Abstract
Catharanthus roseus produces low levels of two dimeric terpenoid indole alkaloids, vinblastine and vincristine, which are widely used in cancer chemotherapy. The dimerization reaction leading to α-3′,4′-anhydrovinblastine is a key regulatory step for the production of the anticancer alkaloids in planta and has potential application in the industrial production of two semisynthetic derivatives also used as anticancer drugs. In this work, we report the cloning, characterization, and subcellular localization of an enzyme with anhydrovinblastine synthase activity identified as the major class III peroxidase present in C. roseus leaves and named CrPrx1. The deduced amino acid sequence corresponds to a polypeptide of 363 amino acids including an N-terminal signal peptide showing the secretory nature of CrPrx1. CrPrx1 has a two-intron structure and is present as a single gene copy. Phylogenetic analysis indicates that CrPrx1 belongs to an evolutionary branch of vacuolar class III peroxidases whose members seem to have been recruited for different functions during evolution. Expression of a green fluorescent protein-CrPrx1 fusion confirmed the vacuolar localization of this peroxidase, the exact subcellular localization of the alkaloid monomeric precursors and dimeric products. Expression data further supports the role of CrPrx1 in α-3′,4′-anhydrovinblastine biosynthesis, indicating the potential of CrPrx1 as a target to increase alkaloid levels in the plant.
Catharanthus roseus, known as the Madagascar periwinkle, accumulates in the leaves the dimeric terpenoid indole alkaloids (TIAs) vinblastine and vincristine, which were the first natural drugs used in cancer therapy and are still among the most valuable agents used in the treatment of cancer. Intensive chemical research has also resulted in the development of semisynthetic derivatives of the dimeric alkaloids showing higher activity and lower toxicity (Sottomayor and Ros Barceló, 2005). The great pharmacological importance of the dimeric alkaloids, allied to its low availability in the plant (approximately half a ton of dry leaves is needed to obtain 1 g of vinblastine for pharmaceutical production; Noble, 1990), stimulated intense research of the TIA pathway, and C. roseus has therefore become one of the most extensively studied medicinal plants (Verpoorte and Memelink, 2002; van der Heijden et al., 2004). The biosynthesis of vinblastine involves more than 20 enzymatic steps, nine of which are now well characterized at the enzyme and gene level, and several regulatory genes of the pathway (ORCAs) have also been cloned. However, considerable parts of the pathway remain relatively hypothetical, and even enzymatic characterization is still lacking for many steps (for review, see Hilliou et al., 2001; El-Sayed and Verpoorte, 2007; Loyola-Vargas et al., 2007; Memelink and Gantet, 2007).
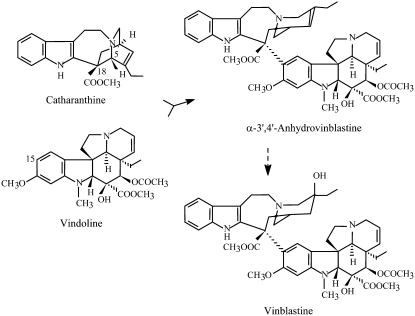
Since early studies on the biosynthesis of vinblastine, the dimerization reaction has attracted much attention due to its regulatory importance and potential application for the semisynthetic production of the dimeric alkaloids. This reaction involves the coupling of the monomeric precursors vindoline and catharanthine into the common precursor of all dimeric alkaloids, α-3′,4′-anhydrovinblastine (AVLB; Fig. 1). The search for the dimerization enzyme in leaf tissue detected a single dimerization activity credited to the single class III plant peroxidase present in the leaves of the plant. The enzyme was purified to homogeneity and a channeling mechanism was proposed for the peroxidase-mediated-vacuolar synthesis of AVLB (Sottomayor et al., 1996, 1998; Sottomayor and Ros Barceló, 2003).
Figure 1.
Biosynthesis of vinblastine from the monomeric precursors catharanthine and vindoline. Anhydrovinblastine is the product of the dimerization reaction and the precursor of the anticancer drugs.
Class III peroxidases are multifunctional enzymes, typical of plants, that catalyze the oxidation of small molecules at the expense of H2O2. They are capable of recognizing a broad range of substrates and also show a remarkable diversity, with the presence of a high number of isoenzymes in a single plant. This diversity is determined to a great extent at the gene level, since the sequencing of the Arabidopsis (Arabidopsis thaliana) genome enabled the identification of 73 class III peroxidase genes and the sequencing of the rice (Oryza sativa) genome enabled the identification of 138 (Tognolli et al., 2002; Welinder et al., 2002; Duroux and Welinder, 2003; Passardi et al., 2004). Peroxidase isoenzymes are targeted by an N-terminal signal peptide to the secretory pathway and have either the cell wall or the vacuole as final destinations. Class III peroxidases have been mostly implicated in key processes determining the architecture and defense properties of the plant cell wall, like lignin and suberin biosynthesis and cross-linking reactions (Barceló et al., 2004; Bernards et al., 2004; Fry, 2004; Ralph et al., 2004; Gabaldón et al., 2005). They have also been implicated in auxin catabolism, in secondary metabolism, in root elongation, in hydrogen peroxide scavenging and production, and they are thought to play important roles in stress resistance and adaptation (Yoshida et al., 2003; Díaz et al., 2004; Kristensen et al., 2004; Mika et al., 2004; Sang et al., 2004; Sottomayor et al., 2004; Passardi et al., 2005, 2006).
In contrast with the high number of studies on cell wall peroxidase functions, much less is known about vacuolar peroxidases (Welinder et al., 2002). In vitro studies have shown the capacity of plant peroxidases to accept as substrates a number of vacuolar metabolites, such as phenols, flavonoids, and alkaloids, and it has been suggested that, in vivo, they assume specific functions in the metabolism of these compounds (Díaz et al., 2004; Kristensen et al., 2004; Sottomayor et al., 2004; Takahama, 2004). However, a consistent amount of evidence supporting a role in planta for a vacuolar peroxidase has seldom been obtained.
Here, we report the full gene structure and cDNA cloning of the major class III peroxidase isoenzyme (CrPrx1) present in the leaves of the medicinal plant C. roseus, where this enzyme is thought to be involved in the production of anticancer alkaloids. Sequence/structural information is analyzed and phylogenetic relations are discussed with insights into the evolution of class III peroxidase functions. CrPrx1 fusions with GFP were used to confirm subcellular localization of the enzyme. The expression pattern of CrPrx1 further supports a role of this enzyme in alkaloid metabolism.
RESULTS
Sequence of the N Terminus and of Trypsin-Derived Fragments of CrPrx1
The major basic peroxidase present in C. roseus leaves was purified to homogeneity (a single band was detected by SDS-PAGE after silver staining; Sottomayor and Ros Barceló, 2003) and was submitted to automatic Edman degradation. The amino acid signals obtained were of 800 fmol, out of 200 pmol of protein loaded, meaning that about 99.6% of the protein molecules had their N terminus blocked. Nevertheless, the high sensitivity of the sequencing apparatus enabled the identification of the first 11 amino acids of the N terminus (Table I). Blocking of the N terminus may have been a consequence of the purification procedure, specifically of the acetone precipitation. The purified protein was digested with trypsin and the N-terminal sequence of four purified peptides was also determined (Table I).
Table I.
Amino acid sequences of the N terminus and peptides resulting from trypsin digestion of pure CrPrx1
| Fragment | Amino Acid Sequence |
|---|---|
| N-terminal fragmenta | PPTVSGLSYTF |
| Peptide 1a | QGLFTSDQDLYTDRR |
| Peptide 2a | IYPNIDPTMDQT |
| Peptide 3a | EVFQNDVEQAAGLLR |
| Peptide 4a | NFXATDVVALSGGHTIG |
| Peptide 5b | IIDDLR |
| Peptide 6b | IVSCSDILALAAR |
| Peptide 7b | DSVFLTGGPDYDIPLGR |
| Peptide 8b | SPNVFDNR |
Sequences obtained by Edman sequencing.
Sequences obtained by MS/MS.
