Abstract
The kinase-associated protein phosphatase (KAPP) is a regulator of the receptor-like kinase (RLK) signaling pathway. Loss-of-function mutations rag1-1 (root attenuated growth1-1) and rag1-2, in the locus encoding KAPP, cause NaCl hypersensitivity in Arabidopsis thaliana. The NaCl hypersensitive phenotype exhibited by rag1 seedlings includes reduced shoot and primary root growth, root tip swelling, and increased lateral root formation. The phenotype exhibited by rag1-1 seedlings is associated with a specific response to Na+ toxicity. The sensitivity to Na+ is Ca2+ independent and is not due to altered intracellular K+/Na+. Analysis of the genetic interaction between rag1-1 and salt overly sensitive1 (sos1-14) revealed that KAPP is not a component of the SOS signal transduction pathway, the only Na+ homeostasis signaling pathway identified so far in plants. All together, these results implicate KAPP as a functional component of the RLK signaling pathway, which also mediates adaptation to Na+ stress. RLK pathway components, known to be modulated by NaCl at the messenger RNA level, are constitutively down-regulated in rag1-1 mutant plants. The effect of NaCl on their expression is not altered by the rag1-1 mutation.
Animal receptor Tyr kinases and receptor Ser/Thr kinases are cell surface enzyme-linked receptors that are activated by peptide ligands and initiate a diverse range of signal transduction pathways, including those that control cell growth, differentiation and survival, defensive responses. and metabolism (Holland and Holland, 2002). Receptor-like kinases (RLKs; a.k.a. plant receptor kinase; Cock et al., 2002) are animal receptor kinase orthologs in plants, so classified because of conserved structures that include an extracellular receptor, a transmembrane domain, and an intracellular kinase domain (Shiu and Li, 2004). The mechanisms by which RLKs activate and regulate downstream components of the signaling pathway resemble those of receptor Tyr kinases and receptor Ser/Thr kinases (Cock et al., 2002; Shiu and Li, 2004). RLK activation occurs upon binding of an extracellular ligand to the plasma membrane-localized heterodimeric receptor form (Morris and Walker, 2003; Tichtinsky et al., 2003; Torii, 2004). Subsequently, the RLK complex undergoes autotransphosphorylation to form an active complex (Trotochaud et al., 1999, 2000; Clark, 2001; Rojo et al., 2002). RLKs are also transcriptionally regulated (Becraft, 2002). Plant RLKs activate diverse signal transduction pathways, including those that control hormone responses (Li and Chory, 1997; Matsubayashi et al., 2002; Montoya et al., 2002; Scheer and Ryan, 2002; Yin et al., 2002; Szekeres, 2003), flower development (Williams et al., 1997; Stone et al., 1998), innate immunity against bacterial pathogens (Gomez-Gomez et al., 2001), self incompatibility (Braun et al., 1997), and root nodule formation (Downie and Walker, 1999; Endre et al., 2002; Krusell et al., 2002; Nishimura et al., 2002; Spaink, 2002; Stracke et al., 2002).
The kinase-associated protein phosphatase (KAPP; Stone et al., 1994) is a cytosolic-oriented, membrane-anchored type 2C protein phosphatase, which binds only to activated (i.e. phosphorylated) forms of RLK via its kinase interaction domain (Braun et al., 1997; Shah et al., 2002) and inactivates the RLKs through dephosphorylation (Tichtinsky et al., 2003). Genetic evidence indicates that KAPP may function as a negative regulator of RLK pathways (Williams et al., 1997; Stone et al., 1998; Gomez-Gomez et al., 2001). KAPP is also essential for RLK internalization via endocytosis (Shah et al., 2002; Vanoosthuyse et al., 2003). In animals, internalization of receptor kinases is an important step for signaling, which leads to degradation and recycling of receptor kinases (Shah et al., 2002). Although the RLK superfamily includes more than 600 members in Arabidopsis (Arabidopsis thaliana; Shiu et al., 2004), KAPP, which binds to many RLKs (Braun et al., 1997; Tichtinsky et al., 2003), is a single copy gene in Arabidopsis as well as in Zea mays (Stone et al., 1994; Braun et al., 1997). Based on these findings and considering its ubiquitous expression (Stone et al., 1994; Braun et al., 1997; Williams et al., 1997), KAPP is thought to play an important role in the RLK pathways. Another type 2C protein phosphatase, ABI1, was shown to negatively regulate abscisic acid-dependent gene repression (Sheen, 1998). Using KAPP overexpression lines, it was demonstrated that KAPP does not affect abscisic acid-dependent gene repression. Some RLKs and the RLK pathway components have been reported to be regulated by NaCl or other abiotic stresses (Piao et al., 2001; Kreps et al., 2002; Ozturk et al., 2002; Seki et al., 2002). However, direct evidence indicating a function of RLK pathway components in salt tolerance has so far not been demonstrated. In addition, plant genes with kinase interaction domain identity to the animal counterparts have not yet been isolated (Braun et al., 1997; Williams et al., 1997).
High salinity is one of the major abiotic stresses that limit land usage and reduce crop yield (Ward et al., 2003). Salinity is becoming more and more problematic for agriculture, as irrigation leads to salt accumulation in the soil (Ward et al., 2003). Recently, molecular genetic approaches aimed at unraveling the complexity of salt stress responses in plants have provided fundamental insights to understanding the biological processes involved in the perception and signal transduction of environmental stimuli (Zhu, 2002, 2003; Chinnusamy et al., 2004). The effects of high salinity on growth and viability of glycophytes are mainly associated with an increased osmolarity of the solution in contact with the roots, a specific Na+ or Cl− ion toxicity, and nutritional imbalance (Zhu, 2000). Sodium (Na) accumulation causes the primary damages due to ionic stress (Tester and Davenport, 2003; Zhu, 2003). Thus, maintenance of low intracellular Na+ concentration is crucial for plant adaptation to saline stress. Toxic effects of Na+ include inhibition of enzyme activity (Serrano et al., 1999; Hasegawa et al., 2000) and disruption of K+ nutrient acquisition (Zhu, 2003). Ca2+ counteracts the negative effects of Na+ by enhancing the selectivity of K+ over Na+ (Epstein, 1998; Zhu, 2002; Tester and Davenport, 2003).
Plants maintain low cytosolic Na+ concentration by controlling compartmentalization, influx, and efflux of Na+ (Zhu, 2003). The Ca2+-dependent SOS (salt overly sensitive) pathway is the only known Na+-specific signal transduction pathway that regulates K+/Na+ homeostasis by controlling Na+ efflux and K+ acquisition. SOS1 (Wu and Zhu, 1996), SOS2 (Zhu et al., 1998), and SOS3 (Liu and Zhu, 1997) were identified in a forward genetic screening (Zhu, 2002) for Na+ hypersensitivity and growth defects under K+ deficiency (Zhu, 2000). The SOS pathway is initiated by a salt stress-induced calcium signal that is sensed by the Ca2+-binding protein SOS3. The activated SOS3 recruits the protein kinase SOS2 to the plasma membrane to form a complex. A conformational change that results from the SOS3-SOS2 complex formation relieves the autoinhibition of the SOS2 kinase activity. Subsequently, the SOS3-SOS2 complex regulates the expression and activity of SOS1, a plasma membrane-localized Na+/H+ exchanger that mediates Na+ efflux across the plasma membrane (Halfter et al., 2000; Qiu et al., 2002; Quintero et al., 2002; Shi et al., 2002b; Zhu, 2003). Unidentified components of the SOS pathway include the phosphatase(s) that dephosphorylates SOS1 and SOS2 as part of the phosphorylation/dephosphorylation-based signal transduction (Ohta et al., 2003).
