Abstract
Phytic acid (PA) contains the major portion of the phosphorus in the soybean (Glycine max) seed and chelates divalent cations. During germination, both minerals and phosphate are released upon phytase-catalyzed degradation of PA. We generated a soybean line (CAPPA) in which an Escherichia coli periplasmic phytase, the product of the appA gene, was expressed in the cytoplasm of developing cotyledons. CAPPA exhibited high levels of phytase expression, ≥90% reduction in seed PA, and concomitant increases in total free phosphate. These traits were stable, and, although resulted in a trend for reduced emergence and a statistically significant reduction in germination rates, had no effect on the number of seeds per plant or seed weight. Because phytate is not digested by monogastric animals, untreated soymeal does not provide monogastrics with sufficient phosphorus and minerals, and PA in the waste stream leads to phosphorus runoff. The expression of a cytoplasmic phytase in the CAPPA line therefore improves phosphorus availability and surpasses gains achieved by other reported transgenic and mutational strategies by combining in seeds both high phytase expression and significant increases in available phosphorus. Thus, in addition to its value as a high-phosphate meal source, soymeal from CAPPA could be used to convert PA of admixed meals, such as cornmeal, directly to utilizable inorganic phosphorus.
In addition to providing protein and oil, soymeal is a source of phosphorus (P). As in other angiosperm seeds, most soybean seed P is in the form of phytic acid (PA; myoinositol-1,2,3,4,5,6-hexakisphosphate). PA is generally indigestible by monogastric animals such as poultry and swine (Erdman, 1979). In addition, in large poultry and swine facilities, PA chelates and lessens the dietary availability of metal ions (iron [Fe], manganese [Mn], magnesium [Mg], zinc [Zn], and copper [Cu]; Vohra et al., 1965). Underutilized PA in animal feeds leads to P pollution of wastewater and runoff from soils to which manure is applied (Sharpley et al., 1994). Feed supplementation with minerals and inorganic P (Pi) is costly and tends to exacerbate the problems of P pollution in the waste stream.
Mutation breeding has lowered seed PA with equivalent increases in seed phosphate in a number of cereal grains (Raboy et al., 2002). Myoinositol-1-phosphate synthase (MIPS) catalyzes the conversion of Glc-6-P to myoinositol-1-phosphate (MIP), which is converted to PA by subsequent phosphorylations. Hitz et al. (2002) identified a missense mutation in the soybean (Glycine max) MIPS structural gene (GmMIPS1). The mips1 line exhibited a 50% reduction in seed PA, with a concomitant increase in Pi. In addition, there was a reduction in galactinol and in raffinosaccharide antinutritionals because MIP can also be dephosphorylated to inositol, essential for their synthesis.
Several other loci are mutational targets for lowering seed PA. Wilcox et al. (2000) identified a mutant line (CX1834) in soybean in which the ratio of Pi to equivalent P in PA increased from about 15% in the progenitor to well over 50%, while total P did not appreciably vary from that of the progenitor (approximately 5 μg P mg−1). The low-PA trait of CX1834 is due to recessive mutations at two interacting, unlinked loci (Walker et al., 2006), neither of which appears to be the seed-expressed GmMIPS1 (Chappell et al., 2006). The maize (Zea mays) low-PA lpa2 and lpa3 mutants have been shown to be defective in MIP kinase (Shi et al., 2003) and myoinositol kinase (Shi et al., 2005), respectively. In barley (Hordeum vulgare), at least four independently induced lpa mutations appear to be nonallelic (V. Raboy, personal communication). Initially, a MIPS defect appeared to be responsible for the low-PA maize mutant Zmlpa1, with reports of genetic colocalization of a maize MIPS gene and the low-PA phenotype (Larson and Raboy, 1999; Raboy et al., 2002). Subsequently, the Zmlpa1 lesion was found to be in an ATP-binding cassette (ABC) transporter, and embryo-specific suppression of the maize lpa1 gene resulted in some lines approaching 100% reduction in PA that lacked some of the deleterious properties of the original lpa1 mutant (Shi et al., 2007). Notably, lines with high germination and yield could be identified among low-PA transgenic selections of ABC transporter-suppressed maize lines (Shi et al., 2007).
Under certain conditions, reduction in seed PA can be deleterious to the plant. Nunes et al. (2006) presented evidence that complete RNA interference (RNAi) knockdown of GmMIPS1 expression resulted in aborted soybean embryos. While it is not clear whether the RNAi construct affected members of the MIPS family expressed in vegetative tissue, transgenic plants appeared normal, while their progeny zygotes inheriting the RNAi construct exhibited early embryo abortion. lpa traits have been associated with yield reductions in maize (Ertl et al., 1998) and in rain-fed barley (Bregitzer and Raboy, 2006).
A commercially feasible, nonmutational strategy to increase utilization of Pi and bound minerals in PA, as well as to reduce P pollution, is to treat soymeal and other animal feeds with commercial phytase, usually a recombinant form encoded by PhyA of Aspergillus niger. Pen et al. (1993) demonstrated that transgenic tobacco seeds could be a source of PhyA phytase expressed under the control of a constitutive promoter (cauliflower mosaic virus 35S) and directed to the extracellular space (apoplast). Plant-expressed PhyA apoplastic phytase has been developed as a patented feed supplement in canola seeds (Phytaseed) and in alfalfa (Medicago sativa; in which phytase is a component of alfalfa juice; for review, see Grabau, 2002). Chen et al. (2007) recently successfully expressed A. niger PhyA2 in maize embryos and obtained measurable seed phytase activity (approximately 2 units/g seed). Even the transgenic maize lines with the highest levels of phytase expression did not have germination or yield defects, although these lines had modest decreases in seed PA.
An alternative transgenic strategy to seed production of active phytase enzyme is to aim for a direct reduction of PA in feed grains. This approach can target PA synthesis as in the RNAi knockdown of GmMIPS1 or the ABC transporter described above (Nunes et al., 2006; Shi et al., 2007). While interference with MIPS appears to have negative consequences for germination, kernel-specific suppression of the ABC transporter in maize produced lines with reduced PA but no germination or yield defects (Shi et al., 2007).