Cloning of the CrPrx1 cDNA and Characterization of the Full-Length Gene
A PCR strategy using degenerated primers designed from peptide 2 (Table I), followed by a screening of a cDNA library and RACE, resulted in a complete CrPrx1 cDNA clone of 1,388 bp, with a perfect sequence match between the deduced amino acid sequence (Fig. 2B) and the sequence of the peptide fragments obtained from the purified CrPrx1 (Table I). To confirm that the isolated cDNA corresponded indeed to the purified leaf peroxidase, the pure protein was further submitted to peptide mass fingerprint after trypsin digestion, followed by peptide sequencing/fragmentation (MS/MS) of peptides different form the ones previously sequenced by Edman degradation. Protein identification of the peptide mass fingerprint obtained from trypsin digestion of the purified CrPrx1 using the National Center for Biotechnology Information (NCBI) database gave a 100% CI protein score identification with the deduced amino acid sequence of the isolated cDNA deposited in GenBank (accession no. AM236087). Four new peptides were sequenced by MS/MS and they all fitted perfectly with the deduced amino acid sequence (Table I and Fig. 2B).
Figure 2.
The gene and protein of peroxidase 1 from C. roseus (CrPrx1). A, Schematic representation of the gene (P, promoter; Int, intron; Ex, exon; UTR, untranslated region). B, Nucleotide and deduced amino acid sequence. The coding region is shown in uppercase letters. The signal peptide and the putative C-terminal propeptide are represented in italics. Sequences highlighted correspond to the peroxidase active site signature (positions 38–49) and to the peroxidase proximal heme-ligand signature (positions 168–178). The single amino acids highlighted are the eight conserved Cys, which allow the formation of four disulfide bridges. Sequences underlined in black match perfectly the sequences determined for the nine sequenced peptides (Table I). Sequences underlined in gray correspond to putative glycosylation sites. Boxes, aataaa polyadenylation signals. Arrowheads, Different positions of the poly(A) tail determined by RACE for seedlings. Arrows, Different positions of the poly(A) tail in two clones isolated from the cDNA library prepared from the first pair of leaves. Accession numbers are: AM236087 (mRNA), AM236088 (promoter region), and AM236089 (intron I).
The 5′ untranslated leader of the cDNA contains an in-frame stop codon. During the isolation of the complete CrPrx1 cDNA, four different positions for the poly(A) tail were found: three in mRNA extracted from seedlings when performing 3′ RACE (arrowheads in Fig. 2B) and two in cDNA library clones obtained from the first pair of leaves of flowering plants (arrows in Fig. 2B).
To determine the intron structure of CrPrx1, we decided to use as a starting point the features of the Arabidopsis Prx gene with the highest homology with CrPrx1. In silico analysis showed that CrPrx1 shares maximum overall amino acid sequence identity (60%) with Arabidopsis AtPrx12 (named after Peroxibase; http://peroxidase.isb-sib.ch/, locus At1g71695). The AtPrx12 gene contains two introns whose flanking regions show an almost complete sequence identity with the corresponding region in CrPrx1. Therefore, we predicted that the CrPrx1 gene could also contain two introns in equivalent positions. The design of primers flanking the putative position of intron I indeed enabled the amplification of an intron by conventional PCR. Intron I of CrPrx1 consists of 828 bp, is inserted in position 300 of the cDNA (in perfect agreement with intron I of the Arabidopsis AtPrx12 gene), and is flanked by the usual GT- and -AG dinucleotides. No obvious sequence similarity was detected between CrPrx1 intron I and the corresponding intron in AtPrx12.
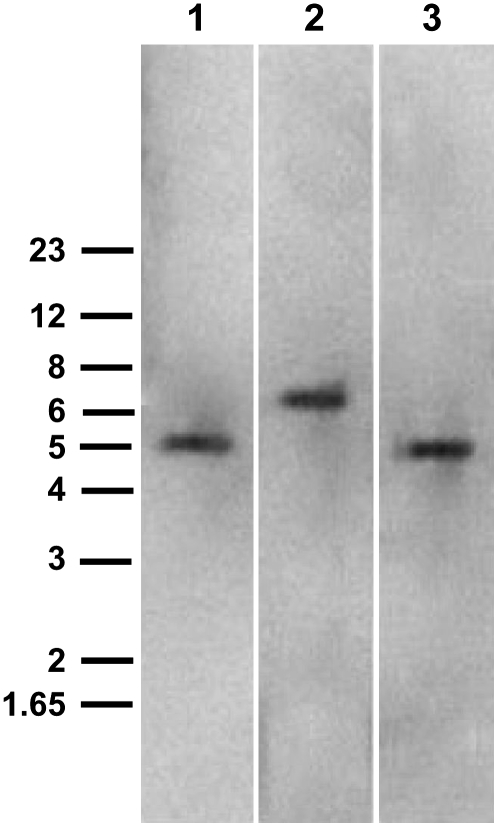
A second intron is also present and likely to be inserted in a position equivalent to intron II of AtPrx12 gene. However, attempts to amplify intron II of CrPrx1 with diverse pairs of oligonucleotide primers specific for its flanking regions repeatedly failed. We anticipated that difficulties in amplifying intron II of CrPrx1 gene could be due to its large size. Southern blots were then performed to estimate both the gene copy number of CrPrx1 and the size of intron II. Genomic DNA was digested with NcoI (an enzyme cutting at positions 41 and 1,058 of the cDNA), with Van91I and EcoRI simultaneously (the former cutting inside the promoter region at position −114, the latter in position 1,361 of the cDNA), or with HincII (an enzyme cutting inside intron I and at position 1,004 of the cDNA). DNA digestions were designed so that a single band should be produced, except if the sequence of intron II contained restriction sites for the enzymes used. Hybridization was performed with an antisense digoxigenin (DIG)-labeled oligonucleotide probe complementary to a 39-nucleotide segment of intron I.
Results are shown in Figure 3. One single band was detectable in all lanes, strongly suggesting that CrPrx1 exists as a single-copy gene in the C. roseus genome. Estimated sizes for the bands in lanes 1 and 2 of Figure 3 (digestions with NcoI and with Van91I and EcoRI, respectively) are 5.35 and 6.38 kb, which give an estimated size for intron II alone of 3.5 to 4.0 kb. Since the oligonucleotide probe used hybridizes upstream of the HincII restriction sites, the result in lane 3 is not informative for an estimation of the size of intron II.
Figure 3.
Southern blot of C. roseus genomic DNA. An antisense DIG-labeled oligonucleotide complementary to a 39-base segment of intron I was used as probe. DNA was digested with NcoI (lane 1), Van91I and EcoRI (lane 2), and HincII (lane 3). Arrows indicate bands, and numbers on the left indicate positions of the DNA Mr markers in kilobase pairs.
For the identification and sequencing of the CrPrx1 promoter region, a strategy involving inverse PCR (IPCR) was used. Different restriction endonucleases were used to digest the DNA before circularization, but only with TaqI was it possible to obtain a PCR amplification product: a single band with an estimated size of 1.35 kb. Cloning and sequencing showed that this PCR product contained 914 nt upstream from the +1 nt of CrPrx1. The precise sequence of the putative TATA box is TATAAA and is located between bases −28 and −33. Analysis of the promoter sequence in databases for recognition of regulatory cis-elements enabled the identification of several regulatory sequences, namely, of several W boxes recognized by the WRKY superfamily of transcription factors, which are involved in the regulation of various physiological programs unique to plants, including pathogen defense and senescence (Eulgem et al., 2000). The capacity of the isolated promoter sequence to direct senescence associated expression is now being investigated in our laboratory.
Analysis of CrPrx1 Deduced Amino Acid Sequence and Predicted Three-Dimensional Structure
The deduced amino acid sequence of the complete cDNA cloned corresponds to a polypeptide with 363 amino acids (Fig. 2B) including the sequence of the tryptic digestion fragments (Table I) and presenting a 100% match with the tryptic mass fingerprint of the pure protein. Thus, the cloned cDNA corresponds to the basic peroxidase isoenzyme present in C. roseus leaves, CrPrx1. The main characteristics of the CrPrx1 mature protein determined for the purified protein and deduced from the amino acid sequence are shown in Table II.
Table II.
Prosequences and consensus sequences detected in CrPrx1
Identification and prediction of specific sequences performed using the PROSITE database (http://www.expasy.org/prosite).
| Putative (P) and Identified (I) Sites | Localization in Protein-Sequence |
|---|---|
| N-terminal signal peptide, ER (I) | −34 to −1 |
| C-terminal propeptide (P) | 305 to 329 |
| Peroxidase proximal heme-ligand signature with invariant H (I) | 168-DVVALSGGHTI |
| Peroxidase active site signature with invariant H and R (I) | 38-AAGLLRLHFHDC |
| Eight conserved Cys (I) establishing four disulfide bridges (P) | 16 and 97, 49 and 54, 103 and 298, 183 and 210 |
| N-glycosylation sites, 6 (P) | 75-NLTL, 151-NTSA, 165-NATDa, 217-NRTF, 266-NQTL, 297-NCSL |
| Glycosaminoglycan attachment sites, 1 (P) | 65-SGPG |
| Protein kinase C phosphorylation sites, 3 (P) | 77-TLR, 252-TDR, 299-SLR |
| Casein kinase II phosphorylation sites, 6 (P) | 102-SCSD, 137-TRAD, 185-SFDE, 219-TFLD, 246-SDQD, 316-SVVE |
| N-myristoylation sites, 4 (P) | 6-GLSYTF, 53-GCDSSV, 132-GLNFAT, 242-GLFTSD |
| Amidation sites, 1 (P) | 127-LGRR |
165 N corresponds to the amino acid not identified in peptide 4 (Table I), which may indicate that this site is indeed glycosylated (prevents peptide sequencing).