Here, we report the isolation and functional characterization of root attenuated growth1 (rag1), a loss-of function mutant of KAPP. rag1 exhibits NaCl sensitivity and is not a component of the SOS pathway. The rag1-1 sos1-14 double mutant exhibits an additive phenotype of both parental mutants, indicating that KAPP is a component of a novel Na+ adaptation pathway, which may be related to the RLK pathway.
RESULTS
Characterization of rag1-1 Salt-Sensitive Phenotype
rag1-1 mutant was isolated by screening of a T-DNA-tagged Arabidopsis population for salt tolerance phenotypes on NaCl-containing medium (Zhu et al., 2002; Koiwa et al., 2003). rag1-1 exhibits substantially reduced primary root growth, root tip swelling, and enhanced lateral root formation on Murashige and Skoog (MS) salt agar medium supplemented with 160 mm NaCl (Fig. 1). However, no substantial differences were observed in the shoot of rag1-1 compared to wild-type plants. In the absence of salt, wild-type and rag1-1 roots were very similar also (Fig. 1A), exhibiting cells of rectangular shape uniformly distributed within the root tissue (Fig. 2, A and B).
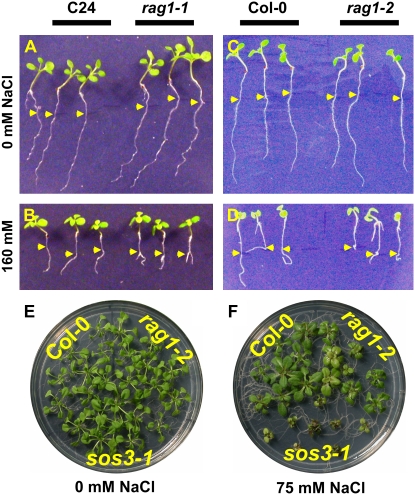
Figure 1.
Growth of rag1 plants. rag1 is almost identical to wild type at optimal growth conditions and is hypersensitive to NaCl. A to D, Root growth reduction of rag1 mutants under high NaCl. Four-day-old seedlings were transferred to MS-agar (1.5%) medium supplemented with 0 mm (A and C) or 160 mm (B and D) NaCl and were allowed to grow for 6 (A and B) or 9 (C and D) additional days. From left to right: wild-type (ecotype C24), rag1-1, wild-type (ecotype Col-0), and rag1-2 seedlings. E and F, Shoot growth reduction of rag1-2 under 75 mm NaCl. Four-day-old seedlings were transferred to MS-agar (0.8%) medium supplemented with 0 mm (E) or 75 mm (F) NaCl and were allowed to grow for 3 additional weeks.
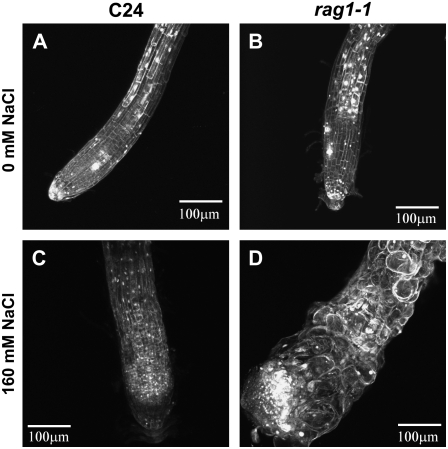
Figure 2.
The root swelling phenotype of rag1-1 is due to cell enlargement. Four-day-old seedlings were transferred to MS-agar medium supplemented with 0 (A and B) or 160 (C and D) mm NaCl and were allowed to grow for 9 additional days. Root tips of wild-type (ecotype C24; A and C) and rag1-1 (B and D) seedlings were observed under a confocal microscope.
In contrast, upon NaCl treatment, the diameter of rag1-1 roots at the maximum swelling position was more than 2 times wider than wild-type roots (Fig. 2, C and D). The swollen region of the NaCl-treated rag1-1 root showed enlarged, inconsistently sized, un-uniform, and round-shaped cells. Deformed cells were observed at the distal elongation zone and at the root cap but not at the differentiation zone (data not shown). One cell layer could not be distinguished from another in the epidermal, cortical, endodermal, and pericycular regions, since deformed cells intruded into each other and no longer formed clear cell layers (Fig. 2D). The boundary of the stele could be observed, but it was not clear if the cells in the stele were affected or not. From these observations, it was concluded that cell enlargement of NaCl-treated rag1-1 appeared to be the primary cause of root swelling.
In wild type, cells of NaCl-treated roots were smaller than that of untreated roots; however, cells retained cell file organization (Fig. 2C). These observations suggest that proper maintenance of the cytoskeleton is defective (Wasteneys and Galway, 2003) in rag1-1 root but is maintained in the wild-type root after salt treatment.
Apparent differences between rag1-1 and the wild type are only observed after NaCl treatment, indicating that the alteration in cell size and cell shape is a specific response to NaCl stress. Despite these morphological changes, however, the root meristem of some rag1-1 retained the ability to regrow after up to 12 d of 160 mm NaCl treatment (data not shown), indicating that neither root tip swelling nor lateral root formation is induced by the death of the primary root meristem. The rescued root retained the already-deformed cells but produced normal-looking cells after being transferred back to control medium (data not shown).
Loss-of-Function Monogenic Mutation in KAPP Causes Salt Sensitivity
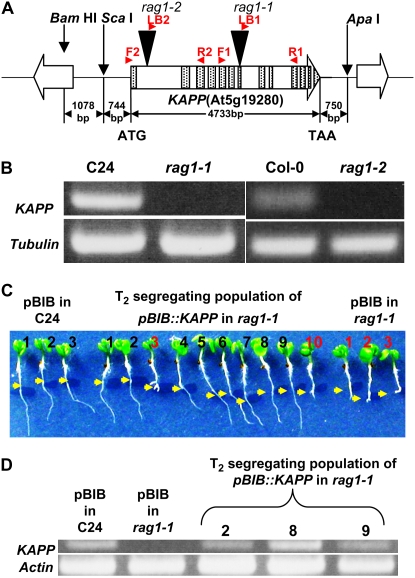
A T-DNA insertion was identified within the seventh exon (2,721 bp downstream of ATG translation start site) of KAPP (Stone et al., 1994) in rag1-1 (Fig. 3A) by thermal asymmetric interlaced-PCR (Liu et al., 1995) analysis. NaCl-treated F2 progenies (245 from 21 F1 lines) derived from backcrossing with wild type segregated to wild-type:rag1-1 phenotype at a 3:1 ratio (187:58; χ2 = 0.23; P > 0.50), indicating that rag1-1 is a monogenic recessive mutation. Genotype was determined for 36 NaCl-sensitive F2 progenies, and all of them were homozygous for the rag1-1 mutation. rag1-2 (SAIL_1255_D05), whose T-DNA insertion is located at the first intron (525 bp downstream of ATG) of KAPP, was isolated through reverse genetic in silico search from SAIL (formerly called GARLIC; Torrey Mesa Research Institute, San Diego; collection ecotype Columbia [Col-0]; http://www.nadii.com/pages/collaborations/garlic_files/GarlicDescription.html). These results indicate that the KAPP mutation is causing the NaCl-sensitive phenotype.
Figure 3.
