found in both the supermatrix analyses and the combined mtDNA supertree) is perhaps not unexpected in hindsight, given that this species has long been considered as having an 'enigmatic taxonomic position' due to its similarity to sea lions in size, skull morphology, and behaviour [64-66].
Several authors [31,32] have recently questioned the status of A. philippii and A. townsendi as separate species (also see [67,68]). Brunner [31] went so far as to suggest that both taxa be removed from Arctocephalus to form subspecies in the previously described genus Arctophoca (Arctophoca philippii philippii and A. p. townsendi [69]). Our results are equivocal on this latter issue, given that these two taxa do form part of the main clade of Arctocephalus, but as sister to the remaining species. The two taxa, however, are indicated to have diverged from one another earlier (0.3 mya; relDate date) than other another pair of undisputed Arctocephalus species (namely A. gazella and A. tropicalis at 0.1 mya), a potential argument in favour of them retaining separate species status (regardless of the generic appellation).
The close genetic relationship we found between A. australis, A. forsteri and A. galapagoensis (also [32]) is also congruent with the morphometric results of Brunner [31], who suggested that A. galapagoensis be considered a subspecies of A. australis (as per [66,67]). Again, the relatively long divergence time between these two taxa (0.7 mya; relDate date) could argue against this arrangement.
Ultimately, relationships within Arctocephalus remain poorly resolved with little agreement between different studies or, as shown in this study, even different analyses of the same base data set. This situation will likely remain at least until additional genes for these taxa are sequenced. We would note that the relationships and divergence times within Arctocephalus presented here are based on the only genetic marker available at the time data were extracted from GenBank (MT-CYB sequences [32]). Additional genetic sequences for these species are desperately required (but see [14,15]).
Divergence dates
The only recent studies to estimate divergence dates for otariids are those of Bininda-Emonds et al. [23] and Arnason et al. [14]. Here, we compare our estimates to those two studies and the available fossil record, which is unfortunately limited. Our relDate estimate of 8.2 ± 2.09 mya for the root of the otariid crown-group is younger than other recent estimates [14,23]. The multidivtime dates (ca. 11–12 mya) agree well with Bininda-Emonds et al. [23], but are still younger than that estimated by Arnason et al. [14]. Thereafter, a series of rapid divergences are inferred to have occurred. The origin of Neophoca was estimated at 6.1 mya based on MT-CYB only (ca. 10 mya using multidivtime), followed by the initial radiation of the remaining species at 5.2 ± 1.09 mya (ca. 9 mya using multidivtime), and the origins of Otaria at 4.5 ± 0.21 mya and Arctocephalus pusillus at 4.3 mya (the latter, again, based only on MT-CYB; both divergences ca. 7 mya in the multidivtime analyses). The oldest known record for the southern hemisphere otariids is established by Hydrarctos lomasiensis from the late Pliocene or early Pleistocene (< 3.4 mya, [70,71]). Fossils from California and Japan suggest that sea lions did not diversify until ca. 3 mya [55,56,72]; however, only the late Pleistocene occurrences (< 0.8 mya) of Otaria bryonia [73] and Neophoca palatine [74] are considered reliable at present [2]. Our date for the origin of the lineage leading to Otaria as a whole is naturally much older than this and almost three times older than that in Bininda-Emonds et al. [23] (which places Otaria in a very different position). Arnason et al. [14] estimated an older divergence time, but also based on a different phylogeny. We infer Phocarctos to have split from the remaining Arctocephalus species 3.4 ± 0.34 mya. Finally, the divergence between Eumetopias and Zalophus was dated as 4.5 ± 0.37 mya, which is considerably older that the earliest known fossils (Pleistocene, 1.64-0.79 mya [56]), but younger than the 8 mya estimate of Arnason et al. [14] (which is still older than the multidivtime estimate of ca. 6 mya).
Our results similarly indicate a rapid radiation within Arctocephalus, with many species originating within the past 1 million years (both dating methods, Tables 2, 3). Overall, the date estimates showed reasonable levels of variation (relDate median CV of 27.5), although some were highly variable. For example, the split between the clades A. gazella + A. tropicalis and A. australis + A. forsteri + A. galapagoensis had a final date estimate of 3.1 mya but a large SE (3.43 my) and 95% confidence intervals on the input date (-2.76–10.68 mya), possibly reflecting weak signal in this area of the tree (see sensitivity analyses). Arctocephaline species are known in the fossil record only from poorly documented records of A. pusillus and A. townsendi from the Pleistocene (< 0.8 mya) [29]. The origin of Arctocephalus + Phocarctos hookeri was estimated here using MT-CYB data at 4.3 mya, which is younger than other recent estimates based on different topologies [14,23]. Although our results lend support to previous suggestions [2,32] that both sea lions and Arctocephalus underwent recent periods of rapid radiation, all the evidence to date tend to be based on a small dataset for most species.
Phocidae
Phylogeny
Compared to otariids, phocid relationships are generally much more agreed upon. The traditional and well-accepted phocid subfamilies Monachinae and Phocinae were both recovered as monophyletic in the supertree and supermatrix analyses (also see [4,11-15,23,26]). Erignathus barbatus was sister to the remaining northern phocids, followed by Cystophora cristata. The next branch of the tree separated Pagophilus groenlandicus and Histriophoca fasciata (= Histriophocina) as the sister group to the remaining taxa (but note the differences in the alternative supertree and the BI supermatrix). Most recent studies [11-15,23,26] have found support for this arrangement among the early branches (i.e., involving the lineages Erignathus, Cystophora, and Histriophocina). Of the six Pusa, Phoca, and Halichoerus species (= Phocina), in the preferred tree Pusa hispida was found to be sister to the remaining species in which Phoca vitulina + Phoca largha formed the sister clade to (Pusa sibirica + (Halichoerus + Pusa caspica)) (again note the alternative arrangements in Figures 2 and 3, indicating poor signal in this part of the pinniped phylogeny). The sister-group relationship between Phoca vitulina and P. largha recovered here in all analyses is consistent among and well supported in numerous studies based on diverse data types [4,11-15,23,26], and reflects early suggestions that the latter species represents a subspecies of the former [68,75].
Arguably the biggest outstanding problem in phocid phylogeny concerns the placement of Halichoerus within Phocina, and there have been long-standing suggestions (e.g., [76]) for taxonomic revision of these taxa. Both Davis et al. [11] and Delisle and Strobeck [12] found the strongest support for Halichoerus as sister to Pusa, with both being sister to Phoca. However, both studies included only Pusa hispida as an exemplar for Pusa. Fulton and Strobeck [15] also recovered a similar result, but did not include Pusa sibirica. Four recent studies have included all three Pusa species [4,14,23,26]. Bininda-Emonds and Russell [4] recovered Halichoerus as sister to Erignathus + Histriophocina + the remaining Phocina using morphological data. Bininda-Emonds et al. [23] resolved an unresolved Pusa as sister to the two Phoca species in their supertree, with Halichoerus being sister to this clade. The molecular results of Arnason et al. [14] and Palo and Väinölä [26] were similar to ours, indicating weak support for a P. caspica + H. grypus clade, and for a basal position for P. hispida within Phocina. Although the precise interrelationships of the species differ slightly, our results support the suggestions of these other recent studies that both Halichoerus and Pusa be included within a redefined Phoca, possibly as subgenera. In fact, Arnason et al. [6] suggested recently that the scientific name for the grey seal be Phoca grypa. This solution also works in light of the continuing uncertainty concerning interrelationships within Phocina (compare Figures 1, 2, and 3 and these with the references above), especially the increasing number of suggestions that Pusa might be paraphyletic (except if it were to be retained as a subgenus).
It is also noteworthy that all the relevant divergences within Phocina apparently occurred in a very short time frame (also see [14,26]), which might make resolution within this group difficult to obtain even with additional markers. By contrast, there were no negative branch lengths in this part of the supertree (although nodes 23 and 24 in Figure 1 were held to be simultaneous initially), indicating relatively good agreement among the sequence data. Also, except for node 25, all the rQS values in this part of the (preferred) tree are > 0, again indicating more agreement than conflict among the set of gene trees (note the rQs values in Fig. 2, the only negative value in the alternative supertree concerns the sister-group relations of the two Histriophocina species).