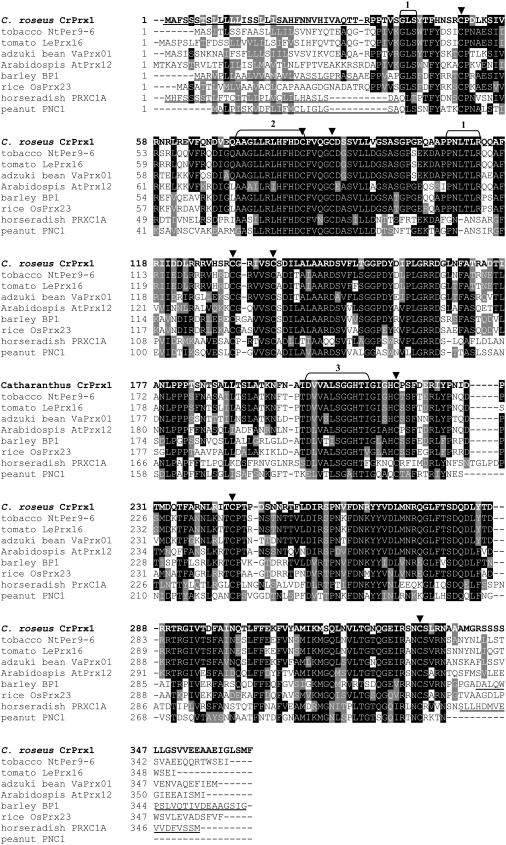
Alignment studies and comparison with consensus peroxidase sequences (Welinder, 1992; Welinder et al., 2002) show that CrPrx1 contains all conserved and highly conserved residues typical of class III peroxidases, including the two His residues interacting with the heme and the eight Cys residues forming four disulfide bridges (Figs. 2B and 4; Table II). The localization of the N-terminal sequence obtained for the purified protein enables the identification of an N-terminal propeptide (NTPP) of 34 amino acids in the full deduced sequence of CrPrx1 (Fig. 2B), putatively corresponding to the endoplasmic reticulum (ER) signal peptide present in all plant peroxidases. Alignment with previously characterized plant peroxidases (Fig. 4) also indicates the presence in CrPrx1 of a C-terminal extension with 23 to 25 amino acids (Fig. 2B).
Figure 4.
Multiple alignment of CrPrx1 with other class III peroxidases. The sequences used include the most similar class III peroxidases (NtPer9-6, LePrx16, and VaPrx01), the most similar class III peroxidases found in Arabidopsis and rice genomes (ApPrx12 and OsPrx23), and the significantly similar and well-characterized barley BP1. The extensively characterized peanut PNC1 and horseradish HrPC (PrxC1A) are also included. Signal peptides and C-terminal propeptides determined experimentally are underlined. Arrowheads, Eight conserved Cys of class II peroxidases. 1, Highly conserved sequences present in all peroxidases of the same phylogenetic branch of CrPrx1 (Fig. 6). 2, Peroxidase active site signature (distal heme-ligand). 3, Peroxidase proximal heme-ligand signature. Accession numbers and percentages of identity with CrPrx1 are: tobacco NtPer9-6, AY032674, 75% identity; tomato LePrx16, TC124085 TIGR, 71% identity; adzuki bean VaPrx01, D11337 NCBI, 66% identity; Arabidopsis AtPrx12, locus At1g71695, 58% identity; barley BP1, TC132499 TIGR, 54% identity; rice OsPrx23, BN000552 NCBI, 53% identity; horseradish PrxC1A, M37156 NCBI, 41% identity; and peanut PNC1, M37636 NCBI, 42% identity (identities determined using the Swiss Institute of Bioinformatics Blast Network Service through Peroxibase, except for PNC1 and PrxC1, which were determined using NCBI Blast 2 sequences). Numbering of sequences starts at first amino acid translated, meaning that CrPrx1 amino acid 1 of Figure 2B corresponds to amino acid 35 in this image due to the 34 amino acids of the signal peptide.
The predicted size of the mature peroxidase is thus 304 amino acids, with a Mr of 33,751. Comparison with the purified protein (Sottomayor et al., 1998) allows the prediction of a carbohydrate moiety with a Mr between 7,000 and 11,000, corresponding to 17% to 24% of the processed protein. This high concentration of sugars may indicate the presence of several glycan chains, as can be predicted from the presence of six putative N-glycosylation sites (Fig. 2B; Table II).
A theoretical three-dimensional (3D) model (Fig. 5) was derived for CrPrx1 using the x-ray crystallography coordinates for the known structure of the homologous barley (Hordeum vulgare) seed peroxidase BP1 (Henriksen et al., 1998) using SWISS-MODEL and the Swiss-Pdb Viewer (Guex and Peitsch, 1997), which are available at www.expasy.ch/spdbv/. The surface charge of CrPrx1 was calculated for the constructed model, indicating the presence of considerable surface areas with positive charge (blue areas in Fig. 5), as expected for a basic protein.
Figure 5.
Hypothetical 3D structural model of CrPrx1. The model was constructed using the x-ray crystallography coordinates for barley peroxidase BP1. Colors represent surface charge: white, neutral; blue, positive; and red, negative. The pore leading to the centrally located heme can be observed on the left. The figure was prepared by homology modeling with barley peroxidase BP1 as the template, using SWISS-MODEL and the Swiss-Pdb Viewer (Guex and Peitsch, 1997), which are available at www.expasy.ch/spdbv/.
Phylogenetic Analysis
BLASTp search analysis of the amino acid sequence deduced for CrPrx1 using class III Peroxibase (Swiss Institute of Bioinformatics Blast Network Service; http://peroxidase.isb-sib.ch) revealed highest homology with a tobacco (Nicotiana tabacum) and a tomato (Lycopersicum esculentum) peroxidase, and with a basic peroxidase from adzuki bean (Vigna angularis) induced by ethylene (Fig. 4). As mentioned above, the Arabidopsis peroxidase protein with the highest identity to CrPrx1 is AtPrx12 (At1g71695). The barley seed basic peroxidase BP1, for which the 3D structure has been determined (Henriksen et al., 1998), also shares significant identity with CrPrx1. Recently, Kumar et al. (2007) characterized another class III peroxidase from C. roseus, but it shares low identity with CrPrx1 (32%) and it has been associated with cell wall synthesis.
Figure 4 shows the multiple alignment of CrPrx1 with identical peroxidases, including the most identical found in Arabidopsis and rice genomes (ApPrx12 and OsPrx23) and the significantly identical barley BP1. The extensively characterized peanut (Arachis hypogaea) PNC1 and horseradish (Armoracia rusticana) HrPC (PrxC1A) were also included. All peroxidases highly similar to CrPrx1 possess a C-terminal extension (Figs. 4 and 2B), which has been found only in vacuolar peroxidases.
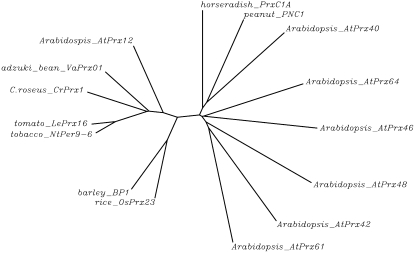
To understand the phylogenetic relations of CrPrx1, an unrooted neighbor-joining phylogenetic tree was constructed relating CrPrx1 with the following peroxidases from Peroxibase: (1) highly identical peroxidases including the most identical present in the genomes of Arabidopsis (AtPrx12) and rice; (2) extensively studied basic peroxidases (barley BP1, horseradish HrPC1, and peanut PNC1); and (3) six more Arabidopsis peroxidases, representing, together with AtPrx12, all seven branches revealed in the phylogenetic tree of Arabidopsis peroxidases constructed by Tognolli et al. (2002). The constructed tree (Fig. 6) shows clearly that CrPrx1 is phylogenetically related to the identical peroxidases, sharing a common ancestor with Arabidopsis AtPrx12, rice OsPrx23, and barley BP1. Together, these cationic peroxidases form an independent branch from all the other Arabidopsis peroxidase groups.