The rag1-1 mutant has a T-DNA insertion within the seventh exon of KAPP. The insertion is homozygous and functionally disrupts the expression of the gene. A, Genetic structure of KAPP and its neighbor genes are shown as block arrows with arrowheads indicating the 3′ terminus. Exons and introns of KAPP are indicated as shaded boxes and the blank regions between them, respectively. T-DNA insertion sites of rag1-1 (isolated by forward genetics, ecotype C24) and rag1-2 (SAIL_1255_D05, Col-0) are shown as black triangles. The red arrowheads and the red letters above them indicate the forward (F1, CATGCACAGATAACATGGAACTCTAC; F2, TTGCTTCTCATCTCCCTCATCA), reverse (R1, CAAGAGAACAGTAGCTGTACAAC; R2, CTTGGCAACATTCACATTGCCT), and left-border (LB1, TTGACCATCATACTCATTGCTG; LB2, TAGCATCTGAATTTCATAACCAATCTCGATACAC) primers used for diagnostic PCR (data not shown) and RT-PCR (B and D). B, mRNA expression of KAPP in 10-d-old C24, rag1-1, Col-0, and rag1-2 seedlings (30 cycles of RT-PCR). Tubulin is used as control to show the equal amount of cDNA applied for RT-PCR. C, Phenotype complementation of three-quarters of T2 progenies of rag1-1. Conditions of the treatment are the same as Figure 1. Red digits indicate the progenies showing the mutant phenotype. Yellow arrows indicate the position of root tips at the time of transfer. D, RT-PCR of the selected seedlings from the T2 progeny. pBIB in C24 and pBIB in rag1-1 seedlings were used as positive and negative control, respectively. DNA samples were extracted from the selected seedlings. 2, 8, and 9, The progenies located at the second, eighth, and ninth positions, respectively, from the left on C. Actin is used as control to show equal amount of cDNA applied for RT-PCR. [See online article for color version of this figure.]
Reverse transcription (RT)-PCR analysis revealed that mRNA accumulation of KAPP is diminished in plants with both mutant alleles to undetectable levels (Fig. 3B). Absence of a shorter transcript in rag1-1 was confirmed using primer sets targeting the upstream region of T-DNA (data not shown). rag1-2 (Fig. 1D) exhibits a root phenotype similar to that of rag1-1 (Fig. 1B) in response to NaCl. However, shoot growth reduction after 3 weeks of 75 mm NaCl treatment was observed in rag1-2 compared with the wild type (Fig. 1, E and F) but not in rag1-1. The phenotype difference between rag1-1 and rag1-2 in the shoot may be due to the ecotype background difference. Col-0 is more sensitive to NaCl treatment than C24, with respect to root growth inhibition, shoot anthocyanin accumulation, and shoot growth reduction at lower salt concentrations (data not shown). A cross between rag1-1 and rag1-2 was made to perform a complementation test. As a result, all F1 progenies (18 seedlings) exhibit a NaCl-sensitive phenotype, indicating that rag1-1 and rag1-2 are indeed allelic (data not shown). Those F1 plants were tested by diagnostic PCR to confirm the heterozygous genotype for both insertions.
Genetic complementation with genomic DNA fragments under the control of the natural promoter further confirmed that a loss-of-function mutation of KAPP caused the salt-sensitive phenotype in rag1 mutants. Two different genomic DNA fragments (6,227-bp ScaI-ApaI and 7,305-bp BamHI-ApaI fragments; Fig. 3A) containing the KAPP open reading frame were digested from the bacterial artificial chromosome F7K24. Interestingly, only the BamHI-ApaI fragment complemented the NaCl-sensitive phenotype (Fig. 3C). This result suggests that there are essential regulatory factors at the region −744 to −1,822 of the KAPP gene. T2 progenies of plants transformed with the BamHI-ApaI fragment exhibited a 3:1 (38:14; χ2 = 0.10; P > 0.70) segregation ratio for NaCl-sensitive phenotype (Fig. 3C). All seedlings shown in Figure 3C were examined for hygromycin sensitivity after the phenotype scoring. Both of two NaCl-sensitive progenies were killed by hygromycin treatment, whereas all of eight NaCl-resistant progenies survived, indicating a tight linkage between recovery of wild-type phenotype and hygromycin resistance. mRNA expression level of KAPP was also recovered in those progenies that survived (Fig. 3D). These results confirmed that the recessive loss-of-function mutation of KAPP is the cause of the NaCl-sensitive phenotype.
rag1-1 NaCl Sensitivity Is Specific to Na+ Ion Toxicity
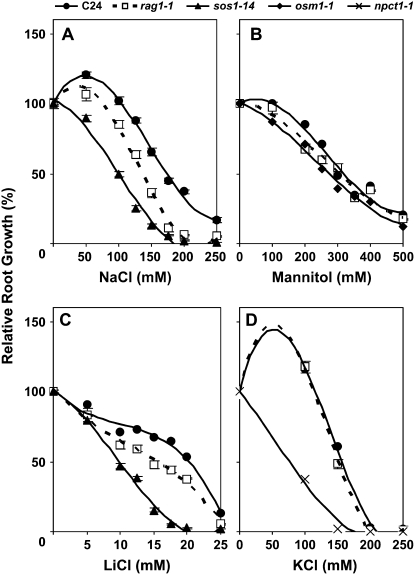
The function of KAPP in ion/osmotic adaptation was further assessed by examining the effects of nonionic and ionic osmotic solutes on root growth and development of wild-type and rag1-1 seedlings. In addition, sos1-14 (Koiwa et al., 2003), osm1-1 (Zhu et al., 2002), and npct1-1 (Y. Nakagawa, B. Cubero, F. Li, K.G. Raghothama, J.M. Pardo, R.A. Bressan, and P.M. Hasegawa, unpublished data) were used as specific controls due to their sensitivity to Na+ and Li+ ions, osmotic stress, and Cl− ion, respectively. Normal root growth of rag1-1 seedlings was inhibited by NaCl (Fig. 4A), but not by mannitol (Fig. 4B) or by KCl (Fig. 4D). These results indicate that rag1-1 seedlings are sensitive to Na+ but not to osmotic stress or Cl− ions. Root growth of rag1-1 was slightly inhibited by LiCl (Fig. 4C). Li+ is a more toxic analog of Na+, with which it presumably shares a transport system and a mechanism of toxicity (Serrano et al., 1999). Small yet statistically significant differences were observed in terms of primary root length. In addition to the primary root growth inhibition, characteristic swollen root tips and lateral root formation (Fig. 1B) were observed only on high NaCl-containing medium and not on high LiCl-containing medium.
Figure 4.
Root growth of the rag1-1 is hypersensitive to Na+. Four-day-old seedlings were transferred onto 1× MS-agar (1.5% agar) plates supplemented with various concentration of NaCl (A), mannitol (B), LiCl (C), and KCl (D) and allowed to grow for 6 additional days (n = 12). Then the plates were scanned, and the length of primary root growth after the transfer was measured by National Institutes of Health's Scion Frame Grabber (Buer et al., 2000). The root growth is shown as relative root length to that of nontreated seedlings. The error bars indicate the se.
These results indicate that KAPP functions in salt adaptation through the control of Na+ homeostasis. The next question arose as to whether this Na+ sensitivity was associated with altered K+ uptake. NaCl-sensitive mutants have been reported to be sensitive to low K+ as well as to high Na+ conditions (Zhu, 2003). There was no substantial difference detected between primary root growth of rag1 and wild-type seedlings treated on 1/20 MS-agar (1.2% agar, 0.15 mm Ca2+) medium supplemented with 0 to 10 (one-half-strength of standard MS medium) mm KCl (Rus et al., 2001), whereas the primary root growth of the positive controls, sos1-14 and sos3-2, was greatly inhibited in low K+ (up to 0.2 mm) medium. When we compared rag1-2, sos3-2, and their relative controls (Col-0 and gl-1), only sos3-2 exhibited substantially more severe growth reduction (about 30%) at lower concentrations of KCl. There was no substantial growth difference among these genotypes at 10 mm KCl. This result indicates that, unlike other known salt-sensitive mutants, the NaCl sensitivity of rag1 is not associated with K+ deficiency. It further suggests that the KAPP-RLK sodium adaptation pathway regulates Na+ toxicity independent of K+ deficiency.