Another popular transgenic strategy is to direct expression of a phytase during embryo development at the site of PA synthesis or storage. Thus, Chiera et al. (2004) achieved the ectopic expression of a soybean seedling phytase (Hegeman and Grabau, 2001) driven by a storage protein (β-conglycinin) promoter, and observed an 8% reduction of PA, 3-fold increase in Pi, and retention of total seed P. These values may underrepresent the impact of the ectopic phytase trait because an HPLC analysis of individual inositol phosphate esters revealed that PA was reduced by as much as 25%. However, 8% to 25% reductions of PA and PA conversion to Pi (3-fold increase) were significantly less than the corresponding values reported for the CX1834 mutant: 50% to 60% PA reduction and a 4- to 17-fold increase in Pi (Wilcox et al., 2000).
We previously transformed Arabidopsis (Arabidopsis thaliana) with an embryo-expressed phytase. We chose APPA, a periplasmic phytase encoded by the Escherichia coli appA gene (Dassa et al., 1990; Greiner et al., 1993). To improve APPA accessibility to seed PA, we employed a construct (Cho et al., 1995) to direct APPA to the vacuole (Coello et al., 2001). While significant reductions in seed PA were observed in two independent transgenic lines, there was not a compensatory increase in Pi. In this study, we employed a cytoplasmic version of APPA in soybean. One transgenic line exhibited high levels of phytase activity, ≥90% reduction in seed PA, and no loss of total seed P. In this article, we demonstrate that cytoplasmic expression of an active phytase enzyme in developing soybean seeds results in the conversion of nearly all seed PA to Pi and produces abundant active enzyme in mature seeds capable of liberation of Pi in admixed meals.
RESULTS
Soybean Seeds Expressing a Recombinant Phytase
Soybean (‘Jack’) was transformed (Zhang et al., 1999; Staswick et al., 2001; Zeng et al., 2004) with constructs containing the E. coli appA open reading frame from which the 22-amino-acid periplasmic signal sequence was removed. Expression was driven by the soybean lectin promoter with either an in-frame soybean lectin signal sequence directing the enzyme to the protein storage vacuole (PSV; VAPPA) or with an ATG replacing the lectin signal sequence (Cho et al., 1995). This latter construct, CAPPA, was predicted to encode a cytoplasmic phytase. The VAPPA construct was previously used to express phytase in transgenic Arabidopsis (Coello et al., 2001).
Of 14 CAPPA and 17 VAPPA independent lines, T1 seeds of only five CAPPA lines revealed individuals with high available phosphate (Raboy et al., 2002) of up to 6 μg P mg−1 compared to the progenitor seed with approximately 0.5 μg P mg−1. The presence of CAPPA seed protein was examined by immunoblot (western) analysis with antibodies to recombinant CAPPA. No cross-reacting protein was detected in lines with normal seed Pi levels or in three of the five lines with higher Pi. However, mature seeds of the two lines with the highest Pi exhibited cross-reacting CAPPA protein (data not shown).
To confirm the immunodetection of CAPPA, phytase enzyme activity was determined in crude protein extracts from at least three mature seeds of each transgenic line. For samples from most of the lines, phytase activity was nearly undetectable (<0.1 μmol Pi min−1 mg protein−1), while five CAPPA lines had enzymatic activity ranging from 0.5 to 326 μmol Pi min−1 mg protein−1. There was a complex relationship among high-Pi phenotype, cross-reacting APPA protein, and phytase enzyme activity in mature seeds. However, high-phytase-specific activity seeds of line CAPPA 14 consistently had high Pi and cross-reacting APPA protein.
Genetic Characterization of CAPPA Event 14
The inheritance of Pi in CAPPA seeds was assessed over multiple generations of selfed plants originally derived from two T1 individuals from CAPPA event 14: 14H and 14N (Table I). Line 14N produced T3 seeds that segregated for the high-Pi phenotype, which, in turn, cosegregated with high-phytase-specific activity; no phytase activity was detected in the seeds with normal Pi levels. In contrast, all seeds tested from selfed line 14H had high Pi and significant phytase enzyme activity. High seed phosphate was stably inherited in each generation between T1 and T5 for line 14H. The inheritance data suggest, therefore, that T2 individuals 14H and 14N were homozygous and hemizygous, respectively, for the functional CAPPA transgene. PCR detection of the CAPPA gene sequence among F2 individuals of a cross of 14H by low-PA line CX1834 (Wilcox et al., 2000) confirmed that 72% carried the CAPPA gene, close to the 75% expected for homozygosity for a single CAPPA insertion in line 14H (data not shown). Further characterization was carried out on progeny of CAPPA line 14H, hereafter referred to as CAPPA.
Table I.
Transmission of high-phosphate phenotype over five generations derived from two seeds (14H and 14N) harvested from CAPPA T0 plant 14
| Seed Isolate | Generation
|
||||
|---|---|---|---|---|---|
| T1 | T2 | T3 | T4 | T5 | |
| μg P mg−1 seed (Pi) | |||||
| 14H (CAPPA/CAPPA) | 6.63a (1) | 2.52a (4) | 4.98a (26) | 4.77a (6) | 6.00b |
| 14N (CAPPA/−) | 4.51a (1) | 3.31a (4) | 5.35c (19); [0.23 (7)] | ND | ND |
Values are averages of determinations of seed chips (n) from given generation.
Value is averaged from determinations of pools of ground whole seeds from T4 plants.
Values separated into averages for high and low (brackets) phosphate from chips of eight to 10 seeds from three T2 plants.
PA Content in CAPPA
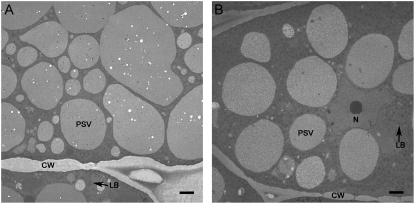
PA content was assessed chemically and by electron microscopy. In soybean seeds, PA is complexed with mineral salts in phytin, located within PSVs. Phytin crystals are electron-dense globoid inclusions (Lott and Buttrose, 1978; Prattley and Stanley, 1982; Chen et al., 1998). Transmission electron micrographs of PSVs in the cotyledons of nontransformed soybean (‘Williams 82’) revealed the presence of numerous globoid crystals. When seeds are subjected to conventional chemical fixation, water-solubilized phytin deposits in the globoid crystals are not well preserved, resulting in the appearance of cavities. Globoid crystals (cavities) in the PSVs of mature T3 generation seeds of CAPPA were significantly less frequent than those of nontransformed seed (Fig. 1).