Figure 6.
Unrooted neighbor-joining phylogenetic tree relating CrPrx1 with other class III peroxidases. The following sequences retrieved from Peroxibase were used: (1) highly identical peroxidases including the most identical present in the genomes of Arabidopsis (AtPrx12) and rice; (2) extensively studied basic peroxidases (barley BP1, horseradish HrPC1, and peanut PNC1); and (3) six further Arabidopsis peroxidases, representing, together with AtPrx12, all the seven branches revealed in the phylogenetic tree of Arabidopsis peroxidases constructed by Tognolli et al. (2002). The tree was constructed using the ClustalW multiple sequence alignment tool of GenomeNet of the Kyoto University Bioinformatics Center, using default parameters (http://align.genome.jp/). See accession numbers of represented genes in legend of Figure 4. Nomenclature of Arabidopsis peroxidase genes according to class III Peroxibase (http://peroxidase.isb-sib.ch).
Subcellular Sorting of CrPrx1 in Vivo
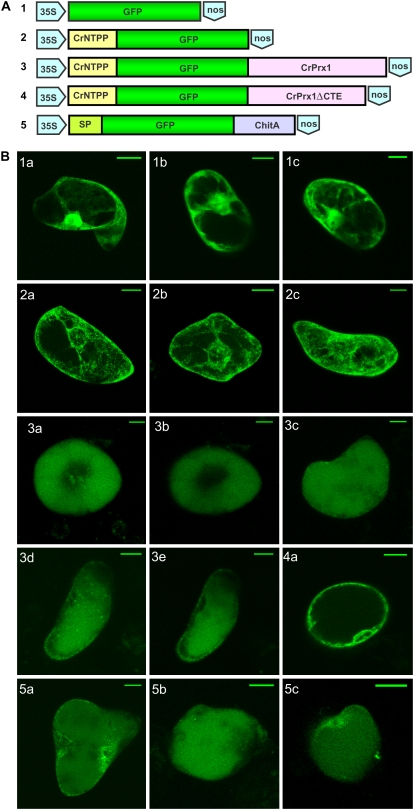
To further characterize the subcellular sorting of CrPrx1, C. roseus suspension culture cells were transformed with GFP-CrPrx1 fusions (Fig. 7A). Constructs were designed to assess the sorting capacity of the CrPrx1 NTPP determined by N-terminal sequencing of the purified CrPrx1 (Fig. 7A, construct 2), and the complementary C-terminal sorting capacity of the rest of the CrPrx1 sequence (Fig. 7A, constructs 3 and 4). Imaging of GFP fluorescence of transformed cells showed that the CrNTPP-GFP fusion resulted in accumulation of GFP fluorescence in the ER (Fig. 7B, 2a–2c), while the CrNTPP-GFP-CrPrx1 fusion resulted in accumulation of the GFP fluorescence in the vacuole (Fig. 7B, 3a–3c). The images observed for this construct were very similar to the ones obtained with the positive control for vacuolar localization of GFP containing the C-terminal propeptide of tobacco chitinase A (Fig. 7B, 5a–5c; Tamura et al., 2003). Exclusion of the C-terminal extension from the CrNTPP-GFP-CrPrx1 fusion resulted in accumulation of GFP fluorescence in the ER and not in the vacuole (Fig. 7B, 4).
Figure 7.
Transient transformation of C. roseus cells with CrPrx1-GFP fusions. A, Scheme of constructs used for transformation. B, Typical GFP fluorescence pattern observed for transformation with each construct. 1a to 1c, 35S∷GFP, control for GFP localization. GFP accumulates in the cytoplasm and nucleus. 2a to 2c, 35S∷CrNTPP-GFP, construct with the NTPP of CrPrx1 (CrNTPP). GFP accumulates in the ER and possibly Golgi. 3a to 3e, 35S∷CrNTPP-GFP-CrPrx1, CrPrx1 does not include the NTPP sequence. GFP accumulates in the central vacuole. 4a, 35S∷CrNTPP-GFP-CrPrx1ΔCTE, the same as 3 but excluding the sequence of the C-terminal extension. GFP accumulates in the ER and possibly Golgi. 5a to 5c, 35S∷SP-GFP-ChitA, positive control for vacuolar localization of GFP, with the signal peptide from pumpkin 2S albumin (SP in image) and the C-terminal propeptide of tobacco chitinase A (ChitA in image), a confirmed vacuolar sorting signal (Tamura et al., 2003). GFP accumulates in the vacuole. Bar = 20 μm.
CrPrx1 and Alkaloid Metabolism
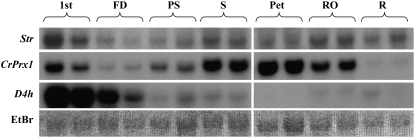
Expression of the CrPrx1 mRNA was observed early during plant development, occurring at least since day 6 after germination (data not shown). In mature flowering plants, CrPrx1 transcripts were detected in all aerial organs of the plant (Fig. 8) but were absent or with low levels in roots.
Figure 8.
mRNA accumulation of CrPrx1, Str, and D4h in different organs and leaf developmental stages of C. roseus. Str codes for an enzyme of the beginning of the indole alkaloid pathway and D4h codes for an enzyme of a later part of the indole alkaloid pathway, near the dimerization step. 1st, First pair of leaves; FD, fully developed leaves; PS, presenescent leaves; S, senescent leaves; Pet, petals; RO, reproductive organs; R, roots. For identification of developmental stages, see Figure 9A. Duplicate results are presented.
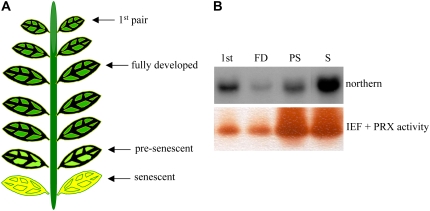
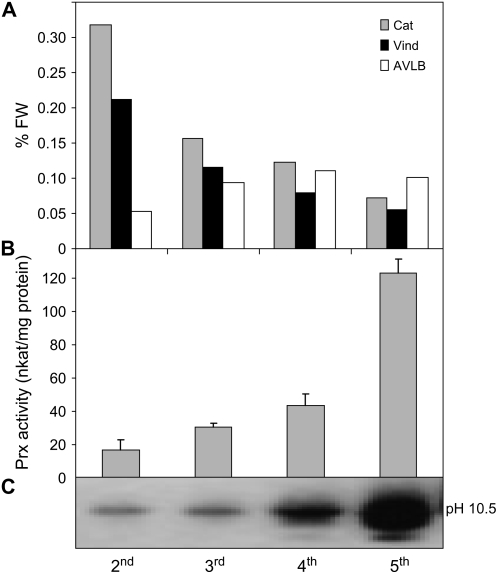
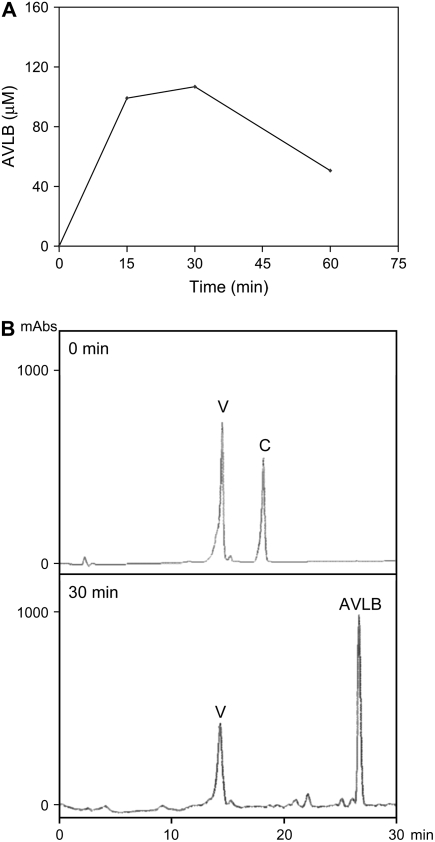
The expression of CrPrx1 in leaves of mature flowering plants follows similar patterns to other genes of the TIA pathway (Fig. 8), with higher expression in very young leaves, decreasing in fully expanded leaves, and eventually increasing again during senescence. The protein/activity levels of CrPrx1 in the same leaf samples were assayed and follow a pattern similar to the respective transcript levels (Fig. 9), indicating prevalence of transcriptional regulation in the determination of the amount of functional CrPrx1. Characterization of alkaloid levels in these samples was also tried, but the levels of individual alkaloids were extremely variable between different samples and did not allow us to obtain coherent results. In view of this heterogeneity, we decided to analyze individual leaves of individual young plants, where alkaloid levels are usually higher. The results showed that peroxidase activity consistently increases with leaf age, in parallel with an increase in AVLB levels and a decrease in the levels of the direct precursors catharanthine and vindoline (Fig. 10, A and B). Peroxidase activity was identified through isoelectric focusing (IEF) as being CrPrx1 (Fig. 10C). In the older leaves, CrPrx1 shows a dramatic increase associated with a decrease of catharanthine and vindoline as before, but this time AVLB also decreases slightly. This may be explained by the fact that AVLB seems to be further oxidized by CrPrx1: a time course of the in vitro synthesis of AVLB from catharanthine and vindoline using purified CrPrx1 shows that AVLB levels reach a peak and decrease subsequently for long incubation times (Fig. 11).