We also assessed the effect of Ca2+ on rag1 NaCl sensitivity. Ca2+ is known to affect Na+ uptake (Pardo and Quintero, 2002); therefore, NaCl sensitivity of some NaCl-hypersensitive mutants (such as sos1 to sos3 and hkt1) can be altered by Ca2+ concentration. Up to 10 mm CaCl2 (1× MS medium contains 3 mm Ca2+) did not affect the NaCl sensitivity of rag1-1 at 100 mm NaCl, while NaCl sensitivity of sos1-14 and sos3 control plants was rescued by high Ca2+ concentration (data not shown).
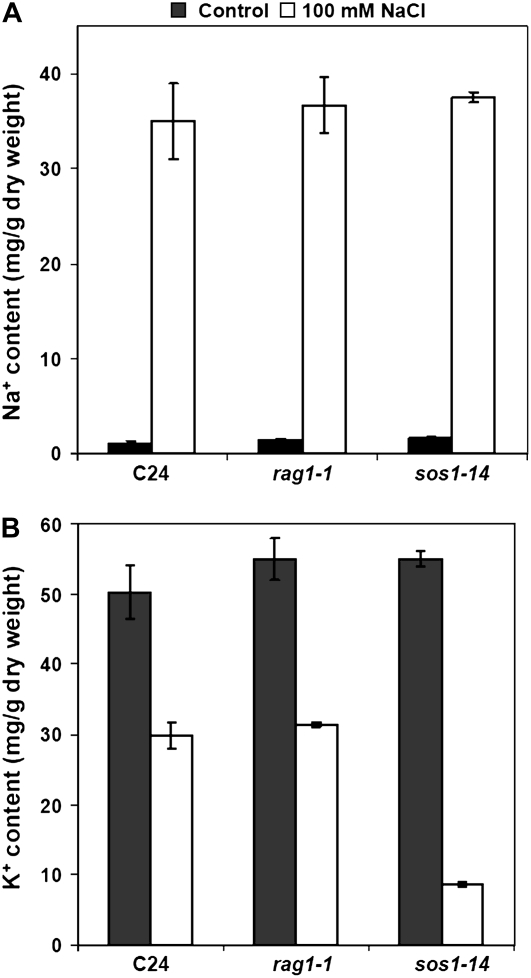
To further determine if KAPP is involved in Na+/K+ homeostasis, intracellular contents of Na+ and K+ ions were measured (Fig. 5) as described by Rus et al. (2001). Whole seedlings of rag1-1, a positive control, sos1-14, and wild type were treated with 100 mm NaCl. There was no substantial difference in Na+ and K+ content among genotypes before the treatment (K+, 53 mg/g dry weight; Na+, 1.3 mg/g dry weight). After the treatment, rag1-1 (K+, 31.5 ± 0.3 mg/g dry weight; Na+, 36.7 ± 3.0 mg/g dry weight) and wild-type seedlings (K+, 28.9 ± 1.9 mg/g dry weight; Na+, 35.0 ± 4.0 mg/g dry weight) exhibited equivalent contents for both Na+ and K+ ions, while sos1-14 (K+, 8.65 ± 0.4 mg/g dry weight; Na+, 37.5 ± 0.5 mg/g dry weight) exhibited much lower K+ content (Fig. 5). Ion contents were also measured on leaves of seedlings treated with 0 to 75 mm NaCl. No substantial differences in Na+ and K+ levels were detected (data not shown).
Figure 5.
K+ and Na+ contents in rag1-1 seedlings exposed to salt stress. Ten-day-old seedlings were transferred into liquid medium 2 d for pretreatment. Then the liquid medium was supplemented with 0 or 100 mm NaCl for 2 additional days of treatment. There was no significant difference observed between wild type and rag1-1 either in presence or in absence of NaCl, while K+ content of the Na+ ion homeostasis mutant, sos1-14, was much lower after NaCl treatment. The value is the average measurement of three individually treated flasks, and error bars indicate se.
Taken together, these results indicate that KAPP is not directly involved in Na+/K+ uptake, yet it may be responsible for sensing high intracellular Na+ concentration or for regulating subcellular Na+ localization. An additional feature of rag1-1 is that it contains higher Ca2+ compared to its relative wild type in the absence of NaCl treatment (D.E. Salt, personal communication).
KAPP Is Involved in a Novel Salt Stress-Responsive Pathway
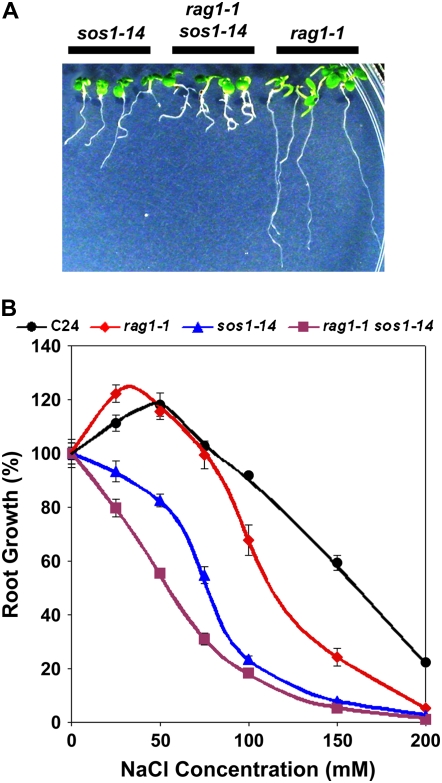
sos1, sos2, and sos3 mutants exhibit both Na+ and Li+ ion-specific sensitivity. Therefore, we generated rag1-1 sos1-14 double mutants to determine the genetic interaction between KAPP and SOS1 and, ultimately, verify whether or not KAPP functions as a part of the SOS pathway. rag1-1 sos1-14 double mutants showed an additive NaCl-sensitive phenotype (Fig. 6). The seedlings of rag1-1 sos1-14 look identical to the wild type at 0 mm NaCl; however, the primary root growth of rag1-1 sos1-14 seedlings was substantially reduced compared to sos1-14 at 25 mm to 75 mm NaCl. In addition to primary root growth inhibition, root morphological alteration (i.e. root tip swelling and lateral root formation) was detectable in rag1-1 sos1-14 double mutants at 50 mm NaCl (Fig. 6), whereas the same phenotype was detectable in rag1-1 only at a much higher concentration (>150 mm NaCl; Fig. 1). This result also supports the interpretation that the phenotype of the rag1-1 mutant is Na+ ion specific. Either a higher Na+ content or an increased Na+ to K+ ratio of rag1-1 sos1-14 double mutant, compared to rag1-1, may have acted as an early trigger for swelling and lateral root formation, which are both typical of the rag1-1 mutation. Additional evidence suggests that KAPP is not involved in the SOS pathway: (1) sos mutants are more sensitive to Li+ ions than Na+ ions, while rag1 is more sensitive to Na+ ions; (2) sos mutants do not exhibit the root tip swelling and lateral root formation phenotype characteristic of rag1; (3) unlike sos mutants, rag1 mutants are not sensitive to K+ deficiency; and (4) unlike sos mutants, the NaCl-sensitive phenotype of rag1 is not affected by Ca2+ availability. These phenotypic differences combined with the genetic analysis lead us to the conclusion that KAPP functions in a novel Na+-responsive pathway.