Figure 1.
Electron microscopic analysis of mature seeds. Cotyledon sections of ‘Williams 82’ (A) and CAPPA (B) represent genotypes with normal and low PA levels, respectively. Phytin globoid cavities are the small white circular areas present in the ‘Williams 82’ PSVs. cw, Cell wall; N, nucleus; LB, lipid bodies.
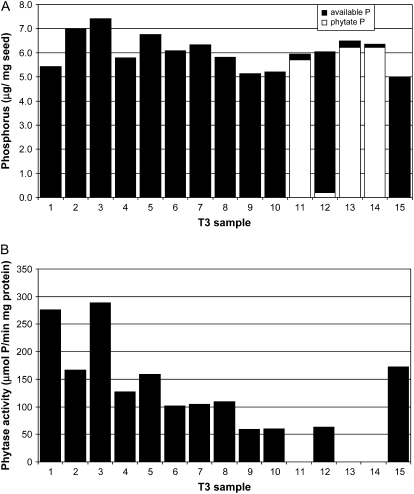
In the T3 and subsequent generations, PA was also quantified by a modification of a high-performance ion chromatography method (HP-IC; Chen and Li, 2003) that distinguishes inositol phosphate metabolism products from intact PA. While PA levels of control seeds typically ranged from 20 to 28 μg PA mg−1, PA in CAPPA samples was consistently at or below the detection limit (2 μg PA mg−1), thus representing ≥90% reduction in seed PA (Fig. 2). Neither the control nor CAPPA soybeans contained detectable IP5 (not shown). The chromatographic profiles agreed with those of CX1834, which did not appear to contain inositol phosphate intermediates (Wilcox et al., 2000). The sum of P in Pi and PA was relatively constant for individual seeds containing either high or low levels of phytase enzyme activity.
Figure 2.
Reduction in PA P correlates with increased Pi and phytase enzyme activity in mature CAPPA seed samples. A, Individual mature seed samples were subjected to three analyses: PA P, Pi P, and phytase enzyme activity. The sum of Pi P and PA P is represented for each T3 seed sample. Samples 1 through 10 were derived from 14H (CAPPA/CAPPA), while samples 11 to 15 were derived from 14N (CAPPA/−) and were segregating for the transgene event. B, Phytase enzyme activity was determined for individual seed samples carried out at 55°C.
Subcellular Localization of CAPPA Phytase
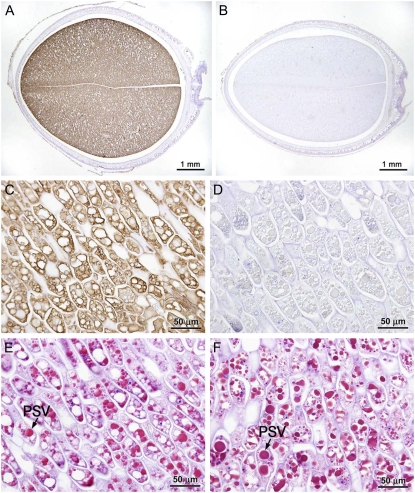
Cotyledon sections of mature seeds were examined by immunohistochemical analysis employing antibodies against recombinant APPA. Seed phytase accumulated throughout the cotyledon of CAPPA seed (Fig. 3A), while none was detected in the progenitor seed (Fig. 3B). Because the CAPPA lines expressed phytase without a signal peptide, we expected phytase to have a cytoplasmic location. Higher magnification observation of CAPPA seeds challenged with phytase antibodies revealed prominent labeling in the cytoplasm (Fig. 3C). Preimmune serum did not show this labeling. Slight, but consistent, background labeling of PSVs by preimmune serum (Fig. 3D) was due probably to antibody against seed storage proteins present in rabbit meal. Parallel sections stained with Schiff's reagent confirmed the anatomical features of CAPPA seed (Fig. 3, E and F).
Figure 3.
Distribution of CAPPA antigen in immature (R6) CAPPA line cotyledons. A and B, Total cotyledon sections of CAPPA (A) and progenitor cultivar ‘Jack’ (B) immunostained with anti-APPA serum and counterstained with haematoxylin. C and D, Photomicrographs of a similarly treated longitudinal cotyledon section of CAPPA (C) at higher magnification and a control section treated with preimmune serum (D). E and F, Parallel sections of CAPPA and ‘Jack’ were stained with Schiff's reagent to observe the anatomical features.
Seed Composition of CAPPA and Progenitor
CAPPA germination and plant development appeared normal in the greenhouse environment. Greenhouse-produced T5 CAPPA and the lines ‘Jack’ (progenitor genotype), ‘Williams 82’, and the low-phytase mutant CX1834 in the ‘Athow’ background (Wilcox et al., 2000) were compared with respect to seed components: protein, oil, Pi, PA, raffinose family oligosaccharides, and phytase enzyme activity (Table II) and mineral composition (Table III). There were minor or no significant differences for the seed parameters unrelated to PA. Total P content was similar for all lines; however, the CAPPA seeds contained no detectable PA and much more Pi. ‘Jack’ and ‘Williams 82’ had significantly higher PA and lower Pi contents. Line CX1834 had intermediate PA and Pi. Seed phytase-specific activity (phytase units per milligram protein) was much higher in the CAPPA seed.
Table II.
Seed composition components of greenhouse-grown CAPPA, CX1834, ‘Williams 82’, and ‘Jack’
| Cultivar/Line | Phytase Activity | PA | Pi P | S:R + Sta | Protein | Oil |
|---|---|---|---|---|---|---|
| μmol P min−1 mg−1 protein | μg/mg seed | g/kg | ||||
| ‘Jack’ | 0.002 | 26.27 | 0.1294 | 1.25 | 38.89 | 20.20 |
| CAPPA | 53.128 | 0.00 | 6.492 | 1.12 | 40.20 | 18.65 |
| CX1834 | 0.092 | 5.32 | 4.683 | 1.21 | 38.30 | 20.25 |
| ‘Williams 82’ | 0.026 | 23.72 | 0.245 | 0.79 | 38.89 | 21.20 |
| lsd0.05 | 7.974 | 1.099 | 0.758 | 0.10 | 1.736 | 0.886 |
Ratio of Suc to raffinose plus stachyose.