Figure 9.
A, Scheme illustrating leaf developmental stages. B, Regulation of CrPrx1 gene expression and CrPrx1 activity levels in different leaf developmental stages. C. roseus plants were 1 year old. [See online article for color version of this figure.]
Figure 10.
Accumulation of alkaloids and CrPrx1 at different leaf developmental stages. A, Levels of catharanthine, vindoline, and AVLB from the different leaf pairs of a single plant. The pattern is representative of four different plants analyzed. B, Activity of CrPrx1 assumed from total peroxidase activity of leaf protein extracts (mean of four plants). C, CrPrx1 band in IEF gels stained for peroxidase activity. Leaf pairs are numbered from the shoot tip. C. roseus plants were 16 weeks old.
Figure 11.
A, Time course of the dimerization reaction catalyzed by CrPrx1. The reaction mixture contained 2 μm CrPrx1, 300 μm vindoline, 400 μm catharanthine, and 330 μm H2O2 in 0.1 m MES, pH 6.8. B, HPLC chromatograms of the substrates of the dimerization reaction (0 min) and of the time point corresponding to a maximum yield of the dimerization reaction (30 min). V, Vindoline; C, catharanthine.
DISCUSSION
Here, we have cloned and characterized the major class III peroxidase present in the leaves of the medicinal plant C. roseus, CrPrx1, which we propose to be involved in the biosynthesis of important anticancer alkaloids.
CrPrx1 is encoded by a single copy gene that includes two introns with sizes of 828 bp (intron I) and approximately 3.5 to 4 kb (intron II; Fig. 2A). The size of the full-length CrPrx1 mRNA fluctuates from 1,216 to 1,370 bp due to the presence of various polyadenylation sites, a feature reported for several genes of plants and other organisms (Proudfoot, 1996).
The CrPrx1 protein is translated as a polypeptide with 363 amino acids containing all conserved and highly conserved residues typical of class III peroxidases (Fig. 2B; Table II). The mature CrPrx1 is predicted to have approximately 304 amino acids after the removal of the NTPP and of the putative C-terminal propeptide (Fig. 2B). The surface charge calculated through 3D modeling indicates the presence of extensive surface areas with positive charge (Fig. 5), in accordance with the rather basic pI of 10.5 previously determined for CrPrx1 (Sottomayor et al., 1998). CrPrx1 shows an N terminus that starts with two Pro residues, the second being invariant in all the peroxidases belonging to the same evolutionary branch (Fig. 4), a feature that has been suggested to protect against proteolysis (Welinder et al., 2002). The motifs GLS and PNLTLR (1 in Fig. 4) are also invariant in the peroxidases of the phylogenetic branch of CrPrx1 and, because they are recognized by the Prosite database as an N-myristoylation motive (GLS) and as an N-glycosylation site (NLTL) or a protein kinase C phosphorylation site (TLR), it is probable that these sequences are important for the processing or regulation of the CrPrx1 branch of peroxidases.
The carbohydrate moiety of CrPrx1 was estimated to account for 17% to 24% of the total molecular mass, near the maximum reported for plant peroxidases (Welinder, 1992).
What Can We Learn from Phylogenetic and Expression Analyses?
Phylogenetic analysis shows that CrPrx1 clusters with tobacco NtPer9-6, tomato LePrx16, adzuki bean VaPrx01, rice OsPrx23, barley BP1, and Arabidopsis AtPrx12, indicating that they share a common ancestor (Fig. 6). Together, these cationic peroxidases form an independent branch from all the other Arabidopsis peroxidase groups, suggesting that the ancestral peroxidase gene has appeared previously to divergence of monocots (rice and barley) and dicots (all the others in the tree). Duroux and Welinder (2003), who made a phylogenetic overview of the peroxidase gene family in plants, actually conclude that the peroxidase lineage corresponding to AtPrx12 (called AtP4 in their article) likely preceded even the divergence between gymnosperms and angiosperms.
Within a large gene family, genes from different species sharing a common ancestor are called orthologous genes, and it is generally considered that they encode proteins with the same function. However, this might not be the case in the class III peroxidase gene family. In fact, the expression pattern recorded for some of the genes that undoubtedly share a close common ancestor with CrPrx1 point to a different conclusion: (1) the mRNA of the adzuki bean VaPrx01 is not constitutively present in leaves and appears only upon wounding or after treatment with ethylene or salicylate (Ishige et al., 1993); (2) barley BP1 is specifically expressed in seeds (Rasmussen et al., 1991); and (3) AtPrx12 expression was not detected by northern blot in flowers, stems, leaves, or roots of Arabidopsis (Tognolli et al., 2002), although it was possible to detect the presence of AtPrx12 transcripts in all organs by the more sensitive reverse transcription-PCR technique (Welinder et al., 2002). All these expression patterns are different from the one observed for the CrPrx1 gene, which encodes the major peroxidase present in C. roseus leaves, and which is expressed everywhere in the plant except in the root, with increased expression in very young and senescent leaves. These expression profiles seem to point to different functions for presumed peroxidase orthologs in different plant species. In Arabidopsis, the main vacuolar peroxidase present in leaves, putatively equivalent to CrPrx1, is AtPrx34 (At3g49120; Zimmermann et al., 2004). This Arabidopsis peroxidase protein is related to AtPrx40 from Figure 6 and does not belong to the same phylogenetic branch of CrPrx1. Taken together, these data suggest that multiplication of peroxidase genes into the different groups present in Arabidopsis has preceded the recruitment of those genes for their present day functions. This agrees with the concept of class III peroxidases as enzymes with high metabolic plasticity, showing largely overlapping reactivity properties, and thus being replaceable during evolution.
Subcellular Sorting and Function of CrPrx1
TIAs are known to accumulate in vacuoles where at least one of the key biosynthetic steps of the pathway has also been localized (Sottomayor and Barceló, 2003; Sottomayor and Ros Barceló, 2005). CrPrx1 was proposed as the enzyme responsible for the synthesis of AVLB because it possesses the single dimerization activity detected in extracts of C. roseus leaves, and biochemical and cytochemical evidence pointed to a vacuolar localization of the enzyme (Sottomayor et al., 1996, 1998; Sottomayor and Ros Barceló, 2003). Here, we have further confirmed the vacuolar nature of CrPrx1, since the fusion CrNTPP-GFP-CrPrx1 clearly resulted in accumulation of green fluorescence in the central vacuole of C. roseus cells, in a manner absolutely identical to the localization observed with a vacuolar targeted GFP control (Fig. 7). This direct proof of the vacuolar localization of CrPrx1 further reinforces its possible involvement in TIA metabolism.
For the investigation of the vacuolar sorting of CrPrx1 using GFP fusions, we first checked the intracellular fate of the fusion CrNTPP-GFP, which resulted in ER accumulation of green fluorescence. This demonstrated that the NTPP identified in CrPrx1 determines the entrance in the secretory system and thus corresponds to an ER signal peptide, as generally assumed for class III peroxidases. The fusion ultimately used to demonstrate the vacuolar sorting included the CrNTPP located at the N terminus of GFP to determine ER entrance, whereas the remainder of CrPrx1 was located at the C terminus of GFP giving CrNTPP-GFP-CrPrx1. This positioning was chosen since it has been postulated that the sorting of vacuolar class III peroxidases is determined by a C-terminal propeptide (Tognolli et al., 2002; Welinder et al., 2002). In fact, all characterized mature peroxidases terminate six to eight residues after the last conserved Cys residue (Fig. 4) and the presence of a C-terminal extension has been interpreted as a putative propeptide with vacuolar sorting capacity. This assumption is due to the fact that, for the extensively characterized vacuolar class III peroxidases BP1, HRPC1A, and HRPE5, this C-terminal extension has been proven to be a propeptide absent from the mature protein (Johansson et al., 1992; Welinder et al., 2002). CrPrx1 also contains this C-terminal extension (Fig. 4) and the results shown here demonstrate that the CrPrx1 polypeptide sequence, when localized in the C terminus of the GFP fusion protein, determines its sorting to the vacuole (Fig. 7B, 3a). Furthermore, exclusion of the CrPrx1 C-terminal extension from the fusion protein prevents sorting to the vacuole, since fluorescence is observed only in the ER (Fig. 7B, 4). This result strongly indicates that the vacuolar sorting information is indeed localized in the C-terminal extension.