Figure 6.
rag1-1 sos1-14 double mutant shows additive phenotype relative to parental monogenic mutants. A, From left to right, sos1-14, rag1-1 sos1-14, and rag1-1 seedlings treated with 75 mm NaCl. B, Primary root growth of rag1-1 sos1-14 along with other relative controls (C24, rag1-1, sos1-14) were measured as reported for Figure 4.
rag1-1 Exhibits Partial De-Etiolation and Root Branching
Both the shoots and roots of rag1-1 seedlings were identical to wild type under optimal growth conditions (1× MS, 3% Suc, and 1.5% agarose), indicating that KAPP is required primarily during salt adaptation (Fig. 1A). Mature rag1-1 plants did not exhibit any of the extreme abnormal growth and development phenotypes associated with RLK pathway mutants (data not shown), such as enlarged siliques (clv1-3; for review, see Clark, 2001), dwarfism, or male sterility (bri1 and bin2; for review, see Clouse, 2002).
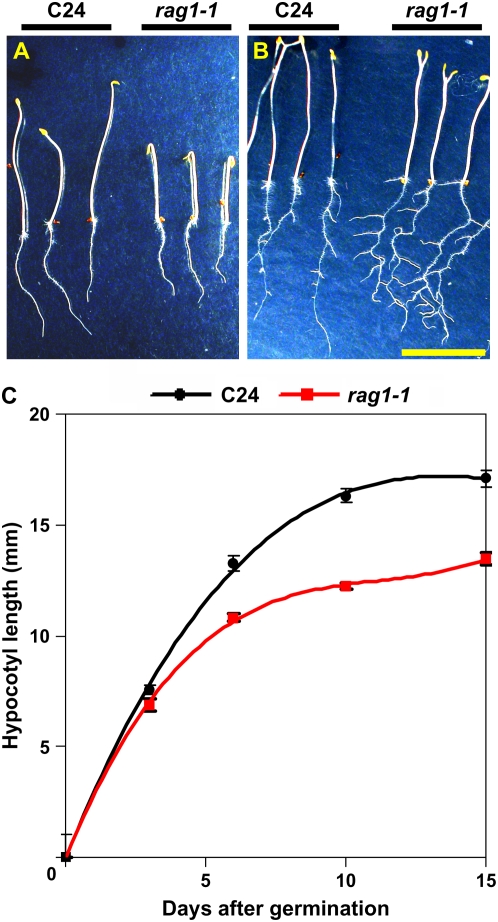
However, dark-grown rag1-1 seedlings exhibit a partial de-etiolation phenotype manifested with short and radially thickened hypocotyls and increased cotyledon size typical of brassinosteroid-deficient or -insensitive mutants (Li et al., 1996; Chory, 1997; Chory and Li, 1997; Bishop et al., 1999; Symons and Reid, 2003; Fig. 7). In addition, hypocotyls of rag1-1 grow more slowly compared to wild-type seedlings (Fig. 7C). These results indicate that the mutation does not arrest growth but may interfere with the regulation of directional cell expansion. Moreover, dark-grown rag1-1 seedlings exhibit greater root branching (Fig. 7B), a phenotype that has not been reported for brassinosteroid-deficient or -insensitive mutants at later growth stages. This difference was most apparent in 12-d-old seedlings. The magnified dark-grown rag1-1 root is deformed and widened compared to wild-type roots (data not shown). These results suggest a possible conditional function of KAPP in brassinosteroid perception.
Figure 7.
Dark-grown rag1-1 seedlings exhibit enhanced lateral root formation and slightly thicker hypocotyls. A, Dark-grown 7-d-old wild-type (left, ecotype C24) and rag1-1 (right) seedlings. Dark-grown seedlings were exposed to light for the first day and then covered with aluminum foil and allowed to grow for additional 6 d on 1× MS (3% Suc, 1.2% agar medium). B, Dark-grown 14-d-old wild-type (left, ecotype C24) and rag1-1 (right) seedlings. C, Hypocotyl length of dark-grown seedlings 3, 6, 10, and 15 d after germination.
The KAPP-RLK Pathway Regulates a Novel Na+-Responsive Pathway
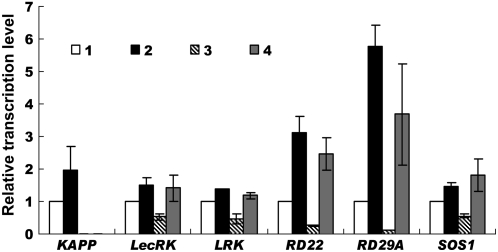
The results so far presented strongly indicate that KAPP is involved in salt adaptation. To confirm this conclusion and to identify possible interactions between stress adaptation pathways, several characterized RLK and RLK pathway components were tested for salt sensitivity and transcriptional regulation. Candidate genes were selected based on their established differential transcriptional regulation in response to salt treatment (Becraft, 2002). LecRK1 (Herve et al., 1996) and LRK1 (Kreps et al., 2002) were tested for transcriptional abundance in rag1 mutants in the absence or presence of NaCl. The level of expression of LecRK1, LRK1, and two other genes included as controls, RD29A and RD22, was constitutively reduced in nonsalinized rag1-1 seedlings, indicating that RAG1 may positively regulate the basal level of these stress-induced genes (Fig. 8). However, upon salt treatment (150 mm NaCl), wild-type and mutant seedlings had similar levels of expression, suggesting that transcription of these genes was possibly induced through a RAG1-independent pathway. Indeed, transcriptional activation of AtLecRK2 in response to salt stress has been reported to be regulated by the ethylene signaling pathway (He et al., 2004). In addition, salt sensitivity of candidate T-DNA mutants was also tested (data not shown). Among the T-DNA mutants tested, only SALK_005054 and SALK_008611 showed altered sensitivity to NaCl in shoot growth but not in root growth (data not shown). Both SALK_005054 and SALK_008611 had a T-DNA insertion in RPK1. These results indicate that RPK1 is possibly involved in this RLK-mediated NaCl adaptation pathway.
Figure 8.
KAPP transcriptionally regulates RLKs and other stress-induced genes. Relative transcription levels of KAPP, LecRK, LRK, RD22, RD29A, and SOS1 in wild-type (1 and 2) and rag1-1 (3 and 4) plants were determined using quantitative RT-PCR. Total RNA was isolated from the untreated seedlings (1 and 3) or the seedlings treated with 150 mm NaCl for 3 h (2 and 4). The gene transcript levels were normalized for the expression of tubulin measured in the same RNA samples. Data are means ± sd of three independent experiments.
DISCUSSION
We report the isolation of two allelic loss-of-function mutations (rag1-1, ecotype C24; and rag1-2, ecotype Col-0) of KAPP as salt-sensitive mutants by forward and reverse genetic identification. Functional characterization revealed that KAPP functions in adaptation to NaCl stress. A unique feature of the rag1 mutant is its specific Na+ hypersensitivity. Salt sensitivity of hkt1 is also Na+ ion specific (Berthomieu et al., 2003). However, there are distinctive phenotypic differences associated to mutations at these two loci. For instance, hkt1 accumulates more sodium into the shoot compared to the wild type, whereas rag1 does not. In addition, hkt1 does not have a seedling root growth attenuation phenotype as seen in rag1 (Fig. 1).
The salt sensitivity of rag1-1 is less severe compared to other reported salt-sensitive mutants isolated through a root-bending assay (Wu and Zhu, 1996; Liu and Zhu, 1997; Zhu et al., 1998; Shi et al., 2002a, 2002b; Zhu et al., 2002; Koiwa et al., 2003). This may explain why rag1 has not been isolated in previous extensive screenings (Zhu et al., 1998; Shi et al., 2003). In this respect, rag1-2 (Col-0 background), for instance, does not exhibit a clear phenotype at 50, 75, or 100 mm NaCl, the concentrations used to isolate the sos mutant series. In addition, the severe sos1, sos2, and sos3 salt-sensitive phenotypes are associated with K+ imbalance. In contrast, Na+-specific hypersensitive mutants such as rag1 may show a less dramatic salt-sensitive phenotype.