Table III.
Major and minor elemental analysis of greenhouse-grown CAPPA, CX1834, ‘Williams 82’, and ‘Jack’
| Cultivar/Line | N | P | K | Ca | Mg | Na | Zn | Fe | Mn | Cu | B | Al | Mo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | ppm | ||||||||||||
| JACK | 5.53 | 0.79 | 2.44 | 0.49 | 0.28 | 0.03 | 71.84 | 37.34 | 37.22 | 6.32 | 35.12 | 1.71 | 2.61 |
| CAPPA | 5.64 | 0.85 | 2.45 | 0.43 | 0.26 | 0.01 | 72.74 | 38.62 | 28.10 | 5.96 | 38.38 | 3.64 | 5.08 |
| CX1834 | 5.31 | 0.80 | 2.49 | 0.48 | 0.24 | 0.01 | 85.78 | 58.52 | 30.26 | 8.64 | 37.06 | 3.32 | 4.98 |
| ‘Williams 82’ | 5.58 | 0.77 | 2.51 | 0.51 | 0.27 | 0.01 | 75.78 | 66.24 | 38.76 | 10.92 | 35.23 | 2.98 | 4.73 |
| lsd0.05 | 0.20 | 0.03 | NS | 0.06 | 0.01 | 0.01 | 4.12 | 10.08 | 2.51 | 1.24 | NS | 1.68 | 0.80 |
Germination, Field Emergence, and Seed Number of CAPPA
Standard laboratory germination tests for field-produced seeds revealed a lower germination percentage for CAPPA compared with the progenitor, ‘Jack’. However, there was no significant difference in field emergence of field- or greenhouse-produced ‘Jack’, CAPPA, ‘Williams 82’, or CX1834 seeds (Table IV). In addition, when grown under noncompetitive field conditions, CAPPA seeds did not differ significantly from ‘Jack’ in seed weight or in seeds per plant (Table IV). CAPPA seed P-related traits were tested in sequentially propagated field generations (next section).
Table IV.
Germination and yield components of CAPPA, CX1834, ‘Williams 82’, and ‘Jack’
| Cultivar/Line | Field Emergence | Standard Germination | Seed Weight | Seed No. |
|---|---|---|---|---|
| % | mg/seed | seeds/plant | ||
| ‘Jack’ | 42.9a | 72.9b | 112.7 | 468 |
| CAPPA | 31.2 | 54.8 | 120.2 | 534 |
| CX1834 | 41.7 | 97.5 | 143.5 | 413 |
| ‘Williams 82’ | 49.4 | 93.4 | 131.9 | 531 |
| lsd0.05 | NSc | 6.9 | NS | NS |
Combined means of 2006 and 2007 field data.
Values derived from four replications of 100 seeds from 2006 and 2007.
No significant differences.
Production of Sequential Generations of CAPPA in a Field Environment
Three generations (T3, T4, and T5) of CAPPA were grown in the same field environment to determine their field phenotypes and phenotypic stability. CAPPA seeds germinated and the plants developed normally in the field environment. Field-produced mature dry seeds, generations T4, T5, and T6, were assayed for PA, Pi, and phytase enzyme activity. Seed phosphate levels were high and PA levels low or undetectable, similar to the seed composition values obtained for greenhouse-grown plants (Table V). Phytase enzyme activity levels remained high in the field-advanced generations. There was no significant reduction in Pi, PA, or phytase activity in seeds from any of the field-advanced generations. Seeds of ‘Jack’ produced in the field environment had values for PA, Pi, and phytase enzyme activity similar to those of ‘Jack’ seeds produced in the greenhouse environment.
Table V.
Seed phytase and phosphorus components of three generations of field-grown CAPPA and ‘Jack’
Means are shown plus or minus 1 sd.
| Cultivar/Line | Phytase Activity | PA | Pi P | PA P | Total Pa |
|---|---|---|---|---|---|
| μmol P min−1 mg−1 protein | μg/mg seed | ||||
| ‘Jack’ | 0.00 | 14.72 ± 2.8 | 0.66 ± 0.7 | 4.15 ± 0.8 | 4.81 |
| CAPPA T4 | 176 ± 17 | 0.00 | 5.09 ± 0.4 | 0.00 | 5.09 |
| CAPPA T5 | 184 ± 7 | 0.00 | 5.08 ± 0.5 | 0.00 | 5.08 |
| CAPPA T6 | 191 ± 16 | 0.00 | 4.55 ± 1.2 | 0.00 | 4.55 |
Sum of Pi P and PA P.
CAPPA Meal as an Alternative to Fungal Phytase for Digesting PA in Normal Meals
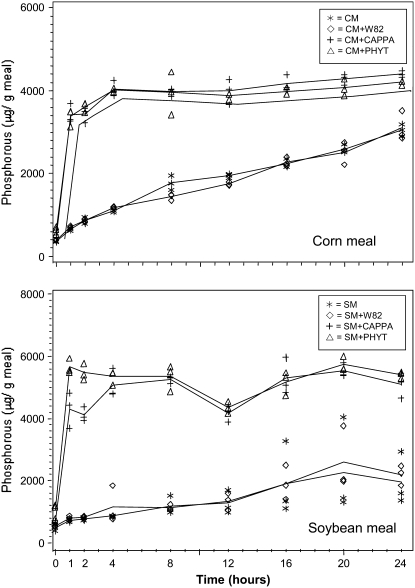
Fortification of animal feeds with phytase enzyme is an accepted practice to reduce PA and increase Pi and minerals in the feed. We evaluated the ability of ground CAPPA seed to provide phytase enzyme activity in an in vitro P liberation assay using commercial cornmeal or soybean meal as the PA P source. Cornmeal and soybean meal were supplemented with a commercial source of phytase, ground CAPPA seed, or ground commodity soybean seed (‘Williams 82’; all added as 4 mg/g meal) and compared to samples containing no additive (Fig. 4). In both the CAPPA and the commercial phytase treatments, Pi was rapidly liberated, to the same extent, from both cornmeal and soybean meal, reaching a plateau within 4 h. Both treatments with CAPPA and commercial phytase additives released significantly more P than control ‘Williams 82’ additive or the no additive controls (P < 0.001). The gradual increase in P liberated in the ‘Williams 82’ mock treatment and in the no-treatment control may have been due to microbial activity because reactions were at 37°C under nonsterile conditions.