CrPrx1 is capable of performing the dimerization of catharanthine and vindoline into AVLB (Fig. 11), and different experimental approaches converge to the conclusion of vacuolar localization of CrPrx1, the exact intracellular location of the substrates and products of the dimerization reaction. All this strongly supports a role of CrPrx1 as the dimerization enzyme in planta. We investigated the expression of CrPrx1 and observed that it correlated with the expression of other genes involved in the TIA pathway, possibly indicating common transcriptional regulation. On the other hand, it was difficult to observe a consistent correlation between the levels of CrPrx1 and the levels of AVLB in mature flowering plants. In fact, although the expression pattern of CrPrx1 was constant in different plants, the alkaloid levels, especially of catharanthine, vindoline, and AVLB, were extremely variable from plant to plant. We could see, however, there was a certain tendency for an increase in AVLB with the age of the leaves, apparently at the expense of a decrease in catharanthine and vindoline, a fact that could be explained by the observed increase of CrPrx1 with leaf age. In fact, analysis of young plants, where alkaloid levels are usually higher, enabled the observation of a clear correlation between the AVLB and CrPrx1 levels (Fig. 10). The alkaloid levels were still variable between plants but there was a constant pattern that was the same in all the plants tested. In any case, it is not surprising that the levels of AVLB do not reflect exactly the levels of CrPrx1 because the dimerization reaction mediated by the basic peroxidase is also dependent on the availability of H2O2, a molecule that is under a complex regulation (Foyer and Noctor, 2005). It is interesting to note that very high levels of CrPrx1 activity as observed in older/senescent leaves can actually be associated with a decrease in AVLB levels, which seems to result from further oxidation of AVLB by CrPrx1 (Fig. 11A). The TIA pathway is thus dependent on a complex network of factors subjected to different regulation programs and does not necessarily match the regulation of a single enzyme.
In conclusion, we have characterized the major class III peroxidase present in the leaves of the medicinal plant C. roseus, CrPrx1. Phylogenetic analysis indicates that CrPrx1 belongs to an evolutionary branch of vacuolar class III peroxidases that likely preceded the divergence between monocots and dicots, and whose members seem to have been recruited for different functions during evolution. CrPrx1 includes an ER signal peptide determining entrance to the secretory pathway and contains further sorting information leading the protein to the vacuole. Results obtained add extra evidence supporting a role of CrPrx1 in the vacuolar dimerization of the indole alkaloids vindoline and catharanthine into the direct precursor of the anticancer indole alkaloids vinblastine and vincristine, indicating the potential of CrPrx1 as a target to increase alkaloid levels in the plant.
MATERIALS AND METHODS
Plant Material
Plants of Catharanthus roseus (‘Little Bright Eye’) were grown at 25°C in a growth chamber under a 16-h white fluorescent light photoperiod at 100 μmol m−2 s−1.
IEF of CrPrx1 in C. roseus Leaf Extracts
IEF of protein extracts followed by staining of peroxidase activity with 4-methoxy-α-naphthol was performed as previously reported (Sottomayor et al., 1996, 1998).
Enzyme Purification
Enzyme purification was carried out using a six-step protocol involving protein precipitation during homogenization in 75% (v/v) acetone at −20°C, followed by ammonium sulfate precipitation and four chromatographic steps, as described before (Sottomayor et al., 1998).
Amino Acid Sequence Analysis and Tryptic Digestion
Amino acid sequencing was carried out by Dr. M.J. Naldrett at the Protein Sequencing Facility of the John Innes Center, Nitrogen Fixation Laboratory. Briefly, 200 pmol of purified protein were analyzed by SDS-PAGE, electroblotted onto a polyvinylidene difluoride membrane (PVDF; Millipore), and stained with Coomassie Blue R250. The membrane piece carrying the protein was used directly for automatic Edman degradation of the N terminus on a Procise protein sequencer (Applied Biosystems).
For trypsin digestion, the heme was removed from 200 pmol of pure protein and the apoenzyme was used for digestion (Kvaratskhelia et al., 1997). Peptides resulting from digestion were separated and sequenced by automatic Edman degradation as described above.
The pure protein was further submitted to peptide mass fingerprint (MALDI-TOF) after trypsin digestion, followed by peptide sequencing/fragmentation (MALDI-TOF/TOF) of peptides different from the ones previously sequenced by Edman degradation (IPATIMUP Proteomics Unit; Applied Biosystems).
Construction of cDNA Library
Total RNA was isolated from the first pair of leaves (counting from the shoot apical meristem) of 60-cm-high flowering plants of C. roseus. Poly(A)+ mRNA was isolated using a Promega kit. The cDNA was synthesized and inserted into λ-ZAP using the ZAP-cDNA kit from Stratagene. A mass excision of an aliquot of the library was performed according to manufacturer's instructions.
Isolation of a Full-Length cDNA Clone Coding for CrPrx1
To isolate the CrPrx1 cDNA, degenerate primers were designed using the information from the amino acid sequence of peptide 2 (Table I). This peptide was selected because of its low similarity with other peroxidase sequences and because of the low degeneracy of the corresponding codon triplets. Four primers were designed from this sequence: AVLB1, AVLB2, AVLB3, and AVLB4 (TAYCCIAAYATHGAYCCIACIATG, AAYATHGAYCCIACIATGGAYC, GTYTGRTCCATIGTIGGRTC, and TCCATIGTIGGRTCDATRTTIGG, respectively, where I is inosine, Y = C + T, R = A + G, D = G + A + T, and H = A + T + C). Seminested PCR amplification of the cDNA library using primers AVLB1 + T7 for the first reaction, followed by primers AVLB2 + T7 for the second reaction, produced a band of 250 bp (fragment 58-2) containing a part of the sequence of the AVLB primers. Seminested PCR amplification of a mass excision of the cDNA library using primers AVLB3 + Rev for the first reaction, followed by primers AVLB4 + Rev for the second reaction, produced a band of 650 bp (fragment 62-6) containing a part of the sequence of the AVLB primers. The deduced amino acid sequence of this band contained the sequence of peptide 1 (Table I).
The design of new primers and two rounds of PCR reactions enabled the very specific amplification of two segments of the cDNA, including sequences encoding the peptide fragments (first PCR reaction, 62-6sense + Rev; second PCR reaction, PERON1 + 62-6sense and 58-2sense + 58-2reverse, in which 58-2reverse = CAATTGGATCTAATTTCAGG, 58-2sense = GGACATTAGATCACCAAACG, 62-6sense = GATTCTTTCGTCGAAAGAAG, and PERON1 = CGGCCACCAACAGTGAGTGG). These products were cloned into pBluescript SK II+, sequenced to confirm their identity, and used to screen the cDNA library.
Three rounds of screening of approximately 750,000 plaques enabled the selection of 13 positive clones. The sequence of CrPrx1 was reconstructed from the overlapping sequences obtained from clones 3, 5, 7, and 11, to produce an incomplete cDNA sequence.
The complete sequence was obtained by RACE (RACE system; GibcoBRL). This was done using two primers close to the 5′ end of the incomplete cDNA together with reverse transcriptase, using poly(A) RNA as template. A fragment containing the start codon AUG was isolated and used to complete the cDNA. PCR amplification of 3′ ends of the CrPrx1 present in the cDNA library was performed and the presence of three differently sized cDNAs varying in the site of polyadenylation was detected (arrowheads in Fig. 2B).