Within Monachinae, all analyses recovered a monophyletic Monachus as sister to Miroungini + Lobodontini. Relationships within Monachus and Mirounga recovered here are consistent among and well supported in numerous studies [4,11-15,23,26] (but see [22] regarding Monachus). Relationships within Lobodontini have traditionally been contentious, although recent studies [11-15] all support the sister relationship between Leptonychotes and Hydrurga recovered here (contra [4,23]). However, the positions of Ommatophoca and Lobodon relative to each other and to the Leptonychotes + Hydrurga clade remain problematic. Many recent studies [11,12,14,15] found the strongest support for an (Ommatophoca, (Lobodon, (Leptonychotes + Hydrurga))) relationship. Our results differed and, similar to Fyler et al. [24], supported Lobodon as being sister to the remaining species. The supermatrix analyses indicated the identical sets of relationships for Monachinae.
Divergence dates
The fossil record suggests that the divergence of the two phocid subfamilies occurred sometime prior to the middle Miocene (> 14.6 mya) [77] and we used 16 mya as a minimum age constraint for the corresponding node (also see [23]). Similarly, Fyler et al. [24] used 15 and 17 mya as calibration points from which to estimate divergence dates in Monachinae. The corresponding molecular estimate of Arnason et al. [14] at 22 mya is older still and in better agreement with our multidivtime dates. The initial divergence in phocines (i.e., the lineage leading to Erignathus) was dated at 13.0 ± 0.90 mya, which is slightly younger than other estimates [14,23,24,26] (the multidivtime dates are again older, ca. 19 mya). Our relDate dates for the origins of Cystophora (8.0 ± 0.42 mya) and Histriophoca + Pagophilus (6.4 ± 0.40 mya) are considerably younger than the corresponding estimates from Bininda-Emonds et al. [23] (which are in closer agreement with the multidivtime dates), but considerably older than the available fossil evidence. Deméré et al. [2] suggested that these basal phocines originated in the Arctic during the Pleistocene and represent the products of a glacioeustatic-forced allopatric speciation event. Arnason et al. [14] estimated a considerably older date (12 mya) for the divergence of Cystophora, again in agreement with both Bininda-Emonds et al. [23] and our multidivtime results, but a comparable 7 mya estimate for the origin of Histriophocina.
The genus Phoca arose 2.2 ± 0.62 mya (using relDate; multidivtime dates ca. 5–6 mya), with both extant species diverging from one another 1.1 ± 0.18 mya. These two nodes were well sampled, with 18 and 12 molecular estimates, respectively. The suggested recent separation and evolution of the two Phoca species (using both dating methods) is in general agreement with other studies [14,23,68,75,78]. Pusa sibirica arose 2.1 ± 0.21 mya, and Halichoerus grypus and Pusa caspica diverged immediately thereafter at 2.0 ± 0.14 mya; the divergence estimates for these last two nodes were each dated by only three genes apiece, and both are considerably older in the multidivtime analyses. Bininda-Emonds et al. [23], by contrast, estimated the origin of Halichoerus to be 7.1 mya, although this was based on a different topology, with Halichoerus in a more basal position. They also dated a Pusa polytomy to 2.8 mya, whereas we estimate here (using relDate) that the three genera Phoca, Halichoerus, and Pusa all arose over a short time span ranging from 2.2 to 1.1 mya (2–6 mya using multidivtime). Palo and Väinölä [26] similarly estimated that the radiation of the five main Phocini mtDNA lineages occurred ca. 2.5–3.1 mya, whereas Arnason et al. [14] estimated that the basal Phocina radiations occurred at 4.5 mya. Sasaki et al. [79] derived considerably younger estimates for divergences within Pusa, although their calibration was based on an incorrect estimate of the general mammalian substitution rate [26]. In addition, the sister-group relationships on which their dates are based conflict with our results and those of other recent studies [14,26]. Regardless of the precise relationships upon which the dates are based, the general consensus is that the diversification within Phocina was both rapid and relatively recent, which agrees with biogeographic evidence suggesting that the evolution of the Halichoerus-Pusa-Phoca complex likely occurred in the Greenland Sea/Barents Sea portion of the Arctic [2], and was possibly related to the closing of the Panama Canal 3.2-2.8 mya, which resulted in the freezing over of the Arctic Ocean [80-82].
Among the southern phocids, most nodes (with the obvious exception of the Monachus schauinslandi and M. tropicalis split) were well sampled, with 12–21 date estimates each. The lineage leading to Monachus split from the remaining species 11.3 ± 0.60 mya, which is slightly younger than other recent estimates [23,24] (and these other estimates are themselves slightly younger than the multidivtime dates). Our relDate estimate of the origin of the lineage leading to M. monachus (9.9 ± 0.28 mya) is considerably older than the 4.8 mya estimate of Bininda-Emonds et al. [23], but in good accord with those of Fyler et al. [24] and Arnason et al. [14]. The multidivtime dates for this node are again older, ca. 15–16 mya. The split between M. schauinslandi and M. tropicalis was interpolated to be 4.9 mya, compared to 2.8 mya estimate from Bininda-Emonds et al. [23] (also based on interpolation from a pure-birth model).
Our results indicate that the Mirounga lineage split from the lobodontine seals 10.0 ± 0.65 mya (ca. 15–16 mya using multidivtime), which accords well with recent genetic studies [14,23,24] and with fossil evidence indicating that the oldest fossils of southern lobodontines are from the late Miocene (6.7-5.2 mya) [71] and suggesting that the divergence occurred sometime before 11 mya [2,83]. Our relDate date for the split between the two Mirounga species (2.3 ± 0.85 mya) was slightly younger than that in other recent studies [14,23,24] (which were all in general agreement with the multidivtime results), but considerably older than the 0.8 mya estimate of Slade et al. [84].
Among the four lobodontine seals, Lobodon diverged first at 7.1 ± 0.34 mya, followed shortly thereafter by Ommatophoca at 6.8 ± 0.26 mya, and finally by Hydrurga + Leptonychotes at 4.3 ± 0.55 mya. The time of origin of the lineage leading to Lobodon is younger than the date estimated by Fyler et al. [24], but older than that of Arnason et al. [14] (who also resolved a different topology). However, both it and time of origin of the lineage leading to Ommatophoca correspond well to the dates of Bininda-Emonds et al. [23]. The divergence dates determined using multidivtime were again considerably older (Table 3).
Conclusion
Our results add to the growing list of studies that highlight the need for a re-evaluation of pinniped taxonomy, with revisions being required for both otariids (with respect to subfamilial classification and the genus Arctocephalus) and phocids (within Phocina, especially as regards Halichoerus and Pusa), ideally based on additional and especially nuclear genetic markers. The divergence-date estimates herein indicate rapid radiations in both families, particularly the southern hemisphere fur seals (Arctocephalus) and the northern phocids (Phocina), a fact which might account for the historical difficulties in assessing the phylogenetic relationships within each group. The historically unusual, but increasingly suggested nesting of Halichoerus within Pusa (see also [6,14,15,26]) highlights both the utility of large molecular datasets with numerous genes and the value of including all relevant species in phylogenetic analysis (see also [4]). We suggest increased genetic sampling throughout the group as the best approaches to further improving our understanding of pinniped phylogenetics. For example, at the time we gathered data, only MT-CYB had been sequenced for most otariid species and only a small number of genes were available for several Pusa species, although additional sequences have since been provided [14,15]. That being said, the problem areas within Phocina and Arctocephalus that were identified by both supertree and supermatrix analyses might prove resistant to resolution even with increased sampling should the apparent rapid branching in these parts of the tree be real.
Phylogenetic comparative methods have become the standard way for comparing aspects of the biology of a group of species while avoiding statistical problems associated with species not being independent due to their shared evolutionary history [85]. Phylogenetic analyses are improved with appropriate reconstruction of ancestral nodes using divergence-date information [86,87], and estimates of divergence dates provide conservation biology with a powerful tool in assessing vulnerability to conservation problems and comparative analysis of extinction risk [88,89]. Our results will allow the use of phylogenetic comparative methods with a robust estimate of pinniped phylogeny and divergence times that includes all species.
Methods
DNA sequence data
The use of large, multigene data sets provides the numerous informative changes required for correct inferences, and may also help to raise weak phylogenetic signals above the noise level [90]. In addition, the best topologies are often resolved when estimates are based on a combination of mitochondrial and nuclear DNA. With these points in mind, we mined GenBank for all available pinniped DNA sequence data to infer a phylogeny based on the largest data set possible. All sequence data were downloaded on January 30, 2006 and mined using the Perl script GenBankStrip v2.0 [91] to retain only those genes that had been sequenced for at least three pinniped species and were longer than 200 bp (except for tRNA genes, which had to be longer than 50 bp). For the 52 genes meeting these criteria (see Table 4), matching sequences for exemplars from Canidae (either Canis lupus or, on one occasion, C. latrans) and/or Ursidae (usually Ursus arctos, but also U. americanus or U. maritimus as needed) were downloaded for outgroup analysis.