Figure 4.
Release of phosphate from soybean meal by CAPPA meal and fungal phytase. Commercial cornmeal (CM) or soybean meal (SM) was incubated without additions or with additions (4 mg/g meal) of ground ‘Williams 82’ soybean (CM+W82; SM+W82), of ground CAPPA soybean (CM+CAPPA; SM+CAPPA), or of commercial phytase (CM+PHYT; SM+PHYT). Lines are plotted means of phosphate determinations for each treatment and time.
DISCUSSION
Several mutational and transgenic strategies to reduce PA in soybean have negative aspects. The CX1834 line of soybean (Wilcox et al., 2000) exhibits a ratio of Pi P to equivalent P in PA of over 50% compared to 15% in the progenitor (‘Athow’). Though considerable, this increase is still less than the much higher ratio observed in CAPPA, in which PA is often undetectable while total P is not reduced. A drawback to the low-PA trait of CX1834 is that it is due to recessive mutations at two interacting, unlinked loci (Walker et al., 2006). Because the superior Pi P to PA P ratio in CAPPA appears to be due to a single active insert, moving the dominant CAPPA gene into a new variety is more efficient than selecting one of 16 F2 individuals in outcrosses of CX1834.
The soybean mips1 line exhibited a 50% reduction in seed PA as well as reductions in galactinol and raffinosaccharides (Hitz et al., 2002). In a thorough study of genotype versus environmental effects, Meis et al. (2003) found that mips seed had a lower field emergence, especially seed produced in a subtropical versus temperate environment. Low germination might be due to lowered PA or to lowered raffinose series sugars. It has been suggested that lowered germination of in vitro-generated zygotic and somatic soybean embryos is due to lowered raffinose series sugars (Chanprame et al., 1998; Obendorf et al., 1998). In our experiments, there was an observed decrease in CAPPA germination, and the field emergence results suggest more investigation is required to understand the effect of such drastic reductions in seed PA. That transgenic maize lines were produced with substantial reductions in PA without defects in germination calls into question the necessity of PA in the germination process (Shi et al., 2007). Recently, several lines of evidence have been reported that would support a model in which high levels of PA are not an absolute requirement for efficient seed germination or emergence (Shi et al., 2007; Spear and Fehr, 2007). Nevertheless, the exact role of PA in soybean germination will require further investigation, especially in additional soybean lines expressing the CAPPA transgene.
CAPPA is superior to lowered PA achieved by ectopic expression of a seedling phytase in developing soybean embryos, as reported by Chiera et al. (2004). They observed an 8% to 25% reduction of PA and only a 3-fold increase in Pi, whereas CAPPA seeds exhibit little or no PA (Table I). The result in CAPPA may be due to one or more factors, including the very high level of phytase expression (>1,000 units g−1 mature CAPPA seed), location of the enzyme in the cytoplasm, presumably the site of PA synthesis, or other unknown factors. It is likely that production of active enzyme at the appropriate location and developmental stage as well as its extremely high expression are critical factors that allow the combination of substantial reductions in PA with high remnant enzyme activity in mature seeds. As an example, the CAPPA line contained approximately 500-fold more enzyme activity than the maize PhyA2 lines (Chen et al., 2007).
The high phytase activity of CAPPA seeds may make its meal a suitable replacement of commercial fungal phytases (product of A. niger PhyA). Ground CAPPA seed as an additive was as effective as commercial phytase in lowering PA in cornmeal and soymeal (Fig. 4). Rodriguez et al. (2000) reported that three site-directed mutations improved both thermostability and kcat in APPA expressed by the methylotrophic yeast, Pichia pastoris. This altered APPA was shown to be an effective addition to swine diets deficient in Pi (Veum et al., 2006). Transgenic expression of this and other altered forms of APPA, forms that have increased acid and heat resistance and improved catalytic properties, should lower dietary PA within digestive systems of monogastric animals, thus obviating the need for pretreatment of meal.
CONCLUSION
The lack of detectable PA in CAPPA seeds had no significant deleterious effect on seed weight, field emergence, or seed number/plant under noncrowded field conditions. Additional germination and field studies, under a variety of conditions, with additional events are needed to test the effect of virtual quantitative conversion of PA to Pi. The high phytase activity of CAPPA seeds was as effective as commercial phytase in lowering PA in cornmeal and soymeal, making CAPPA meal a suitable replacement for commercial fungal phytases (product of A. niger PhyA). The unique combination in CAPPA of nearly quantitative conversion of PA to Pi and high levels of remnant phytase activity in mature seed meal is a distinct improvement over other lines developed via transgenic or mutagenic strategies which, respectively, either lower seed PA or produce seed phytase as single traits.
MATERIALS AND METHODS
Transformation of Soybean
DNA fragments containing soybean (Glycine max) seed lectin promoter-phytase constructs, with or without the lectin signal sequence, were excised from the plasmids pBinGLPH7-5 (Coello et al., 2001) and pBinGlep5, respectively, and subcloned into the XbaI site of the binary vector pZY101 (http://plantsci.missouri.edu/muptcf/pzy101.html). The resulting vectors, designated as pVAPPA and pCAPPA, respectively, were mobilized into the Agrobacterium strain EHA101 (Hood et al., 1986) by the freeze-thaw method (Holsters et al., 1978). Soybean ‘Jack’ was subjected to Agrobacterium-mediated cotyledonary node transformation using the bar gene as a selectable marker and the herbicide glufosinate as a selection agent, following the procedure of Zeng et al. (2004).
Fourteen CAPPA and 17 VAPPA independent T0 plants were recovered, and mature T1 seeds were screened for a high “available” phosphate (Pi), a trait that has been inversely associated with seed PA levels (Raboy et al., 2002). Seeds of all of the VAPPA lines and of nine of 14 CAPPA lines retained the phosphate levels (≤0.5 μg P mg−1) typical of the progenitor. In contrast, analysis of samples of the remaining five CAPPA lines revealed individual seeds with high Pi containing up to 6 μg P mg−1.