IPCR
To obtain the promoter region of CrPrx1, a modification of an IPCR method (Ochman et al., 1988; Triglia et al., 1988) was performed as follows: TaqI- or Sau3AI-digested plant DNA (0.2 μg) was diluted to 1 ng/μL in T4 DNA ligase buffer (New England Biolabs), incubated at 65°C for 5 min, and then placed on ice for another 5 min. T4 DNA ligase was added (2 units) and the reaction mixture incubated for 16 h at 4°C. DNA was then ethanol-precipitated and finally resuspended in 20 μL of DNase-free ddH2O. PCR was subsequently performed on 7.5 μL of the ligated DNA preparation using an enzyme mixture of the DNA polymerases with proofreading activity Taq and Tgo, according to the manufacturer's instructions (Expand Long Template PCR System; Roche Molecular Biochemicals). Forward and reverse primers were 367-CAGAAATCACTATGCAAACATGC and 146-GGCCGAGTTGTTTGAGCTAC, respectively (forward primer was specific for a sequence in the first intron).
Intron Analysis
The CrPrx1 cDNA sequence was aligned with the sequence of AtPrx12 using NCBI BLAST 2 SEQUENCES (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html), the putative sites of two introns were determined, and primers flanking those sites were designed. For intron I, forward and reverse primers were CCACCAACAGTGGACTT and ATGATCCTGAAAGCCTGCTG, respectively. PCR of this region was performed using routine PCR protocols. Amplification products containing intron I were electrophoresed, extracted from 1% agarose gels, and sequenced. For the region containing intron II, due to difficulties found in amplifying this region, a collection of different primers was tested and long-PCR protocols were employed, including extension times up to 6 min per cycle. PCR experiments were performed in both cases with the Expand Long Template PCR system.
Southern Blot
DNA extracted from C. roseus radicles was digested overnight with the restriction enzymes NcoI, Van91I and EcoRI, and HincII. Electrophoresis was performed in agarose gels loaded with 5 μg of digested DNA/lane. A DIG Easy Hyb system was used, and all gel, transfer, and filter procedures were based on the DIG application manual for filter hybridization from Roche Molecular Biochemicals. DNA was fixed to the nylon membranes by UV cross-linking. Membranes were prehybridized for 30 min with DIG Easy Hyb at 42°C, and then hybridized with DIG Easy Hyb containing the oligonucleotide probe INT1.39.antisense, CCAGGATTTACTAAGTGCATGTTTGCATAGTGATTTCTG, labeled with DIG (MWG Biotech) at its 3′-end, at 5 pmol/mL, for 3 h at 42 °C. Probe-target hybrid molecules were detected using an alkaline phosphatase-conjugated anti-DIG antibody (1:10,000) and a chemiluminescent substrate.
Construction of GFP Fusion Proteins and Expression in C. roseus Cells
The constructs below were designed to study the subcellular localization of CrPrx1. The CrPrx1 sequences used in fusions were amplified by PCR with primers including the required restriction sites. The amplified CrPrx1 NTPP was cloned in frame in pTH-2 (Chiu et al., 1996; Niwa et al., 1999) with SalI/NcoI to generate the construct 35S-CrNTPP-GFP, and in pTH2-BN (a derivative lacking the GFP stop codon). The latter construct was used to further insert the amplified CrPrx1 sequence (excluding the NTPP) with BglII/XhoI to generate the construct 35S-CrNTPP-GFP-CrPrx1. The sequences of the individual primers are as follows (engineered restriction sites are underlined): NTPP, 5′-ACGCGTCGACAAAATGGCTTTTTCTTCTTCAACTTCTCTGC-3′ and 5′-CATGCCATGGGCCGAGTTGTTTGAGCTACAATATGG-3′; and CrPrx1 (excluding the NTPP), 5′-GAAGATCTTACCACCAACAGTGAGTGGACTTTC-3′ and 5′-CCGCTCGAGTTAAAACATAGACAAGCCAATTTCAGC-3′.
C. roseus cell line MP183L was grown in LS medium (Linsmaier and Skoog, 1965) with 3% Suc, 2 mg/L 1-naphthaleneacetic acid, and 0.2 mg/L kinetin at 25°C (16:8 light/dark cycle) and subcultured every 7 d by transfer of 5 to 7.5 mL of cell culture into 50 mL of fresh medium. For transformation, about 6 mL of cells from a 4-d-old culture were collected on a paper filter disc (Ø 42.5 mm, Whatman No. 4) and placed onto medium solidified with 0.7% plant tissue culture agar. Filters with cells were bombarded using a homemade helium gun and tungsten particles (1.8 μm) coated with 10 μg of plasmid DNA according to van der Fits and Memelink (1997). The filters with bombarded C. roseus cells were incubated for 2 d on the solidified medium, in the dark, at 25°C, before observation (the dark treatment is essential for observation of GFP fluorescence in the vacuole; Tamura et al., 2003). For GFP analysis, cells were placed on slides in a drop of water and GFP fluorescence was examined using an Axioplan upright microscope (Zeiss) equipped with a Bio-Rad MRC1024ES scanhead with a krypton/argon laser. For visualization of GFP the excitation wavelength was 488 nm and the emission wavelength was 522 nm ± 16 nm.
Northern Blot
RNA extraction and northern-blot analysis were performed as described before (Menke et al., 1999), loading 20-μg RNA samples onto the gels. Hybridization was performed with 32P-labeled DNA probes. For CrPrx1, a 360-bp fragment including part of the C-terminal propeptide and part of the 3′ UTR was used as a probe. DNA probes for strictosidine synthase (Str) and desacetoxyvindoline 4-hydroxylase (D4h) have been described previously (Menke et al., 1999; van der Fits and Memelink, 2000).
Extraction and Quantification of Alkaloids
Alkaloids of each leaf were extracted and analyzed as described in Sottomayor and Ros Barceló (2003).
Determination of AVLB Synthase Activity
The assay of this enzymatic activity was performed as described in Sottomayor and Ros Barceló (2003).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: CrPrx1, AM236087 (mRNA), AM236088 (promoter region), and AM236089 (intron I); tobacco NtPer9-6, AY032674; tomato LePrx16, TC124085 TIGR; adzuki bean VaPrx01, D11337 NCBI; Arabidopsis AtPrx12, locus At1g71695; barley BP1, TC132499 TIGR; rice OsPrx23, BN000552 NCBI; horseradish PrxC1A, M37156 NCBI; and peanut PNC1, M37636 NCBI.
Acknowledgments
The authors would like to thank Dr. Mike Naldrett (Biological Chemistry Department, John Innes Centre, UK) for the excellent work in N-terminal sequencing of the protein and digestion peptides, Dr. Patrice Simon (Laboratoire de Biochimie et Physiologie Végétales, Université de Genève, Switzerland) for helpful discussions and for the 3D prediction model of CrPrx1, and Dr. Pieter Ouwerkerk (Institute of Biology, Leiden University, The Netherlands) for providing the pTH2-BN expression vector.
This work was supported by grants from Fundação para a Ciência e Tecnologia, Portugal (PRAXIS/P/BIA/10267/1998 and POCTI/BIO/38369/2001) and Fundación Séneca, Spain (project no. 00545/PI/04).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Mariana Sottomayor (msottoma@ibmc.up.pt).
Some figures in this article are displayed in color online but in black and white in the print edition.