Table 4.
Genetic sequences used in this study with their inferred models of evolution.
| Phylogeny estimation | Fitting to supertree topology | ||||||||||
| Gene | Number of taxa | Number of bps | Model selected | Nonclock ln L | Model selected | Nonclock ln L | Clock ln L | Chi-squared | df | LRT P-value | Clock? |
| ALDOA | 7 | 120 | K80 * | 228.5901 | K80 | 228.5901 | 228.98093 | 0.78166 | 5 | 0.9782 | yes |
| ALDOC | 7 | 129 | K3P * | 213.1006 | K81+I | 216.4168 | 218.7521 | 4.6706 | 5 | 0.4574 | yes |
| APOB (editing region) | 5 | 175 | TVM * | 317.7049 | TVM+G | 317.3427 | 319.90041 | 5.1154 | 3 | 0.1635 | yes |
| APOB (exon 26) | 6 | 963 | HKY | 1701.6729 | HKY | 1701.673 | 1705.76283 | 8.1797 | 4 | 0.08521 | yes |
| APOB (exon 29) | 3 | 621 | n/a | n/a | TVM+G | 1380.1937 | 1381.96031 | 3.5332 | 1 | 0.06015 | yes |
| CYP1A1 | 5 | 1560 | HKY | 2641.2015 | HKY+I | 2913.9797 | 3546.2776 | 1264.6 | 3 | 0 | |
| CYP1A2 | 5 | 1539 | TVM+G | 2859.5616 | TVM+I | 2861.4082 | 2863.05456 | 3.2927 | 3 | 0.3487 | yes |
| H2AFZ | 7 | 52 | TrNef * | 95.5468 | TrNef | 97.6302 | 99.00264 | 2.7449 | 5 | 0.7392 | yes |
| HLA-DOA | 5 | 399 | HKY * | 598.9182 | HKY | 598.9182 | 599.15301 | 0.46962 | 3 | 0.9255 | yes |
| LEP | 5 | 504 | GTR * | 1110.6539 | GTR | 1110.6539 | 1126.3367 | 31.366 | 3 | 7.12E-07 | |
| LYZ | 3 | 447 | n/a | n/a | K80 | 677.5543 | 677.71442 | 0.32024 | 1 | 0.5715 | yes |
| MHC-DQA1 | 7 | 162 | TVMef * | 393.3731 | TVMef | 393.7065 | 395.79033 | 4.1677 | 5 | 0.5255 | yes |
| MHC-DQA2 | 7 | 230 | HKY+G * | 455.7005 | HKY+I | 458.4345 | 460.01745 | 3.1659 | 5 | 0.6744 | yes |
| MHC-DQB1 | 5 | 141 | K3Puf+G * | 307.2022 | TVM+I | 306.0257 | 306.25311 | 0.45482 | 3 | 0.9287 | yes |
| MT-ATP6 | 22 | 681 | GTR+I+G * | 4821.8578 | TVM+I+G | 4823.9243 | 4837.61057 | 27.373 | 20 | 0.1251 | yes |
| MT-ATP8 | 22 | 204 | HKY+I+G * | 1598.4148 | TVM+I+G | 1597.1221 | 1609.01567 | 23.787 | 20 | 0.2518 | yes |
| MT-CO1 | 25 | 1545 | GTR+I+G * | 10168.6228 | TVM+I+G | 10171.9424 | 10194.06882 | 44.253 | 23 | 0.004901 | yes |
| MT-CO2 | 25 | 687 | HKY+I+G * | 4427.0054 | HKY+I+G | 4428.0015 | 4446.36885 | 36.735 | 23 | 0.03465 | yes |
| MT-CO3 | 22 | 784 | HKY+I+G * | 4896.904 | TVM+I+G | 4893.5391 | 4909.02367 | 30.969 | 20 | 0.0556 | yes |
| MT-CYB | 35 | 1140 | TrN+I+G * | 8835.9098 | GTR+I+G | 8836.9434 | 8872.23839 | 70.59 | 33 | 0.0001522 | |
| MT-ND1 | 22 | 957 | HKY+I+G * | 5866.8206 | TVM+I+G | 5863.5283 | 5882.62305 | 38.19 | 20 | 0.008394 | yes |
| MT-ND2 | 24 | 1044 | TrN+I+G * | 7689.7091 | TIM+I+G | 7698.3613 | 7720.88041 | 45.038 | 22 | 0.002625 | yes |
| MT-ND3 | 22 | 350 | HKY+G * | 2365.8095 | TVM+I+G | 2363.5559 | 2374.43717 | 21.763 | 20 | 0.3535 | yes |
| MT-ND4 | 21 | 1378 | GTR+I+G * | 9587.3198 | TVM+I+G | 9586.7549 | 9609.42621 | 45.343 | 19 | 0.0006135 | |
| MT-ND4L | 22 | 300 | HKY+I+G * | 1900.4839 | HKY+I+G | 1897.8574 | 1909.16103 | 22.607 | 20 | 0.3085 | yes |
| MT-ND5 | 22 | 1836 | GTR+I+G * | 13444.3742 | TVM+I+G | 13450.124 | 13482.66844 | 65.089 | 20 | 1.13E-06 | |
| MT-ND6 | 13 | 528 | HKY+I+G * | 2457.8525 | HKY+I+G | 2457.8848 | 2466.38674 | 17.004 | 11 | 0.1078 | yes |
| MT-RNR1 | 18 | 984 | GTR+G * | 4033.2182 | GTR+I+G | 4033.6277 | 4047.13902 | 27.023 | 16 | 0.04123 | yes |
| MT-RNR2 | 14 | 1608 | GTR+I+G | 5722.1966 | GTR+I+G | 5710.8447 | 5730.04437 | 38.399 | 12 | 0.0001321 | |
| MT-TA | 7 | 69 | HKY+G * | 193.586 | HKY+G | 192.1818 | 197.11463 | 9.8657 | 5 | 0.07913 | yes |
| MT-TC | 7 | 69 | K80+G * | 190.9928 | K80+G | 190.9817 | 192.76403 | 3.5647 | 5 | 0.6136 | yes |
| MT-TD | 7 | 68 | HKY+G * | 177.8663 | HKY+G | 177.7805 | 178.50213 | 1.4433 | 5 | 0.9195 | yes |
| MT-TE | 9 | 72 | HKY+G * | 205.1879 | HKY+I | 204.3486 | 208.84935 | 9.0015 | 7 | 0.2525 | yes |
| MT-TF | 7 | 72 | TrNef * | 201.267 | TrN+G | 197.1401 | 202.84986 | 11.42 | 5 | 0.04366 | yes |
| MT-TG | 7 | 72 | TrNef+G * | 269.0899 | HKY+G | 267.3327 | 271.41557 | 8.1657 | 5 | 0.1473 | yes |
| MT-TH | 7 | 69 | HKY+G * | 201.8982 | HKY+G | 203.5126 | 205.11176 | 3.1983 | 5 | 0.6694 | yes |
| MT-TI | 7 | 71 | HKY+G * | 141.7316 | HKY+I | 142.4105 | 145.12331 | 5.4256 | 5 | 0.3662 | yes |
| MT-TK | 7 | 70 | HKY+G * | 219.0096 | HKY+G | 221.4528 | 228.78366 | 14.662 | 5 | 0.01191 | yes |
| MT-TM | 7 | 71 | K80+G * | 140.5878 | TrNef+I+G | 135.8801 | 145.67015 | 19.58 | 5 | 0.001498 | yes |
| MT-TN | 7 | 73 | K80+G * | 204.0605 | HKY+I | 201.8885 | 205.49416 | 7.2113 | 5 | 0.2054 | yes |
| MT-TP | 8 | 68 | HKY+G * | 200.9591 | HKY+G | 202.8423 | 204.98693 | 4.2893 | 6 | 0.6376 | yes |
| MT-TQ | 7 | 76 | HKY+G * | 202.9076 | TrN+G | 203.4861 | 222.92371 | 38.875 | 5 | 2.52E-07 | |
| MT-TR | 8 | 71 | HKY+G * | 197.8915 | K81uf+G | 196.7379 | 201.8366 | 10.197 | 6 | 0.1166 | yes |
| MT-TT | 9 | 74 | K80+G * | 226.9842 | HKY+I | 224.7003 | 230.34978 | 11.299 | 7 | 0.1261 | yes |
| MT-TV | 8 | 76 | K80+G * | 231.1146 | HKY+G | 230.1562 | 233.04494 | 5.7775 | 6 | 0.4486 | yes |
| MT-TW | 7 | 68 | K80+I+G * | 215.6285 | HKY+I+G | 211.9622 | 218.25694 | 12.589 | 5 | 0.02755 | yes |
| MT-TY | 7 | 70 | K80+G * | 225.9578 | HKY+I | 222.7287 | 230.7563 | 16.055 | 5 | 0.006689 | yes |
| MX1 | 4 | 1980 | TrN+G | 3887.1331 | TrN+G | 3887.1331 | 3892.79094 | 11.316 | 2 | 0.003489 | yes |
| RAG1 | 5 | 741 | HKY+G | 1713.0793 | TVM+I | 1709.8627 | 1712.53176 | 5.3381 | 3 | 0.1486 | yes |
| RHO | 5 | 1077 | HKY+I+G | 2055.8504 | HKY+I | 2056.0244 | 2056.56021 | 1.0716 | 3 | 0.7839 | yes |
| SERPINA7 | 7 | 442 | TrNef * | 869.6311 | TrNef | 869.631 | 870.82096 | 2.3799 | 5 | 0.7945 | yes |
| SRY | 21 | 231 | HKY * | 488.8158 | HKY | 488.3143 | 494.82962 | 13.031 | 19 | 0.837 | yes |
| TOTAL | 26818 | ||||||||||
Models followed by an asterisk were determined using AICc; all others were determined using AIC. LYZ and APOB (exon 29) were not used to construct the supertree. Gene symbols follow Wain et al. [126].