Plant Growth
T1 through T5 seeds were produced in either greenhouse with supplemental lighting or in growth chamber environments. Greenhouse settings were 16-h daylength and 30°C/18°C day/night temperatures. Growth chamber settings included 14.5-h daylength and 28°C/22°C day/night temperatures. Plants were grown in 6-inch or 2-gallon pots in PRO-MIX (Premier Horticulture) medium and fertilized with Miracle-Gro or Osmocote Plus (Scotts) according to the manufacturer's recommendations.
Independent field studies were conducted consisting of a field emergence study and yield component study. Four soybean entries, ‘Williams 82’, ‘Jack’, CAPPA, and CX1834-1-3, were grown at the Bradford Research and Extension Center, near Columbia, MO, in 2006 and 2007. The emergence study plot was disced and cultivated using typical agricultural methods. The experimental design was a randomized complete block with four replications and rows on 0.8-m centers. Seeds were hand planted into rows at a depth of 3 cm every 30 cm to maximize stand count accuracy and minimize seedling competition and spread of disease. Emerged seedlings were counted at 2 weeks after planting (TeKrony and Egli, 1977).
The yield component study was cultivated in 2006 and 2007 as described previously. Experimental design was a randomized complete block with rows as previously described. Seeds inoculated with Rhizobium japonicum were hand planted at a depth of 3 cm. Seedlings were later thinned to obtain one plant every 0.8 m. Plants were harvested at the R8 stage (Fehr and Caviness, 1977) by single plant threshing and stored at 4°C until analysis. Seeds per plant and 100 seed weights were recorded. Standard germination tests were conducted based on the standards of the Association of Official Seed Analysts (1993).
In addition, T4 through T6 CAPPA seeds were produced at a research field location in Columbia, MO in the summer of 2006. Seeds were inoculated and hand planted in rows on 11.8-cm centers, and plants were irrigated as needed.
Immunostaining of Paraffin Sections
The APPA open reading frame was PCR amplified with Taq polymerase (Promega) so that it lacked 27 N-terminal amino acids, and the 28th codon was converted from CTG (Leu) to ATG (Met), thus creating an NdeI site. Cloning of the amplified product into NdeI, NotI-digested pET28a (Novagen-Merck) created an in-frame N-terminal His(6×) tag with APPA (N-His-APPA). Expression in mid-log cells of the Escherichia coli HMS174 host was induced with 1 mm β-isopropyl thiogalactoside and further growth for 2 h. Cells were disrupted by sonication in 6 m urea, 500 mm NaCl, 5 mm imidazole in 20 mm Tris-HCl, pH 7.9, as recommended by the manufacturer of the Ni2+ affinity column (iminodiacetic acid-substituted His-Bind Resin; Novagen). After removing insolubles, the supernatant was poured over the column that was subsequently washed in the sonication buffer. Bound N-His-APPA was eluted with 1 m imidazole in the same buffer and dialyzed against 10% glycerol, 1 m urea in phosphate-buffered saline. Approximately 0.5 mL (0.16 mg N-His-APPA) was injected subcutaneously into each of two New Zealand white rabbits. Two weeks after a booster injection, rabbits were exsanguinated; the serum fraction, collected and preserved at −20°C in 50% glycerol, was used in immunostaining.
Field-grown soybean seeds at R6 stage (Fehr and Caviness, 1977) were separated into cotyledonary halves and immediately fixed in 50% ethyl alcohol, 5% glacial acetic acid, and 10% formaldehyde for 24 h at 4°C. The seed tissue was dehydrated sequentially in a graded ethanol/xylene series and infiltrated with paraffin. Sections were cut with a microtome and collected on poly-l-lysine-coated slides. Following the removal of paraffin with xylene, the sections were processed for immunostaining. To inactivate endogenous peroxidase activity, the deparaffinized sections were first treated with methanol-hydrogen peroxide. After incubating with 5% goat serum and 1% bovine serum albumin, the sections were further incubated 20 min in 1:500 diluted E. coli phytase antiserum. Sections were then treated sequentially with biotinylated linker, streptavidin conjugated to horseradish peroxidase, and substrate-chromogen solution according to the manufacturer's recommendation (DAKO). The sections were counterstained with hematoxylin and observed with bright field optics.
Transmission Electron Microscopy
Dormant soybean seeds were imbibed in water and incubated at 30°C for 12 h. Seeds were cut into small pieces (1–2 mm) with a double-edged razor blade and fixed immediately in 2.5% glutaraldehyde buffered with 50 mm sodium phosphate, pH 7.2. Fixation was carried out at room temperature for 4 h. The tissue samples were washed four times at 15-min intervals with 50 mm phosphate buffer, pH 7.2, and post-fixed, 1 h at room temperature, with 2% aqueous osmium tetroxide. After extensive rinses in distilled water, the samples were dehydrated in a graded acetone series and infiltrated with Spurr's resin essentially as described (Krishnan et al., 1986). Thin sections were cut with a diamond knife and collected on uncoated copper grids. Sections were stained with 0.5% aqueous uranyl acetate and 0.4% aqueous lead citrate and viewed with a JEOL JEM 100B electron microscope at 100 kV.
Analytical Techniques
Pi was quantified by slight modifications of the method described by Wilcox et al. (2000). Single seeds were either ground to a powder in liquid nitrogen or chipped with a scalpel to remove an approximately 10-mg portion distal to the axis. Subsequent immunohistochemical analysis revealed a uniform distribution of CAPPA throughout the embryo (Fig. 3), validating the values obtained from seed chips. As a further control, single seed chip analysis was compared to that of the remaining seed that was processed by grinding. No differences in P values were detected. Ground samples (8–15 mg) or intact seed chips were extracted overnight by shaking at 4°C in 0.5 mL of extraction buffer [12.5% (w/v) TCA, 25 mm MgCl2]. Particulates were allowed to settle at room temperature for 30 min to 1 h. Ten microliters of sample supernatant was combined with 90 μL of water and 100 μL of colorimetric reagent [1 volume of 3 m H2SO4, 1 volume of 0.02 n ammonium molybdate, 1 volume of 10% (v/v) ascorbic acid, and 2 volumes of water] in a 96-well plate well, allowed to incubate at room temperature 1.5 h, and then analyzed at 825 nm in a Versamax tunable microplate reader (Molecular Devices). A standard curve was generated with K2HPO4 standards. Results were converted to μg P mg seed−1.