The NaCl-Induced Root Tip Swelling and the Dark-Grown Phenotype of rag1 Are Similar to Mutants with Impaired Cellulose Biosynthesis
Constitutive root swelling has been reported for several other salt-sensitive mutants (Liu and Zhu, 1997; Shi et al., 2002a, 2003; Koiwa et al., 2003). Nevertheless, all the mutants showing this phenotype also present additional distinctive features with respect to other disrupted/altered functions associated with the salt-sensitive phenotype. Unlike sos5, swelling and lateral root formation in rag1 was not observed upon an extended period of culture (data not shown; Shi et al., 2003). The swollen root tip observed in rag1, as well as in some other salt-sensitive mutants, resembles the temperature-sensitive phenotype of rsw mutants (Baskin and Wilson, 1997; Wiedemeier et al., 2002), which have defects in cellulose synthesis or microtubule organization. These results indicate that KAPP may contribute to the regulation of cellulose synthesis and/or microtubule organization only under high Na+ concentration.
Relationship between KAPP and the SOS Pathway
Analysis of the double mutant, rag1-1 sos1-14, revealed that KAPP is not a component of the SOS pathway. Unlike sos mutants, the rag1-1 mutant is only slightly more sensitive to Li+, which is thought to share transport systems and toxicity target with Na+ (Serrano et al., 1999). Two possible explanations may be provided for the hypersensitivity to Na+ observed in rag1-1 sos1-14 double mutant. The first possibility is that a diffusive signal produced in the shoot, due to an altered higher Na+ to K+ ratio resulting from the sos1-14 mutation, is transported to the root where it initiates an RLK pathway, which in turn leads to the branching phenotype. The second possibility is that either the higher Na+ concentration or any other damage caused by the sos1 mutation in the root induces the branching phenotype.
The Function of KAPP in Planta
Because KAPP is predicted to have a promiscuous function in down-regulating multiple RLK pathways (Braun et al., 1997; Williams et al., 1997), it was surprising to find that rag1-1 did not have an apparent phenotype at optimum growth conditions. Although overexpression of KAPP is shown to mimic the phenotype of CLV and FLS (Williams et al., 1997; Gomez-Gomez et al., 2001), the lack of a developmental phenotype in rag1-1 indicates that KAPP may not be a central component of these pathways in planta. Stone et al. (1998) showed that inhibition of KAPP could suppress the clv-like phenotype. Gene suppression may cause a cosuppression of other genes, and, consequently, the resulting phenotype may diverge from that of a single gene knockout. Our preliminary results indicate that loss of function of KAPP does not suppress the clv phenotype of either clv2-1 or clv3-2 (data not shown).
One possible explanation is that there is another functionally redundant gene(s), which is not similar in the overall structure but has partial homology, such as POLTERGEIST (Yu et al., 2003). Another possibility is that KAPP becomes active only in response to certain environmental cues such as salinity or lack of light. In this case it is likely that, in addition to the described salt and light sensitivity of rag1, other undetermined phenotypes related to RLK may exist, which can only be observed in certain conditions or in response to certain stimuli.
RLK pathways regulate a broad range of signaling involved in either development or defense (Dievart and Clark, 2004). Those pathways involved in development regulate the balance between cell division and cell expansion or differentiation. rag1 mutants have a defect in maintaining the correct ratio of cell division and expansion under NaCl-stressed or dark-grown conditions. Therefore, rag1 exhibits a radially expanded root or shoot under those conditions. Shah et al. (2002) proposed that KAPP is an integral part of the endocytosis mechanism of RLKs. In this respect, KAPP may function as a positive regulator by mediating salt-induced endocytosis. This could be an important function of KAPP in the regulation of salt adaptation.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) C24RD29A:LUC was transformed (mutagenized) with pSKI015 (Weigel et al., 2000). The T2 T-DNA population was screened for NaCl-sensitive root growth (Zhu et al., 2002; Koiwa et al., 2003). rag1-2, sos1-14, osm1-1, and npct1-1 were identified through this screen. rag1-2 (Col-0) was identified through an in silico search of the SAIL T-DNA insertional mutant collection (Sessions et al., 2002).
For dose response experiments and histochemistry, seeds were sown on cellophane membranes and grown for 4 d as described previously (Zhu et al., 2002). Four-day-old seedlings were transferred to treatment medium (1× MS, pH 5.7, 1.5% agar, 3% Suc) supplemented with various concentrations of salts or osmolyte and allowed to grow for 6 or 9 additional days. Treatment media for low potassium and low calcium contained (1/20× MS macro elements, 1× MS micro elements) as described previously (Liu and Zhu, 1997; Rus et al., 2001). Seedlings grown on plates were placed in a controlled environment (16 h of light at 22°C and 8 h of darkness at 18°C), and plants grown on soil were placed in the greenhouse.
Growth Measurements
To measure root growth, the position of the root tip was marked at the bottom of petri dishes at the time of transfer and scanned by a flatbed scanner (Epson Perfection 1200U) at 300 pixels per inch after the treatments. The scanned images were saved as TIF format and were measured using the National Institutes of Health's Scion Frame Grabber as described (Buer et al., 2000).
Genetic Analysis of rag1 T-DNA Insertion Alleles
Genomic sequence flanking T-DNA in rag1-1 was determined using thermal asymmetric interlaced-PCR as described (Koiwa et al., 2003). To confirm the cosegregation of T-DNA with the salt-sensitive phenotype, homozygous rag1-1 plants were backcrossed to wild type (C24 × rag1-1), and F2 progenies were tested for salt sensitivity. DNA was extracted from F2 progenies exhibiting a salt-sensitive phenotype. Then, diagnostic PCR was performed using primers F1, R1, and LB1 (shown in Figure 3) as described previously (Koiwa et al., 2003).
T-DNA insertion of rag1-2 was confirmed by diagnostic PCR using primers F2, R2, and LB2 (shown in Fig. 3).
RNA was isolated from 10-d-old seedlings that were grown on cellophane membrane and treated on filter paper soaked with one-half-strength MS medium, pH 5.7, supplemented with 0 mm (control) or 175 mm (NaCl treatment) using the RNAeasy total RNA isolation kit (Qiagen) as described (Yokoi et al., 2002). First-strand cDNA was synthesized using the Superscript III kit (Gibco BRL) from total RNA (2 mg) as recommended in the manual protocol. The PCR reaction was carried out using the primers described in Figure 3A.
Genetic Complementation
Two different genomic DNA fragments (6,227-bp ScaI-ApaI and 7,305-bp BamHI-ApaI fragments; Figure 3A) containing the KAPP open reading frame were digested from the bacterial artificial chromosome F7K24. The ScaI-ApaI fragment contains 740 bp of the 5′-untranslated region, and the BamHI-ApaI fragment includes a sequence region of the putative 3′-untranslated region of the next upstream open reading frame. These fragments were subcloned into a shuttle vector, pBluescript SK+ (Stratagene); the 7,305-bp fragment into the XbaI site, and the 6,227-bp fragment blunt-end ligated into the HindIII site. Both fragments were cloned subsequently into the KpnI site of the pBIB binary vector that contains a gene for hygromycin resistance in planta as selection marker (Becker, 1990). The pBIB:KAPP and pBIB (without insert as a control) vectors were introduced into Agrobacterium GV3101 to transform rag1-1 and wild type (C24) using the floral infiltration method (Bechtold et al., 1993) as modified by Koiwa et al. (2002).