Sequences in each data set were aligned using ClustalW [92] or with transAlign [93] in combination with ClustalW for the protein-coding sequences, and improved manually where needed. Thereafter, each aligned data set was passed through the Perl script seqCleaner v1.0.2 [91] to standardize the species names, to eliminate inferior sequences (i.e., those with > 5% Ns), and to ensure that all sequences overlapped pairwise by at least 100 bps (or 25 bps for the tRNA genes). Note that although species names were standardized according to Wilson and Reeder [94] for the analyses, those used in the text for Phocini follow the currently accepted International Commission of Zoological Nomenclature (ICZN) taxonomy, which recognizes the five genera Halichoerus, Histriophoca, Pagophilus, Phoca, and Pusa.
The final data set of 52 genes (Table 4) comprises 26818 bps in total, or an average of 515.7 bp per gene (range = 68–1980 bps). On average, each gene was sampled for 11.2 species (range = 3–35); however, only an average of 5.5 species per nuclear gene were available for study. Two genes, LYZ and exon 29 of APOB, contained fewer than three pinniped species and, as such, were uninformative for resolving pinniped interrelationships. However, they were still retained to determine times of divergence. Accession numbers for all sequences used in the final data set are provided as supplementary material (Additional file 1).
The final data set is dominated by mitochondrial genes, which forms a single locus due to its common inheritance and general lack of recombination. As such, it must be kept in mind that all the resulting topologies (be they derived in a supertree or supermatrix framework) and divergence times could be biased by any peculiarities related to mitochondrial sequence data (e.g., introgression or linkage) or simply the disproportionately large amount of mitochondrial data. However, the data set represents the "current systematic database" for pinnipeds and so the best possible current data source for which to infer their phylogenetic relationships. However, to assess the impact of this potential source of bias, we performed a second supertree analysis where all mtDNA genes were combined to form a single source tree (yielding 12 source trees in total). Nevertheless, the collection of additional nuclear markers is desperately needed for this group.
The final data set used for the phylogenetic analyses, together with the supertree and supermatrix trees is freely available from TreeBASE [95] (study accession number S1911, matrix accession numbers M3516-M3518).
Phylogeny reconstruction and supertree analysis
Our general approach to infer the phylogeny of the pinnipeds involved a divide-and-conquer strategy in which individual genes trees were determined using the best possible methodology for each and then combined as a supertree. Compared to a simultaneous analysis of the multigene "supermatrix", this procedure has been argued to potentially account better for the differential models of evolution that might be present [96] and, for extremely large matrices, looks to be a faster analytical method without any appreciable loss of accuracy [97]. Although the use of mixed models is possible in both maximum likelihood (ML, [98]) and Bayesian frameworks, the accuracy of the resulting tree, at least in a Bayesian framework, has recently been called into question [99], especially when reasonable levels of conflict exist between the different data partitions [100]. Furthermore, Jeffroy et al. [101] have also recently argued that trees derived from multigene, phylogenomic data sets should be treated more cautiously than those from single-gene analyses given that the systematic biases inherent to phylogeny reconstruction become more apparent with larger data sets. Nevertheless, in light of the fierce criticism that the supertree approach has attracted (e.g., [102,103], but see [104,105]), we also conduct ML and Bayesian inference (BI) analyses of the concatenated supermatrix to help identify especially problematic regions of the pinniped tree as part of a global congruence framework [50] and to add to the growing body of studies comparing phylogenetic inference under these two frameworks (e.g., [15,106]).
For the supertree analyses, we used PHYML [106] to determine the ML tree for each of the 50 phylogenetically informative genes after determining their optimal model of evolution according to either AIC or AICc (as appropriate, the latter being a version of the AIC corrected for small sample sizes) using MrAIC [107] and PHYML [106] (Table 4). The 50 gene trees were then used to build a weighted supertree of the group using matrix representation with parsimony (MRP, [48,49]). In so doing, we have assumed that each gene tree forms an independent unit in our preferred supertree, something that is admittedly debatable for the mitochondrial genes and especially the very small tRNA genes. However, in the absence of any robust linkage information, this assumption seemed more justifiable and objective than the defining of gene partitions based on assumed linkage or for purely practical considerations (e.g., concatenating all the tRNA genes because of their small size). Nonetheless, the sensitivity of these assumptions was assessed using the second supertree in which all mtDNA genes formed a single source tree.
All gene trees were encoded for the MRP analysis using semi-rooted coding [108], whereby only those trees with either a canid and/or ursid outgroup taxon and where the pinnipeds were reconstructed as being monophyletic were held to be rooted. Furthermore, the individual MRP characters, which correspond to a particular node on a gene tree, were weighted according to the bootstrap frequency [109] of that node, as determined using PHYML and based on 1000 replicates. This procedure has been demonstrated to increase the accuracy of MRP supertree construction in simulation [110]. The weighted parsimony analysis of the resulting MRP matrix was accomplished using a branch-and-bound search in PAUP* v4.0b10 [111], with Canidae and Ursidae being specified as a paraphyletic outgroup. Monachus tropicalis, for which no molecular data exist, was added to the supertree manually as the sister species of M. schauinslandii (following [4,23]).
Support for both supertrees and the relationships in them were quantified with the supertree-specific rQS index [51,52], which compares the topology of the supertree to that of each of the source trees contributing to it. As such, it is preferable to such conventional, character-based support measures such as Bremer support [112] and the bootstrap, which are invalid in this context given that MRP characters for a given source tree are non-independent. Values for rQS range from + 1 to -1, with the two values indicating that a given node is directly supported or directly contradicted by all source trees, respectively. The rQS value for the entire tree is simply the average of all the nodal rQS values. Previous applications of the rQS index show that it often tends to negative values [51,52,113], indicating that more conflict than agreement generally exists among a set of source trees for a given node. As such, positive values of rQS can be taken to indicate good support in the sense that more source trees support the relationship than contradict it.
The individual gene data sets were also concatenated to form a single supermatrix that was analyzed using both partitioned ML and BI methods. ML analyses used RAxML VI-HPC v2.2.3 [114]. A GTR + G model was assumed for the data using the CAT approximation of the gamma distribution, with the model parameters being allowed to vary independently for each gene. CAT is both a fast approximation of the gamma model (due to its lower computational and memory costs) and one that appears to yield better log likelihood scores even when calculated under a real gamma model [115], and therefore is ideally suited to large, computationally intensive data matrices such as ours. The ML tree was taken to be the optimal tree over 100 replicates, for which nodal support was estimated using the bootstrap with 1000 replicates and search parameters matching those for the optimality search.