PA was quantified by modified HP-IC method (Chen and Li, 2003). Seed samples (0.025 g), powdered in liquid nitrogen, were extracted 1 h by shaking at room temperature in 0.5 mL of 0.5 n HCl. After centrifugation, 15 min at 15,000g, supernatants were filtered through a 0.22-micron filter, and 100 μL of filtrate was analyzed. PA and inositol polyphosphate separations were performed by a linear gradient elution program on a Dionex CarboPac PA-100 guard column and a CarboPac PA-100 analytical column on an Agilent 1100 series HPLC system. The elution gradient was effected by a mixture of two eluents: water and 0.5 n HCl; time 0 min, 8% 0.5 n HCl; time 30, 100% 0.5 n HCl; time 35, 100% 0.5 n HCl; time 35.1, 100% 0.5 n HCl; time 40, 8% 0.5 n HCl. A post-column derivitization was achieved with a solution of 1 g L−1 Fe(NO3)3 in 0.33 m HClO4 using a 750-μL knitted coil and was followed by detection of A295. Flow rates of eluent and post-column solution were 1.0 and 0.4 mL min−1, respectively. PA standard (PA dipotassium salt; Sigma) eluted at 30 min, while a myoinositol-1,3,4,5,6-pentakisphosphate (Sigma) standard eluted at 23 min. Standard curves were calculated from dilutions of PA standards. Results were converted to μg PA mg seed−1 or PA P mg seed−1.
Protein, oil, and moisture concentration were determined by near infra-red spectroscopy according to standard procedures summarized by Dyer (2004). Chemometric analysis included modified partial least squares regression as described by Westerhaus et al. (2004).
For micro- and macronutrient analyses, dried ground seed (0.5 g) was ashed 5 h at 500°C and dissolved in 10 mL of 6 n HCl. Digested samples were diluted with water, filtered, and analyzed for potassium, boron, calcium, Mg, Zn, sodium, Fe, Mn, aluminum, molybedenum, and Cu by ICP-OES (Varian), nitrogen by thermal conductivity of nitrogenous gases with a Leco model FP-428 nitrogen analyzer (Leco), and P by colorimetric analysis (Murphy and Riley, 1962).
Oligosaccharides were determined by HP-IC with pulsed amperometric detection employing Agilent 1100 series HPLC and an ESA Coulochem III detector (Agilent Technologies). A 12.5-mg ground seed sample was extracted with 0.5 mL of 50% ETOH at 70°C for 30 min. Samples were then centrifuged 15 min at 16,000g. The supernatant was passed through a 0.2-μm filter. Sugars were separated on a Dionex Carbo Pac PA 10 analytical column (250 mm × 4 mm, 10 μm) connected to a Carbo Pac PA 10 guard column (50 mm × 4 mm). The mobile phase was 90 mm NaOH with a flow rate of 1.5 mL min−1 maintained at 30°C. Detection settings were: time 0, 0.1 v; time 0.41, −2.0 v; time 0.42, 0.6 v; and time 0.44, −0.1 v.
Phytase Assay
Soybeans were powdered in liquid nitrogen, and 25 mg of either fresh or lyophilized sample was extracted in 1 mL of enzyme buffer (0.1 m sodium acetate, pH 5.5, 1 mm CaCl2, and 0.1 g L−1 Tween 20) by thorough vortex mixing, followed by incubation on ice 10 min. Samples were centrifuged 5 min at 15,000g, 4°C. The supernatants were then either diluted up to 50-fold in enzyme buffer or sampled without further dilution. With solutions cooled on ice, a 40-μL aliquot of sample was combined with 680 μL of reaction buffer (0.1 m sodium acetate, pH 4.5) and 80 μL of 12.5 mm PA. For the zero time point, 125 μL of the mixture was removed and mixed with 125 μL of cold 20% TCA, vortexed, and placed on ice. The remaining reactions were incubated 15 min in a 55°C or 37°C water bath. Reactions (125 μL) were stopped as for the zero time point. All samples were centrifuged 5 min at 15,000g, 4°C. Pi content was quantified as above with 100 μL of sample combined with 100 μL of colorimetric reagent, except that standards were assayed in 10% TCA rather than water. Extracted protein was quantified against bovine serum albumin standard according to the recommendations of the manufacturer of the Bio-Rad protein assay. Results were converted to phytase specific activity: μmol Pi released min−1 mg protein−1.
In Vitro Phosphorus Liberation from Meals Mixed with CAPPA Meal and Commercial Phytase
Coarse-ground whole seed of CAPPA (T4 generation) and ‘Williams-82’ were ground to fine powders in a small grinder (SmartGrind; Black & Decker). Commercial phytase (Natuphos 5,000 units g−1; BASF) was ground to a fine powder in a mortar. Fine-ground CAPPA, ‘Williams 82’ seed, or commercial phytase (40 mg) was mixed with 10 g commercially available soybean meal and cornmeal (MFA). One-gram samples of mixed and control meals were suspended in 10 mL of reaction buffer (0.1 m sodium acetate, pH 4.5) in 15-mL screw-top plastic tubes and incubated at 37°C on a rotary shaker. One tube in each treatment was randomly sampled at 0, 1, 2, 4, 8, 12, 16, 20, or 24 h and 400 μL was centrifuged, 200 μL of the supernatant was transferred to a fresh tube, mixed with 200 μL 20% TCA, and stored on ice prior to storage at −20°C. Samples were thawed and centrifuged 5 min at 15,000g, 4°C and 10 μL of supernatant was combined with 90 μL of 10% TCA in 0.1 m sodium acetate, pH 4.5, and 100 μL of colorimetric reagent (see above). Pi was quantified as above, except that standards were assayed in 0.05 m sodium acetate, pH 4.5, containing 10% TCA rather than water. Results were converted to μg P g meal−1. Treatments and controls were done in triplicate.