The progeny of segregating T2 populations derived from hygromycin-resistant T1 lines (plants obtained from seed of plants directly after floral transformation) were evaluated for cosegregation of KAPP expression-dependent salt tolerance and hygromycin resistance (χ2 analysis for one or multiple insertions).
Histochemical Analysis
Seedling roots, treated in the same way as described for the dose response tests, were stained with 10 μg/mL propidium-iodide for 10 min to visualize cell walls and washed with distilled water. Then samples were imaged in distilled water using a confocal laser microscope (Bio-Rad MRC1024; Bio-Rad Laboratories). Illumination was provided at 568-nm wavelength and red emission was collected for 5 min. The figures are projection of 52 to 69 optimal dissections (Fig. 2).
Quantitative RT-PCR Analysis
The expression of the related genes was analyzed by real-time quantitative RT-PCR using the fluorescent intercalating dye SYBR-Green and ABI PRISM 7000 Real-Time system. Tubulin was used as a standard control in the RT-PCR reactions. A two-step RT-PCR procedure was performed in all experiments. Total RNA was isolated from seedlings treated by one-half-strength MS or one-half-strength MS supplemented with 150 mm NaCl. The cDNA were used as template in real-time PCR reactions with gene-specific primers (Table I). The RNA RT and real-time PCR reaction were performed using SYBR PrimeScript RT-PCR kit (TaKaRa) according to manufacturer's instruction. PCR amplification was done in two steps: DNA denaturation at 95°C for 10 s and elongation at 60°C for 40 s. Fluorescence was evaluated at the end of the elongation. PCR reactions were maintained for 40 cycles. The amplification of the target genes was monitored for every cycle by SYBR-Green fluorescence. The Ct, defined as the PCR cycle at which a statistically significant increase of reporter fluorescence is first detected, is used as a measure for the starting copy numbers of the target gene.
Table I.
Primers used for quantitative RT-PCR analysis (Fig. 8)
| Primer | Sequence |
|---|---|
| KAPP-F | CAAAATTGGTGTGGCCTCAGA |
| KAPP-R | AAGCGGCCACTTGTAATGACA |
| SOS1-f | CCAAAATTGAGCGACATGATCA |
| SOS1-r | GTGACACCACGCAGTTTCAT |
| Tubulin-f | AGGCAAAATGAGCACGAAAGA |
| Tubulin-r | TCAGACCTGTTGGTGGAATGTCAC |
| LRK-f | GTTTCGCATGAGTCTCGTCAAG |
| LRK-r | GAAGCTCACCTCTCCTACGACAA |
| LecRK1-f | TAGGTGTGTTGTGTTCGCATCA |
| LecRK1-r | TCCATTGATGTCTCAGGCCA |
| RD22-f | TACCCATTCGCGGTGTTCTACT |
| RD22-r | CTTTAGCTCGCATCCCGTTCT |
| RD29A-f | GAGACCCCGATAACGTTGGA |
| RD29A-r | CAATCTCCGGTACTCCTCCA |
Acknowledgments
We thank Jennie Sturgis and Terry Kirk for their support with the confocal microscope and the atomic absorption spectrophotometer. We thank the Salk Institute and the Torrey Mesa Research Institute (Syngenta) for providing T-DNA inserted Arabidopsis lines and the Arabidopsis Biological Resource Center for providing the BAC clone.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Albino Maggio (albino.maggio@unina.it).
Some figures in this article are displayed in color online but in black and white in the print edition.
References
- Baskin TI, Wilson JE (1997) Inhibitors of protein kinases and phosphatases alter root morphology and disorganize cortical microtubules. Plant Physiol 113 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In-planta agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Ser III Life Sci 316 1194–1199 [Google Scholar]
- Becker D (1990) Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res 18 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW (2002) Receptor kinase signaling in plant development. Annu Rev Cell Dev Biol 18 163–192 [DOI] [PubMed] [Google Scholar]
- Berthomieu P, Conejero G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F, et al (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na(+) recirculation by the phloem is crucial for salt tolerance. EMBO J 22 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GJ, Nomura T, Yokota T, Harrison K, Noguchi T, Fujioka S, Takatsuto S, Jones JD, Kamiya Y (1999) The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc Natl Acad Sci USA 96 1761–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DM, Stone JM, Walker JC (1997) Interaction of the maize and Arabidopsis kinase interaction domains with a subset of receptor-like protein kinases: implications for transmembrane signaling in plants. Plant J 12 83–95 [DOI] [PubMed] [Google Scholar]
- Buer CS, Masle J, Wasteneys GO (2000) Growth conditions modulate root-wave phenotypes in Arabidopsis. Plant Cell Physiol 41 1164–1170 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Schumaker K, Zhu JK (2004) Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot 55 225–236 [DOI] [PubMed] [Google Scholar]
- Chory J (1997) Light modulation of vegetative development. Plant Cell 9 1225–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Li J (1997) Gibberellins, brassinosteroids and light-regulated development. Plant Cell Environ 20 801–806 [Google Scholar]
- Clark SE (2001) Cell signalling at the shoot meristem. Nat Rev Mol Cell Biol 2 276–284 [DOI] [PubMed] [Google Scholar]
- Clouse SD (2002) Brassinosteroid signaling: novel downstream components emerge. Curr Biol 12 R485–R487 [DOI] [PubMed] [Google Scholar]
- Cock JM, Vanoosthuyse V, Gaude T (2002) Receptor kinase signalling in plants and animals: distinct molecular systems with mechanistic similarities. Curr Opin Cell Biol 14 230–236 [DOI] [PubMed] [Google Scholar]
- Dievart A, Clark SE (2004) LRR-containing receptors regulating plant development and defense. Development 131 251–261 [DOI] [PubMed] [Google Scholar]
- Downie JA, Walker SA (1999) Plant responses to nodulation factors. Curr Opin Plant Biol 2 483–489 [DOI] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kalo P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417 962–966 [DOI] [PubMed] [Google Scholar]
- Epstein E (1998) How calcium enhances plant salt tolerance. Science 280 1906–1907 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Bauer Z, Boller T (2001) Both the extracellular leucine-rich repeat domain and the kinase activity of FLS2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13 1155–1163 [PMC free article] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu JK (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97 3735–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51 463–499 [DOI] [PubMed] [Google Scholar]
- He JX, Zhang JG, Yan DQ, Zhang JS, Chen SY (2004) A salt-responsive receptor-like kinase gene regulated by ethylene signaling pathway encodes a plasma membrane serine/threonine kinase. Theor Appl Genet 109 377–383 [DOI] [PubMed] [Google Scholar]
- Herve C, Dabos P, Galaud JP, Rouge P, Lescure B (1996) Characterization of an Arabidopsis thaliana gene that defines a new class of putative plant receptor kinases with an extracellular lectin-like domain. J Mol Biol 258 778–788 [DOI] [PubMed] [Google Scholar]
- Holland KA, Holland IB (2002) Transmembrane signalling. In Nature Encyclopedia of Life Sciences. Nature Publishing Group, London, pp 1–8
- Koiwa H, Barb AW, Xiong L, Li F, McCully MG, Lee BH, Sokolchik I, Zhu J, Gong Z, Reddy M, et al (2002) C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc Natl Acad Sci USA 99 10893–10898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwa H, Li F, McCully MG, Mendoza I, Koizumi N, Manabe Y, Nakagawa Y, Zhu J, Rus A, Pardo JM, et al (2003) The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell 15 2273–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K, Duc G, Kaneko T, Tabata S, de Bruijn F, et al (2002) Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420 422–426 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90 929–938 [DOI] [PubMed] [Google Scholar]