BI used MrBayes v3.1.2 [116], with the individual models specified for each individual gene matching the optimal model determined in the gene-tree analyses as closely as possible. Otherwise, flat priors were used in all cases. Searches employed a MCMC algorithm of two separate runs, each with four chains that were run for 10000000 generations and with the first 5000000 generations being discarded as burn-in. Trees were sampled every 5000 generations to derive the final BI tree and estimates of the posterior probabilities.
Divergence date estimations
Following Bininda-Emonds et al. [117], divergence times on the supertree only were determined using a combination of fossil calibration points and molecular dates under the assumption of a local molecular clock (see [118]). As a first step, the optimal model of evolution for all 52 genes was (re)determined using an AIC in ModelTEST v3.6 [119] in combination with PAUP*, with the appropriately pruned supertree topology being used as the reference tree in place of the default NJ tree. This combination was used here in place of the previous MrAIC/PHYML combination largely because it can be used to test for the applicability of a molecular clock (through PAUP*) using a likelihood-ratio test. The small taxonomic distribution meant that all but six genes (CYP1A1, MT-ND4, MT-ND5, MT-RNR2, OB, and MT-TQ) evolved according to a molecular clock.
Thereafter, we used PAUP* to fit the sequence data for each gene to the (pruned) supertree topology under the optimal model in a ML framework. In line with Purvis' [118] local-clock model, the relative branch lengths for each gene tree relative to the topology of the supertree were determined using the Perl script relDate v2.2.1 [91]. Only the gene trees for the clock-like genes were considered to be rooted and relative branch lengths were calculated with respect to ancestral nodes only (and not also with respect to daughter nodes).
Divergence times were then determined by calibrating the relative branch lengths for each gene tree using a set of fossil dates (Table 5). For a given node, the initial divergence date was taken to be the maximum of 1) the median of all fossil plus molecular estimates and 2) the fossil estimate. In this way, the fossil estimate acts as a minimum age constraint that can overrule the molecular estimates. Upper and lower bounds on any given date estimates took the form of the 95% confidence interval derived from all individual gene and/or fossil estimates for that node. Although error in the branch-length estimation for the individual gene trees can also contribute to uncertainty in the final date estimates [120], it is likely to be less important than the variation present between the different genes themselves. However, together with uncertainties in the fossil dates, it cannot be excluded that our confidence intervals are underestimates of the true values.
Table 5.
Fossil calibrations used to anchor molecular date estimates.
| Divergence | Date | Source(s) | Node |
| Canids from arctoid carnivores | 43.5 | [127] | 1 |
| Pinniped and ursid split | 19.5 | [17] | 2 |
| Split between Phocidae and Otariidae + Odobenidae | 23 | [2] | 3 |
| Odobenoids first evolved | 18 | [2] | 4 |
| Monachinae-Phocinae split | 16 | [1, 55, 128] | 18 |
| Split between Monachus and other southern phocids | 8 | [71, 128] | 28 |
| Split between Mirounga + Lobodontini | 7 | [71, 128] | 29 |
| Origin of Callorhinus | 6 | [56] | 5 |
All dates (in millions of years ago) also represent minimum age constraints.
Finally, the Perl script chronoGrapher v1.3.3 [91] was used to correct for any negative branch lengths and simultaneously to derive a divergence-time estimate for the single node lacking an initial estimate (that linking Monachus schauinslandi and M. tropicalis). The date for this latter node was interpolated from the dates of up to five of its ancestral nodes based on the relative number of species descended from each node, assuming a constant birth model (see [117]).
More details regarding this dating procedure, including its strengths and weaknesses with respect to other relaxed molecular clock methods (recently reviewed in [121]) can be found in Bininda-Emonds et al. [117].
The Bayesian relaxed molecular clock method implemented by multidivtime [122,123] was also used to calculate divergence dates from the supermatrix data fitted to the preferred supertree topology. General methodology followed Rutschman [124], with maximum likelihood parameters estimated using PAML version 3.15 [125]. Incomplete overlap of sequences between taxa (in particular the outgroup sequence(s) not being represented in every partition) meant that model partitioning by gene was impossible; instead, a single F84 + gamma model was applied to the entire supermatrix. The root prior rttm (the mean of the prior distribution for the time from the ingroup root to the tips; in other words, the age of the ursid-pinniped split) was specified as 19.5 mya, with the remaining constraints the same as in the supertree dating analysis (Table 5). Other multidivtime parameters were calculated following the recommendations of Rutschmann [124]: rtrate (mean of prior distribution for the rate at the root node) = X/rttm, where X is the median amount of evolution from the root to tips; rtratesd (standard deviation of rtrtate) = 0.5 × rtrate; brownmean (mean of the prior distribution for the autocorrelation parameter, v) = 1/rttm; brownsd (standard deviation of brownmean) = brownmean. Three independent multidivtime analyses were run for 1 × 106 cycles, with samples taken every 100 cycles after a burn-in period of 1 × 105 cycles. The dates presented here are mean values for the three runs. The multidivtime analyses were then repeated using only the mitochondrial genes to investigate whether the inclusion of nuclear genes greatly altered the estimated divergence dates.
Authors' contributions
JWH collected data, conceived of and coordinated the study, assisted with analyses and drafted the manuscript. OB-E collected data; conducted the supertree, supermatrix, and dating analyses. RB conducted the multidivtime analyses and helped draft the manuscript. SHF and OB-E participated in the design of the study and helped draft the manuscript, and all authors read and approved the final manuscript.
Supplementary Material
GenBank accession numbers. GenBank accession numbers for all pinniped, canid and ursid sequences used in this study.
Acknowledgments
Acknowledgements
This research was supported by ArcticNet, NSERC, Fisheries and Oceans Canada (JH and SF), a Heisenberg Scholarship of the DFG (Germany) and the BMBF (Germany) through the "Bioinformatics for the Functional Analysis of Mammalian Genomes" project (OB-E). A review by L. Postma and three anonymous reviewers greatly improved this manuscript.
Contributor Information
Jeff W Higdon, Email: higdonj@dfo-mpo.gc.ca.
Olaf RP Bininda-Emonds, Email: olaf.bininda@uni-jena.de.
Robin MD Beck, Email: robin.beck@student.unsw.edu.au.
Steven H Ferguson, Email: fergusonsh@dfo-mpo.gc.ca.