References
- Barceló AR, Gómez Ros LV, Gabaldón C, López-Serrano M, Pomar F, Carrión JS, Pedreño MA (2004) Basic peroxidases: the gateway for lignin evolution? Phytochem Rev 3 61–78 [Google Scholar]
- Bernards MA, Summerhurst DK, Razem FA (2004) Oxidases, peroxidases and hydrogen peroxide: the suberin connection. Phytochem Rev 3 113–126 [Google Scholar]
- Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6 325–330 [DOI] [PubMed] [Google Scholar]
- Díaz J, Pomar F, Bernal A, Merino F (2004) Peroxidases and the metabolism of capsaicin in Capsicum annuum L. Phytochem Rev 3 141–157 [Google Scholar]
- Duroux L, Welinder KG (2003) The peroxidase gene family in plants: a phylogenetic overview. J Mol Evol 57 397–407 [DOI] [PubMed] [Google Scholar]
- El-Sayed M, Verpoorte R (2007) Catharanthus terpenoid indole alkaloids: biosynthesis and regulation. Phytochem Rev 6 277–305 [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5 199–206 [DOI] [PubMed] [Google Scholar]
- Foyer C, Noctor G (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC (2004) Oxidative coupling of tyrosine and ferulic acid residues: intra- and extra-protoplasmic occurrence, predominance of trimers and larger products, and possible role in inter-polymeric cross-linking. Phytochem Rev 3 97–111 [Google Scholar]
- Gabaldón C, López-Serrano M, Pedreño MA, Ros Barceló A (2005) Cloning and molecular characterization of the basic peroxidase isoenzyme from Zinnia elegans, an enzyme involved in lignin biosynthesis. Plant Physiol 139 1138–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-Pdb viewer: an environment for comparative protein modeling. Electrophoresis 18 2714–2723 [DOI] [PubMed] [Google Scholar]
- Henriksen A, Welinder KG, Gajhede M (1998) Structure of barley grain peroxidase refined at 1.9-angstrom resolution. A plant peroxidase reversibly inactivated at neutral pH. J Biol Chem 273 2241–2248 [DOI] [PubMed] [Google Scholar]
- Hilliou F, van der Fits L, Memelink J (2001) Molecular regulation of monoterpenoid indole alkaloid biosynthesis. In JT Romeo, JA Saunders, BF Matthews, eds, Regulation of Phytochemicals by Molecular Techniques, Vol 35. Elsevier Science, Amsterdam, pp 275–295
- Ishige F, Mori H, Yamazaki K, Imaseki H (1993) Identification of a basic glycoprotein induced by ethylene in primary leaves of azuki-bean as a cationic peroxidase. Plant Physiol 101 193–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A, Rasmussen SK, Harthill JE, Welinder KG (1992) cDNA, Amino-acid and carbohydrate sequence of barley seed-specific peroxidase BP-1. Plant Mol Biol 18 1151–1161 [DOI] [PubMed] [Google Scholar]
- Kristensen BK, Burhenne K, Rasmussen SK (2004) Peroxidases and the metabolism of hydroxycinnamic acid amides in Poaceae. Phytochem Rev 3 127–140 [Google Scholar]
- Kumar S, Dutta A, Sinha AK, Sen J (2007) Cloning, characterization and localization of a novel basic peroxidase gene from Catharanthus roseus. FEBS J 274 1290–1303 [DOI] [PubMed] [Google Scholar]
- Kvaratskhelia M, Winkel C, Thorneley RNF (1997) Purification and characterization of a novel class III peroxidase isoenzyme from tea leaves. Plant Physiol 114 1237–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsmaier EM, Skoog F (1965) Organic growth factor requirements of tobacco tissue cultures. Physiol Plant 18 100–127 [Google Scholar]
- Loyola-Vargas VM, Galaz-Ávalos RM, Kú-Cauich R (2007) Catharanthus biosynthetic enzymes: the road ahead. Phytochem Rev 6 307–339 [Google Scholar]
- Memelink J, Gantet P (2007) Transcription factors involved in terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Phytochem Rev 6 353–362 [Google Scholar]
- Menke FL, Parchmann S, Mueller MJ, Kijne JW, Memelink J (1999) Involvement of the octadecanoid pathway and protein phosphorylation in fungal elicitor-induced expression of terpenoid indole alkaloid biosynthetic genes in Catharanthus roseus. Plant Physiol 119 1289–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika A, Minibayeva F, Beckett R, Lüthje S (2004) Possible functions of extracellular peroxidases in stress-induced generation and detoxification of active oxygen species. Phytochem Rev 3 173–193 [Google Scholar]
- Niwa Y, Hirano T, Yoshimoto K, Shimizu M, Kobayashi H (1999) Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J 18 455–463 [DOI] [PubMed] [Google Scholar]
- Noble RL (1990) The discovery of the vinca alkaloids—chemotherapeutic agents against cancer. Biochem Cell Biol 68 1344–1351 [PubMed] [Google Scholar]
- Ochman H, Gerber AS, Hartl DL (1988) Genetic applications of an inverse polymerase chain-reaction. Genetics 120 621–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi F, Cosio C, Penel C, Dunand C (2005) Peroxidases have more functions than a Swiss army knife. Plant Cell Rep 24 255–265 [DOI] [PubMed] [Google Scholar]
- Passardi F, Longet D, Penel C, Dunand C (2004) The class III peroxidase multigenic in land plants family in rice and its evolution. Phytochemistry 65 1879–1893 [DOI] [PubMed] [Google Scholar]
- Passardi F, Tognolli M, De Meyer M, Penel C, Dunand C (2006) Two cell wall associated peroxidases from Arabidopsis influence root elongation. Planta 223 965–974 [DOI] [PubMed] [Google Scholar]
- Proudfoot N (1996) Ending the message is not so simple. Cell 87 779–781 [DOI] [PubMed] [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, et al (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem Rev 3 29 [Google Scholar]
- Rasmussen SK, Welinder KG, Hejgaard J (1991) cDNA cloning, characterization and expression of an endosperm-specific barley peroxidase. Plant Mol Biol 16 317–327 [DOI] [PubMed] [Google Scholar]
- Sang S, Yang CS, Ho C-T (2004) Peroxidase-mediated oxidation of catechins. Phytochem Rev 3 229–241 [Google Scholar]
- Sottomayor M, Barceló AR (2003) How to address the function of a member of a prolific gene family encoding a multifunctional enzyme? The case of a Catharanthus roseus class III peroxidase involved in alkaloid biosynthesis. In Proceedings of the XII International Congress on Genes, Gene Families, and Isozymes. Medimond Publishing, Bologna, Italy, pp 217–221
- Sottomayor M, De Pinto MC, Salema R, DiCosmo F, Pedreño MA, Ros Barcelo A (1996) The vacuolar localization of a basic peroxidase isoenzyme responsible for the synthesis of α-3′,4′-anhydrovinblastine in Catharanthus roseus (L.) G. Don leaves. Plant Cell Environ 19 761–767 [Google Scholar]
- Sottomayor M, Lopes Cardoso I, Pereira LG, Ros Barceló A (2004) Peroxidase and the biosynthesis of terpenoid indole alkaloids in the medicinal plant Catharanthus roseus (L.) G. Don. Phytochem Rev 3 159–171 [Google Scholar]
- Sottomayor M, Lopez-Serrano M, DiCosmo F, Ros Barceló A (1998) Purification and characterization of alpha-3′,4′-anhydrovinblastine synthase (peroxidase-like) from Catharanthus roseus (L.) G. Don. FEBS Lett 428 299–303 [DOI] [PubMed] [Google Scholar]
- Sottomayor M, Ros Barceló A (2003) Peroxidase from Catharanthus roseus (L.) G. Don and the biosynthesis of alpha-3′,4′-anhydrovinblastine: a specific role for a multifunctional enzyme. Protoplasma 222 97–105 [DOI] [PubMed] [Google Scholar]
- Sottomayor M, Ros Barceló A (2005) The vinca alkaloids: from biosynthesis and accumulation in plant cells, to uptake, activity and metabolism in animal cells. In Atta-ur-Rahman, ed, Studies in Natural Products Chemistry (Bioactive Natural Products), Vol 33. Elsevier Science, Amsterdam, pp 813–857
- Takahama U (2004) Oxidation of vacuolar and apoplastic phenolic substrates by peroxidase: physiological significance of the oxidation reactions. Phytochem Rev 3 207–219 [Google Scholar]
- Tamura K, Shimada T, Ono E, Tanaka Y, Nagatani A, Higashi S, Watanabe M, Nishimura M, Hara-Nishimura I (2003) Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J 35 545–555 [DOI] [PubMed] [Google Scholar]
- Tognolli M, Penel C, Greppin H, Simon P (2002) Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288 129–138 [DOI] [PubMed] [Google Scholar]
- Triglia T, Peterson MG, Kemp DJ (1988) A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res 16 8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L, Memelink J (1997) Comparison of the activities of CaMV 35S and FMV 34S promoter derivatives in Catharanthus roseus cells transiently and stably transformed by particle bombardment. Plant Mol Biol 33 943–946 [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J (2000) ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289 295–297 [DOI] [PubMed] [Google Scholar]
- van der Heijden R, Jacobs DI, Snoeijer W, Hallard D, Verpoorte R (2004) The Catharanthus alkaloids: pharmacognosy and biotechnology. Curr Med Chem 11 607–628 [DOI] [PubMed] [Google Scholar]
- Verpoorte R, Memelink J (2002) Engineering secondary metabolite production in plants. Curr Opin Biotechnol 13 181–187 [DOI] [PubMed] [Google Scholar]
- Welinder KG (1992) Superfamily of plant, fungal and bacterial peroxidases. Curr Opin Struct Biol 2 388–393 [Google Scholar]
- Welinder KG, Justesen AF, Kjaersgard IV, Jensen RB, Rasmussen SK, Jespersen HM, Duroux L (2002) Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur J Biochem 269 6063–6081 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Kaothien P, Matsui T, Kawaoka A, Shinmyo A (2003) Molecular biology and application of plant peroxidase genes. Appl Microbiol Biotechnol 60 665–670 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]