- Li JM, Nagpal P, Vitart V, McMorris TC, Chory J (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272 398–401 [DOI] [PubMed] [Google Scholar]
- Liu JP, Zhu JK (1997) An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proc Natl Acad Sci USA 94 14960–14964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8 457–463 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Ogawa M, Morita A, Sakagami Y (2002) An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296 1470–1472 [DOI] [PubMed] [Google Scholar]
- Montoya T, Nomura T, Farrar K, Kaneta T, Yokota T, Bishop GJ (2002) Cloning the tomato curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/SR160 in plant steroid hormone and peptide hormone signaling. Plant Cell 14 3163–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris ER, Walker JC (2003) Receptor-like protein kinases: the keys to response. Curr Opin Plant Biol 6 1–4 [DOI] [PubMed] [Google Scholar]
- Nishimura R, Hayashi M, Wu GJ, Kouchi H, Imaizumi-Anraku H, Murakami Y, Kawasaki S, Akao S, Ohmori M, Nagasawa M, et al (2002) HAR1 mediates systemic regulation of symbiotic organ development. Nature 420 426–429 [DOI] [PubMed] [Google Scholar]
- Ohta M, Guo Y, Halfter U, Zhu JK (2003) A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci USA 100 11771–11776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk ZN, Talame V, Deyholos M, Michalowski CB, Galbraith DW, Gozukirmizi N, Tuberosa R, Bohnert HJ (2002) Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol Biol 48 551–573 [DOI] [PubMed] [Google Scholar]
- Pardo JM, Quintero FJ (2002) Plants and sodium ions: keeping company with the enemy. Genome Biol 3 REVIEWS1017 [DOI] [PMC free article] [PubMed]
- Piao HL, Lim JH, Kim SJ, Cheong GW, Hwang I (2001) Constitutive over-expression of AtGSK1 induces NaCl stress responses in the absence of NaCl stress and results in enhanced NaCl tolerance in Arabidopsis. Plant J 27 305–314 [DOI] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi HZ, Zhu JK, Pardo JM (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E, Sharma VK, Kovaleva V, Raikhel NV, Fletcher JC (2002) CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A, Yokoi S, Sharkhuu A, Reddy M, Lee BH, Matsumoto TK, Koiwa H, Zhu JK, Bressan RA, Hasegawa PM (2001) AtHKT1 is a salt tolerance determinant that controls Na(+) entry into plant roots. Proc Natl Acad Sci USA 98 14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer JM, Ryan CA (2002) The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc Natl Acad Sci USA 99 9585–9590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31 279–292 [DOI] [PubMed] [Google Scholar]
- Serrano R, Mulet J, Rios G, Marquez J, de Larrinoa I, Leube M, Mendizabal I, Pascual-Ahuir A, Proft M, Ros R, et al (1999) A glimpse of the mechanisms of ion homeostasis during salt stress. J Exp Bot 50 1023–1036 [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K, Russinova E, Gadella TWJ, Willemse J, de Vries SC (2002) The Arabidopsis kinase-associated protein phosphatase controls internalization of the somatic embryogenesis receptor kinase 1. Genes Dev 16 1707–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J (1998) Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci USA 95 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Xiong L, Stevenson B, Lu T, Zhu JK (2002. a) The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell 14 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi HZ, Quintero FJ, Pardo JM, Zhu JK (2002. b) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16 1220–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Li WH (2004) Origins, lineage-specific expansions, and multiple losses of tyrosine kinases in eukaryotes. Mol Biol Evol 21 828–840 [DOI] [PubMed] [Google Scholar]
- Spaink HP (2002) Plant-microbe interactions: a receptor in symbiotic dialogue. Nature 417 910–911 [DOI] [PubMed] [Google Scholar]
- Stone JM, Collinge MA, Smith RD, Horn MA, Walker JC (1994) Interaction of a protein phosphatase with an Arabidopsis serine-threonine receptor kinase. Science 266 793–795 [DOI] [PubMed] [Google Scholar]
- Stone JM, Trotochaud AE, Walker JC, Clark SE (1998) Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interactions. Plant Physiol 117 1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417 959–962 [DOI] [PubMed] [Google Scholar]
- Symons GM, Reid JB (2003) Hormone levels and response during de-etiolation in pea. Planta 216 422–431 [DOI] [PubMed] [Google Scholar]
- Szekeres M (2003) Brassinosteroid and systemin: two hormones perceived by the same receptor. Trends Plant Sci 8 102–104 [DOI] [PubMed] [Google Scholar]
- Tester M, Davenport R (2003) Na+ tolerance and Na+ transport in higher plants. Ann Bot (Lond) 91 503–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichtinsky G, Vanoosthuyse V, Cock JM, Gaude T (2003) Making inroads into plant receptor kinase signalling pathways. Trends Plant Sci 8 231–237 [DOI] [PubMed] [Google Scholar]
- Torii KU (2004) Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int Rev Cytol 234 1–46 [DOI] [PubMed] [Google Scholar]
- Trotochaud AE, Hao T, Wu G, Yang Z, Clark SE (1999) The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and Rho-related protein. Plant Cell 11 393–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud AE, Jeong S, Clark SE (2000) CLAVATA3, a multimeric ligand for the CLAVATA1 receptor-kinase. Science 289 613–617 [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V, Tichtinsky G, Dumas C, Gaude T, Cock JM (2003) Interaction of calmodulin, a sorting nexin and kinase-associated protein phosphatase with the Brassica oleracea S locus receptor kinase. Plant Physiol 133 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JM, Hirschi KD, Sze H (2003) Plants pass the salt. Trends Plant Sci 8 200–201 [DOI] [PubMed] [Google Scholar]
- Wasteneys GO, Galway ME (2003) Remodeling the cytoskeleton for growth and form: an overview with some new views. Annu Rev Plant Biol 54 691–722 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al (2000) Activation tagging in Arabidopsis. Plant Physiol 122 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemeier AM, Judy-March JE, Hocart CH, Wasteneys GO, Williamson RE, Baskin TI (2002) Mutant alleles of Arabidopsis RADIALLY SWOLLEN 4 and 7 reduce growth anisotropy without altering the transverse orientation of cortical microtubules or cellulose microfibrils. Development 129 4821–4830 [DOI] [PubMed] [Google Scholar]
- Williams RW, Wilson JM, Meyerowitz EM (1997) A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway. Proc Natl Acad Sci USA 94 10467–10472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Zhu JK (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wu D, Chory J (2002) Plant receptor kinases: systemin receptor identified. Proc Natl Acad Sci USA 99 9090–9092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi S, Quintero FJ, Cubero B, Ruiz MT, Bressan RA, Hasegawa PM, Pardo JM (2002) Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J 30 529–539 [DOI] [PubMed] [Google Scholar]
- Yu LP, Miller AK, Clark SE (2003) POLTERGEIST encodes a protein phosphatase 2C that regulates CLAVATA pathways controlling stem cell identity at Arabidopsis shoot and flower meristems. Curr Biol 13 179–188 [DOI] [PubMed] [Google Scholar]
- Zhu JH, Gong ZZ, Zhang CQ, Song CP, Damsz B, Inan G, Koiwa H, Zhu JK, Hasegawa PM, Bressan RA (2002) OSM1/SYP61: a syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell 14 3009–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2000) Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol 124 941–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol 53 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6 441–445 [DOI] [PubMed] [Google Scholar]
- Zhu JK, Liu J, Xiong L (1998) Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition. Plant Cell 10 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]