References
- Berta A, Adam P. The evolutionary biology of pinnipeds. In: de Buffrenil V, Mazin J-M, editor. Secondary Adaptation of Tetrapods to Life in the Water. Germany: Verlag Dr. Frederich Pfeil; 2001. pp. 235–260. [Google Scholar]
- Deméré TA, Berta A, Adam PJ. Pinnipedimorph evolutionary biogeography. Bull Amer Mus Nat Hist. 2003;279:32–76. doi: 10.1206/0003-0090(2003)279<0032:C>2.0.CO;2. [DOI] [Google Scholar]
- Wozencraft WC. Classification of the recent Carnivora. In: Gittleman JL, editor. Carnivore Behaviour, Ecology and Evolution. London, UK: Chapman and Hall; 1989. pp. 569–593. [Google Scholar]
- Bininda-Emonds ORP, Russell AP. A morphological perspective on the phylogenetic relationships of the extant phocid seals (Mammalia: Carnivora: Phocidae) Bonner Zool Monogr. 1996;41:1–256. [Google Scholar]
- Vrana PB, Milinkovitch MC, Powell JR, Wheeler WC. Higher level relationships of arctoid Carnivora based on sequence data and "total evidence". Mol Phylogenet Evol. 1994;3:47–58. doi: 10.1006/mpev.1994.1006. [DOI] [PubMed] [Google Scholar]
- Arnason U, Bodin K, Gullgerg A, Ledje C, Mouchaty S. A molecular view of pinniped relationships with a particular emphasis on the true seals. J Mol Evol. 1995;40:78–85. doi: 10.1007/BF00166598. [DOI] [PubMed] [Google Scholar]
- Lento GM, Hickson RE, Chambers GK, Penny D. Use of spectral analysis to test hypotheses on the origin of pinnipeds. Mol Biol Evol. 1995;12:28–52. doi: 10.1093/oxfordjournals.molbev.a040189. [DOI] [PubMed] [Google Scholar]
- Ledje C, Arnason U. Phylogenetic analysis of complete cytochrome b genes of the order Carnivora with emphasis on the Caniformia. J Mol Evol. 1996;42:135–144. doi: 10.1007/BF02198839. [DOI] [PubMed] [Google Scholar]
- Ledje C, Arnason U. Phylogenetic relationships within caniform carnivores based on analysis of the mitochondrial 12S rRNA gene. J Mol Evol. 1996;43:641–649. doi: 10.1007/BF02202112. [DOI] [PubMed] [Google Scholar]
- Arnason U, Adegoke JA, Bodin K, Born EW, Esa YB, Gullberg A, Nilsson M, Short RV, Xu X, Janke A. Mammalian mitogenomic relationships and the root of the eutherian tree. PNAS. 2002;99:8151–8156. doi: 10.1073/pnas.102164299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CS, Delisle I, Stirling I, Siniff DB, Strobeck C. A phylogeny of the extant Phocidae inferred from complete mitochondrial DNA coding regions. Mol Phylogenet Evol. 2004;33:363–377. doi: 10.1016/j.ympev.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Delisle I, Strobeck C. A phylogeny of the Caniformia (order Carnivora) based on 12 complete protein-coding mitochondrial genes. Mol Phylogenet Evol. 2005;37:192–201. doi: 10.1016/j.ympev.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Flynn JJ, Finarelli JA, Zehr S, Hsu J, Nedbal MA. Molecular phylogeny of the Carnivora (Mammalia): assessing the impact of increased sampling on resolving enigmatic relationships. Syst Biol. 2005;54:317–337. doi: 10.1080/10635150590923326. [DOI] [PubMed] [Google Scholar]
- Arnason A, Gullberg A, Janke A, Kullberg M, Lehman N, Petrov EA, Väinölä R. Pinniped phylogeny and a new hypothesis for their origin and dispersal. Mol Phylogenet Evol. 2006;46:345–354. doi: 10.1016/j.ympev.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Fulton TL, Strobeck C. Molecular phylogeny of the Arctoidea (Carnivora): effect of missing data on supertree and supermatrix analyses of multiple gene data sets. Mol Phylogenet Evol. 2006;41:165–181. doi: 10.1016/j.ympev.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Wyss AR. The walrus auditory region and monophyly of pinnipeds. Am Mus Novitates. 1987;2871:1–31. [Google Scholar]
- Wyss AR, Flynn JJ. A phylogenetic analysis and definition of the Carnivora. In: Szalay FZ, Novacek MJ, McKenna MC, editor. Mammal phylogeny: Placentals. New York: Springer-Verlag; 1993. pp. 32–52. [Google Scholar]
- Berta A, Wyss AR. Pinniped phylogeny. Proc San Diego Soc Nat Hist. 1994;29:33–56. [Google Scholar]
- Rice DW. Marine Mammals of the World: Systematics and Distribution. Lawrence, KA: Society for Marine Mammalogy, Special Publication No. 4; 1998. [Google Scholar]
- Repenning CA, Ray CE. The origin of the Hawaiian monk seal. Proc Biol Soc Washington. 1977;89:667–688. [Google Scholar]
- de Muizon C. Phocid phylogeny and dispersal. Ann S Afr Mus. 1982;89:175–213. [Google Scholar]
- Wyss AR. Evidence from flipper structure for a single origin of pinnipeds. Nature. 1988;334:427–428. doi: 10.1038/334427a0. [DOI] [Google Scholar]
- Bininda-Emonds ORP, Gittleman JL, Purvis A. Building large trees by combining phylogenetic information: a complete phylogeny of the extant Carnivora (Mammalia) Biol Rev. 1999;74:143–175. doi: 10.1017/S0006323199005307. [DOI] [PubMed] [Google Scholar]
- Fyler CA, Reeder TW, Berta A, Antonelis G, Aguilar A, Androukaki E. Historical biogeography and phylogeny of monachine seals (Pinnipedia: Phocidae) based on mitochondrial and nuclear DNA data. J Biogeog. 2005;32:1267–1279. doi: 10.1111/j.1365-2699.2005.01281.x. [DOI] [Google Scholar]
- King JE. Relationships of the hooded and elephant seals (genera Cystophora and Mirounga) J Zool London. 1966;148:385–398. [Google Scholar]
- Palo JU, Väinölä R. The enigma of the landlocked Baikal and Caspian seals addressed through phylogeny of phocine mitochondrial sequences. Biol J Linnean Soc. 2006;88:61–72. doi: 10.1111/j.1095-8312.2006.00607.x. [DOI] [Google Scholar]
- Reidman M. The Pinnipeds: Seals, Sea Lions, and Walruses. Berkley, CA: University of California Press; 1990. [Google Scholar]
- Reynolds JE, III, Odell DK, Rommel SA. Marine mammals of the world. In: Reynolds JE III, Rommel SA, editor. Biology of marine mammals. Washington, DC: Smithsonian Institution Press; 1999. pp. 1–14. [Google Scholar]
- Repenning CA, Tedford RH. Otarioid seals of the Neogene: classification, historical zoogeography, and temporal correlation of the sea lions and walruses from the North Pacific region. US Geol Surv Prof Pap. 1977;992:1–93. [Google Scholar]
- Brunner S. Cranial morphometrics of fur seals and sea lions (Family: Otariidae) – systematics, geographic variation and growth. Sydney, Australia: Veterinary Anatomy and Pathology, University of Sydney; 2000. [Google Scholar]
- Brunner S. Fur seals and sea lions (Otariidae): identification of species and taxonomic review. Syst Biodiv. 2003;1:339–439. doi: 10.1017/S147720000300121X. [DOI] [Google Scholar]
- Wynen LP, Goldsworthy SD, Insley SJ, Adams M, Bickman JW, Francis J, Gallo JP, Hoelzel AR, Majluf P, White RWG, Slade S. Phylogenetic relationships within the eared seals (Otariidae: Carnivora): implications for the historical biogeography of the family. Mol Phylogenet Evol. 2001;21:270–284. doi: 10.1006/mpev.2001.1012. [DOI] [PubMed] [Google Scholar]
- Goldsworthy SD, Boness DJ, Fleischer RC. Mate choice among sympatric fur seals: female preference for conphenotypic males. Behav Ecol Sociobiol. 1999;45:263–267. doi: 10.1007/s002650050560. [DOI] [Google Scholar]
- Brunner S. A probable hybrid sea lion – Zalophus californianus X Otaria bryonia. J Mamm. 2002;83:135–144. doi: 10.1644/1545-1542(2002)083<0135:APHSLZ>2.0.CO;2. [DOI] [Google Scholar]
- Bininda-Emonds ORP, Gittleman JL, Kelly CK. Flippers versus feet: comparative trends in aquatic and non-aquatic carnivores. J Animal Ecol. 2001;70:86–400. [Google Scholar]
- Diniz-Filho JAF, Tôrres NM. Phylogenetic comparative methods and the geographic range size-body size relationship in new world terrestrial Carnivora. Evol Ecol. 2002;16:351–367. doi: 10.1023/A:1020210321776. [DOI] [Google Scholar]
- Ferguson SH, Larivière S. Can comparing life histories help conserve carnivores? Animal Conserv. 2002;5:1–12. [Google Scholar]
- Ferguson SH, Larivière S. Are long penis bones an adaptation to high latitude snowy environments? Oikos. 2004;105:255–267. doi: 10.1111/j.0030-1299.2004.13173.x. [DOI] [Google Scholar]
- Larivière S, Ferguson SH. On the evolution of the mammalian baculum: vaginal friction, prolonged intromission, and induced ovulation? Mamm Rev. 2002;32:283–294. doi: 10.1046/j.1365-2907.2002.00112.x. [DOI] [Google Scholar]
- Larivière S, Ferguson SH. Evolution of induced ovulation in North American carnivores. J Mamm. 2003;84:937–947. doi: 10.1644/BME-003. [DOI] [Google Scholar]
- Nunn CL, Gittleman JL, Antonovics J. A comparative study of white blood cell counts and disease risk in carnivores. Proc Royal Soc B Biol Sci. 2003;270:347–356. doi: 10.1098/rspb.2002.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardillo M, Purvis A, Sechrest W, Gittleman JL. Human population density and extinction risk in the world's carnivores. PLoS Biol. 2004;2:909–914. doi: 10.1371/journal.pbio.0020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bininda-Emonds ORP, Gittleman JL. Are pinnipeds functionally different from fissiped carnivores? The importance of phylogenetic comparative analyses. Evolution Int J Org Evolution. 2000;54:1011–1023. doi: 10.1111/j.0014-3820.2000.tb00100.x. [DOI] [PubMed] [Google Scholar]
- Lindenfors P, Tullberg BS, Biuw M. Phylogenetic analyses of sexual selection and sexual size dimorphism in pinnipeds. Behav Eco Sociobiol. 2002;52:188–193. doi: 10.1007/s00265-002-0507-x. [DOI] [Google Scholar]
- Schulz TM, Bowen WD. The evolution of lactation strategies in pinnipeds: a phylogenetic analysis. Ecol Monogr. 2005;75:159–177. doi: 10.1890/04-0319. [DOI] [Google Scholar]
- Ferguson SH. The influences of environment, mating habitat, and predation on evolution of pinniped lactation strategies. J Mamm Evol. 2006;13:63–82. doi: 10.1007/s10914-005-9003-1. [DOI] [Google Scholar]
- Ferguson SH, Higdon JW. How seals divide up the world: environment, life history, and conservation. Oecologia. 2006;150:318–329. doi: 10.1007/s00442-006-0489-x. [DOI] [PubMed] [Google Scholar]
- Baum BR. Combining trees as a way of combining data sets for phylogenetic inference, and the desirability of combining gene trees. Taxon. 1992;41:3–10. doi: 10.2307/1222480. [DOI] [Google Scholar]
- Ragan MA. Phylogenetic inference based on matrix representation of trees. Mol Phylogenet Evol. 1992;1:53–58. doi: 10.1016/1055-7903(92)90035-F. [DOI] [PubMed] [Google Scholar]
- Lapointe F-J, Kirsch JAW, Hutcheon JM. Total evidence, consensus, and bat phylogeny: a distance based approach. Mol Phylogenet Evol. 1999;11:55–66. doi: 10.1006/mpev.1998.0561. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds ORP. Novel versus unsupported clades: assessing the qualitative support for clades in MRP supertrees. Syst Biol. 2003;52:839–848. doi: 10.1080/10635150390252242. [DOI] [PubMed] [Google Scholar]
- Price SA, Bininda-Emonds ORP, Gittleman JL. A complete phylogeny of the whales, dolphins and even-toed hoofed mammals (Cetartiodactyla) Biol Rev. 2005;80:445–473. doi: 10.1017/S1464793105006743. [DOI] [PubMed] [Google Scholar]
- Berta A, Sumich JL. Marine mammals: evolutionary biology. San Diego: Academic Press; 1999. [Google Scholar]
- Repenning CA. Adaptive evolution of sea lions and walruses. Syst Zool. 1976;25:375–390. doi: 10.2307/2412512. [DOI] [Google Scholar]
- Repenning CA, Ray CE, Grigorescu D. Pinniped biogeography. In: Gray J, Boucot AJ, editor. Historical Biogeography, Plate Tectonics, and the Changing Environment. Corvallis, OR: Oregon State University Press; 1979. pp. 357–369. [Google Scholar]
- Miyazaki S, Horikawa H, Kohno N, Hirota K, Kimura M, Hasegawa Y, Tomida Y, Barnes LG, Ray CE. Summary of the fossil record of pinnipeds from Japan, and comparisons with that from the eastern North Pacific. Island Arc. 1995;3:361–372. doi: 10.1111/j.1440-1738.1994.tb00120.x. [DOI] [Google Scholar]
- Mitchell ED, Tedford RH. The Enaliarctinae: a new group of extinct aquatic Carnivora and a consideration of the origin of the Otariidae. Bull Am Mus Nat Hist. 1973;151:201–284. [Google Scholar]
- Berta A. New Enaliarctos (Pinnipedimorpha) from the Oligocene and Miocene of Oregon and the role of "enaliarctids" in pinniped phylogeny. Smithson Contrib Paleobiol. 1991;69:1–33. [Google Scholar]
- Koretsky IA, Sanders AE. Pinniped bones from the Late Oligocene of South Carolina: the oldest known seal (Carnivora: Phocidae) [abstract] J Vert Paleo. 1997;17:58A. [Google Scholar]
- Koretsky IA, Sanders AE. Paleontology of the Late Oligocene Ashley and Chandler Bridge Formations of South Carolina, 1: Paleogene pinniped remains; the oldest known seal (Carnivora: Phocidae) In: Emry RJ, editor. Cenozoic Mammals of Land and Sea: Tributes to the Career of Clayton E Ray Smith Cont Paleo. Vol. 93. 2002. pp. 179–183. [Google Scholar]
- Barnes LG, Domning DP, Ray CE. Status of studies on fossil marine mammals. Mar Mammal Sci. 1985;1:15–53. doi: 10.1111/j.1748-7692.1985.tb00530.x. [DOI] [Google Scholar]
- Costa DP. The relationship between reproductive and foraging energetics and the evolution of the Pinnipedia. Symp Zool Soc London. 1993;66:293–314. [Google Scholar]
- Repenning CA, Peterson RS, Hubbs CL. Contributions to the systematics of the southern fur seals, with particular reference to the Juan Fernandez and Guadalupe species. In: Burt WH, editor. Antarctic Pinnipedia. Washington, DC: American Geophysical Union; 1971. pp. 1–52. [Google Scholar]
- Stirling I, Warneke RM. Implications of a comparison of the airborne vocalizations and some aspects of the behaviour of the two Australian fur seals, Arctocephalus spp., on the evolution and present taxonomy of the genus. Australian J Zool. 1971;19:227–241. doi: 10.1071/ZO9710227. [DOI] [Google Scholar]
- Trillmich F, Majluf P. First observations on colony structure, behaviour and vocal repertoire of the South American fur seal (Arctocephalus australis Zimmermann, 1783) in Peru. Zeitschrift Saugetierkunde. 1981;46:310–322. [Google Scholar]
- Goldsworthy SD, Pemberton D, Warneke RM. Field identification of Australian and New Zealand fur seals, Arctocephalus ssp., based on external features. In: Hindell M, Kemper K, editor. Marine Mammal Research in the Southern Hemisphere. Sydney, Australia: Surrey Beatty and Sons; 1997. pp. 63–71. [Google Scholar]
- King JE. The otariid seals of the Pacific coast of America. British Mus (Nat Hist) Zool Publ. 1954;2:309–337. [Google Scholar]
- Scheffer VB. Seals, Sea Lions and Walruses – A Review of the Pinnipedia. Stanford, CA: Stanford University Press; 1958. [Google Scholar]
- Sivertsen E. Scientific results of the Norwegian Antarctic expeditions 1927–1928 (et seq) Oslo: Det Norske Videnskaps – Akademii Oslo; 1954. A survey of the eared seals (family Otariidae) with remarks on the Antarctic seals collected by M/K "Norvegia" in 1928–1929. [Google Scholar]
- de Muizon C. Arctocephalus (Hydrarctos) lomasiensis, subgen, nov. et nov. sp., un nouvel Otariidae du Mio-Pliocene de Sacaco (Perou) Bull de l'Institut Francais des Etudes Andines. 1978;7:169–188. [Google Scholar]
- de Muizon C, De Vries TJ. Geology and paleontology of late Cenozoic marine deposits in the Sacaco area (Peru) Sonderdruck aus Geologische Rundschau. 1985;74:547–563. doi: 10.1007/BF01821211. [DOI] [Google Scholar]
- Kim KC, Repenning CA, Morejohn GV. Specific antiquity of the sucking lice and evolution of otariids seals. Rapp P-V Reun Con Internat Explor Mer. 1975;169:544–549. [Google Scholar]
- Drehmer CJ, Ribeiro AM. A temporal bone of an Otariidae (Mammalia: Pinnipedia) from the late Pleistocene of Rio Grandedo Sul State, Brazil. Geociencias. 1998;3:39–44. [Google Scholar]
- King JE. Seals of the World. St. Lucia: University of Queensland Press; 1983. [Google Scholar]
- Burns JJ. Remarks on the distribution and natural history of pagophilic pinnipeds in the Bering and Chukchi seas. J Mammal. 1970;51:445–454. doi: 10.2307/1378386. [DOI] [Google Scholar]
- McLaren IA. A speculative overview of phocid evolution. Rapp P-v Réun Cons Int Explor Mer. 1975;169:43–48. [Google Scholar]
- Ray CE. Phoca wymani and other Tertiary seals (Mammalia: Phocidae) described from the Eastern Seaboard of North America. Smith Cont Paleo. 1977;28:1–36. [Google Scholar]
- Arnason U, Xu XF, Gullberg A. The 'Phoca standard': an external molecular reference for calibrating recent evolutionary divergences. J Mol Evol. 1996;43:41–45. doi: 10.1007/BF02352298. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Numachi K, Grachev MA. The origin and genetic relationships of the Baikal Seal, Phoca sibirica, by restriction analysis of mitochondrial DNA. Zool Sci. 2003;20:1417–1422. doi: 10.2108/zsj.20.1417. [DOI] [PubMed] [Google Scholar]
- Haug GH, Tiedemann R. Effect of the formation of the Isthmus of Panama on Atlantic Ocean thermohaline circulation. Nature. 1998;393:673–676. doi: 10.1038/31447. [DOI] [Google Scholar]
- Mercer JM, Roth VL. The effects of Cenozoic global change on squirrel phylogeny. Science. 2003;299:1568–1572. doi: 10.1126/science.1079705. [DOI] [PubMed] [Google Scholar]
- Miya M, Nishida M. Speciation in the open ocean. Nature. 1997;389:803–804. doi: 10.1038/39774. [DOI] [Google Scholar]
- Ray CE. Geography of phocid evolution. Syst Zool. 1976;25:391–406. doi: 10.2307/2412513. [DOI] [Google Scholar]
- Slade RW, Moritz C, Hoelzel AR, Burton HR. Molecular population genetics of the southern elephant seal Mirounga leonina. Genetics. 1998;149:1945–1957. doi: 10.1093/genetics/149.4.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey PH, Pagel MD. The comparative method in evolutionary biology. Oxford, UK: Oxford University Press; 1991. [Google Scholar]
- Garland T, Jr, Midford PE, Ives AR. An introduction to phylogenetically based statistical methods, with a new method for confidence intervals on ancestral values. Am Zool. 1999;39:374–388. [Google Scholar]
- Polly PD. Paleontology and the comparative method: ancestral node reconstruction versus observed node values. Am Nat. 2001;157:596–609. doi: 10.1086/320622. [DOI] [PubMed] [Google Scholar]
- Fisher DO, Owens IPF. The comparative method in conservation biology. Trends Ecol Evol. 2004;19:391–398. doi: 10.1016/j.tree.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Isaac NJB, Turvey ST, Collen B, Waterman C, Baillie JEM. Mammals on the EDGE: conservation priorities based on threat and phylogeny. PLoS ONE. 2007;2:e296. doi: 10.1371/journal.pone.0000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Huelsenbeck JP, Cunningham CW, Swofford DL, Waddel PJ. Partitioning and combining data in phylogenetic analysis. Syst Biol. 1993;42:384–397. doi: 10.2307/2992473. [DOI] [Google Scholar]
- Bininda-Emonds ORP. Programs distributed by the author. Institut für Spezielle Zoologie und Evolutionsbiologie, University of Jena; 2007. Perl scripts GenBankStrip, seqCleaner, relDate, ChronoGrapher.http://www.personal.uni-jena.de/~b6biol2/ProgramsMain.html [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bininda-Emonds ORP. transAlign: using amino acids to facilitate the multiple alignment of protein-coding DNA sequences. BMC Bioinform. 2005;6:156. doi: 10.1186/1471-2105-6-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DE, Reeder DM, (Editors) Mammal species of the world: a taxonomic and geographic reference. Washington, DC: Smithsonian Institution Press; 1993. [Google Scholar]
- Sanderson MJ, Donoghue MJ, Piel W, Eriksson T. TreeBASE: a prototype database of phylogenetic analyses and an interactive tool for browsing the phylogeny of life. Amer J Bot. 1994;81:183. doi: 10.2307/2445299. [DOI] [Google Scholar]
- Daubin V, Gouy M, Perrière G. Bacterial molecular phylogeny using supertree approach. Genome Inform. 2001;12:155–164. [PubMed] [Google Scholar]
- Bininda-Emonds ORP, Stamatakis A. Taxon sampling versus computational complexity and their impact on obtaining the Tree of Life. In: Hodkinson TR, Parnell JAN, editor. Reconstructing the Tree of Life: Taxonomy and Systematics of Species Rich Taxa. Boca Raton, FL: CRC Press; 2006. pp. 77–95. [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Mossel E, Vigoda E. Phylogenetic MCMC algorithms are misleading on mixtures of trees. Science. 2005;309:2207–2209. doi: 10.1126/science.1115493. [DOI] [PubMed] [Google Scholar]
- Kubatko LS, Degnan JH. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst Biol. 2007;56:17–24. doi: 10.1080/10635150601146041. [DOI] [PubMed] [Google Scholar]
- Jeffroy O, Brinkmann H, Delsuc F, Phillippe H. Phylogenomics: the beginning of incongruence? Trends Genet. 2006;22:225–231. doi: 10.1016/j.tig.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Gatesy J, Baker RH, Hayashi C. Inconsistencies in arguments for the supertree approach: supermatrices versus supertrees of Crocodylia. Syst Biol. 2004;53:324–355. doi: 10.1080/10635150490423971. [DOI] [PubMed] [Google Scholar]
- Gatesy J, Matthee CA, DeSalle R, Hayashi C. Resolution of a supertree/supermatrix paradox. Syst Biol. 2002;51:652–664. doi: 10.1080/10635150290102311. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds ORP, Jones KE, Price SA, Grenyer R, Cardillo M, Habib M, Purvis A, Gittleman JL. Supertrees are a necessary not-so-evil, a comment on Gatesy et al. Syst Biol. 2003;52:724–729. doi: 10.1080/10635150390235647. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds ORP. Trees versus characters and the supertree/supermatrix "paradox". Syst Biol. 2004;53:356–359. doi: 10.1080/10635150490440396. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gsacuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Nylander JAA. Program distributed by the author. Evolutionary Biology Centre, Uppsala University; 2004. MrAIC.pl. [Google Scholar]
- Bininda-Emonds ORP, Beck RMD, Purvis A. Getting to the roots of matrix representation. Syst Biol. 2005;54:668–672. doi: 10.1080/10635150590947113. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evol. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds ORP, Sanderson MJ. Assessment of the accuracy of matrix representation with parsimony supertree construction. Syst Biol. 2001;50:565–579. doi: 10.1080/106351501750435112. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Phylogenetic analysis using parsimony (*and other methods) v.4. Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- Bremer K. The limits of amino acid sequence data in angiosperm phylogenetic reconstruction. Evol. 1988;42:795–803. doi: 10.2307/2408870. [DOI] [PubMed] [Google Scholar]
- Beck RDM, Bininda-Emonds ORP, Cardillo M, Liu F-GR, Purvis A. A higher-level MRP supertree of mammals. BMC Evol Biol. 2006;6 doi: 10.1186/1471-2148-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinform. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. Phylogenetic models of rate heterogeneity: a high performance computing perspective. Proceedings of 20th IEEE/ACM International Parallel and Distributed Processing Symposium (IPDPS2006), High Performance Computational Biology Workshop, Proceedings on CD, Rhodos, Greece. 2006.
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinform. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds ORP, Cardillo M, Jones KE, MacPhee RDE, Beck RMD, Grenyer R, Price S, Vos RA, Gittleman JL, Purvis A. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Purvis A. A composite estimate of primate phylogeny. Philos Trans R Soc Lond B. 1995;348:405–421. doi: 10.1098/rstb.1995.0078. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall K. MODELTEST: testing the model of DNA substitution. Bioinform. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Graur D, Martin W. Reading the entrails of chickens: molecular timescales of evolution and the illusion of precision. Trends Genet. 2004;20:80–86. doi: 10.1016/j.tig.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Renner SS. Relaxed molecular clocks for dating historical plant dispersal events. Trends Pl Sci. 2005;10:550–558. doi: 10.1016/j.tplants.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Kishino H, Thorne JL, Bruno WJ. Performance of a divergence time estimation method under a probabilistic model of rate evolution. Mol Biol Evol. 2001;18:352–361. doi: 10.1093/oxfordjournals.molbev.a003811. [DOI] [PubMed] [Google Scholar]
- Thorne JL, Kishino H, Painter IS. Estimating the rate of evolution of the rate of molecular evolution. Mol Biol Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- Rutschmann F. Bayesian molecular dating using PAML/multidivtime: a step-by-step manual. Switzerland: University of Zurich; 2005. [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Wain HM, Lush M, Ducluzeau F, Povey S. Genew: the human gene nomenclature database. Nucleic Acids Res. 2002;30:169–171. doi: 10.1093/nar/30.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna MC, Bell SK. Classification of Mammals above the Species Level. New York: Columbia University Press; 1997. [Google Scholar]
- de Muizon C. Phocid phylogeny and dispersal. Annals S Afr Mus. 1982;89:175–213. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GenBank accession numbers. GenBank accession numbers for all pinniped, canid and ursid sequences used in this study.