Data were analyzed by PROC Mixed routine (SAS, 1990) for a split plot design with three replications. Treatment means per sampling time were analyzed by Fisher's lsd tests to determine significant differences among means (P < 0.05). Graphs of P plotted against sampling time were generated by GPLOT routine.
Acknowledgments
We thank Sean Pfaff, Paul Little, and Justin Kleffner for excellent technical assistance.
This work was supported by the National Center of Soybean Biotechnology (University of Missouri).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Joe C. Polacco (polaccoj@missouri.edu).
Open Access articles can be viewed online without a subscription.
References
- Association of Official Seed Analysts (1993) Rules for testing seeds. Journal of Seed Technology 16 1–113 [Google Scholar]
- Bregitzer P, Raboy V (2006) Effects of four independent low-phytate mutations on barley agronomic performance. Crop Sci 46 1318–1322 [Google Scholar]
- Chanprame S, Kuo TM, Widholm JM (1998) Soluble carbohydrate content of soybean (Glycine max (L.) Merr.) somatic and zygotic embryos during development. In Vitro Cell Dev Biol Plant 34 64–68 [Google Scholar]
- Chappell AS, Scaboo AM, Wu X, Nguyen H, Pantalone VR, Bilyeu KD (2006) Characterization of the MIPS gene family in Glycine max. Plant Breed 125 493–500 [Google Scholar]
- Chen QC, Li BW (2003) Separation of phytic acid and other related inositol phosphates by high-performance ion chromatography and its applications. J Chromatogr A 1018 41–52 [DOI] [PubMed] [Google Scholar]
- Chen R, Xue G, Chen P, Yao B, Yang W, Ma Q, Fan Y, Zhao Z, Tarczynski MC, Shi J (October 12, 2007) Transgenic maize plants expressing a fungal phytase gene. Transgenic Res http://dx.doi.org/10.1007/s11248-007-9138-3 [DOI] [PubMed]
- Chen Z, Ilarslan H, Palmer RG, Shoemaker RC (1998) Development of protein bodies and accumulation of carbohydrates in a soybean (Leguminosae) shriveled seed mutant. Am J Bot 85 492–499 [PubMed] [Google Scholar]
- Chiera JM, Finer JJ, Grabau EA (2004) Ectopic expression of a soybean phytase in developing seeds of Glycine max to improve phosphorus availability. Plant Mol Biol 56 895–904 [DOI] [PubMed] [Google Scholar]
- Cho MJ, Widholm JM, Vodkin LO (1995) Cassettes for seed-specific expression tested in transformed embryogenic cultures of soybean. Plant Mol Biol Rep 13 255–269 [Google Scholar]
- Coello P, Maughan JP, Mendoza A, Philip R, Bollinger DW, Veum TL, Vodkin LO, Polacco JC (2001) Generation of low phytic acid Arabidopsis seeds expressing an E. coli phytase during embryo development. Seed Sci Res 11 285–292 [Google Scholar]
- Dassa J, Marck C, Boquet P (1990) The complete nucleotide sequence of the Escherichia coli gene appA reveals significant homology between pH 2.5 acid phosphatase and glucose 1-phosphate. J Bacteriol 172 5497–5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer D (2004) Analysis of oilseeds and coarse grains. In CA Roberts, J Workman Jr, JB Reeves, eds, Near-Infrared Spectroscopy in Agriculture (#44). ASA, CSSA, and SSSA Agronomy Monographs, Madison, WI, pp 321–344
- Erdman JW (1979) Oilseed phytates: nutritional implications. J Am Oil Chem Soc 56 736–741 [Google Scholar]
- Ertl DS, Young KA, Raboy V (1998) Plant genetic approaches to phosphorus management in agricultural production. J Environ Qual 27 299–304 [Google Scholar]
- Fehr WR, Caviness CE (1977) Stages of Soybean Development. Special Report 80. Iowa State University, Ames, IA
- Grabau EA (2002) Phytase expression in transgenic plants. In NR Reddy, SK Sathe, eds, Food Phytates. CRC Press, Boca Raton, FL, pp 85–105
- Greiner R, Konietzny U, Jany KD (1993) Purification and characterization of two phytases from Escherichia coli. Arch Biochem Biophys 303 107–113 [DOI] [PubMed] [Google Scholar]
- Hegeman CE, Grabau EA (2001) A novel phytase with sequence similarity to purple acid phosphatases is expressed in cotyledons of germinating soybean seedlings. Plant Physiol 126 1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitz WD, Carlson TJ, Kerr PS, Sebastian SA (2002) Biochemical and molecular characterization of a mutation that confers a decreased raffinosaccharide and phytic acid phenotype on soybean seeds. Plant Physiol 128 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsters M, de Waele D, Depicker A, Messens E, van Montague M, Schell J (1978) Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet 163 181–187 [DOI] [PubMed] [Google Scholar]
- Hood EE, Helmer GL, Fraley RT, Chilton MM (1986) The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J Bacteriol 168 1291–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HB, Franceschi VR, Okita TW (1986) Immunocytochemical studies on the role of the Golgi complex in protein body formation in rice seeds. Planta 169 471–480 [DOI] [PubMed] [Google Scholar]
- Larson SR, Raboy V (1999) Linkage mapping of maize and barley myo-inositol 1-phosphate synthase DNA sequences: correspondence with a low phytic acid mutation. Theor Appl Genet 99 27–36 [Google Scholar]
- Lott JN, Buttrose MS (1978) Globoids in protein bodies of legume seed cotyledons. Aust J Plant Physiol 5 89–111 [Google Scholar]
- Meis SJ, Fehr WR, Schnebly SR (2003) Seed source effect on field emergence of soybean lines with reduced phytate and raffinose saccharides. Crop Sci 43 1336–1339 [Google Scholar]
- Murphy J, Riley JP (1962) A modified single solution method for determination of phosphate in natural waters. Anal Chim Acta 27 31–36 [Google Scholar]
- Nunes A, Vianna G, Cuneo F, Amaya-Farfán J, de Capdeville G, Rech E, Aragão F (2006) RNAi-mediated silencing of the myo-inositol-1-phosphate synthase gene (GmMIPS1) in transgenic soybean inhibited seed development and reduced phytate content. Planta 224 125–132 [DOI] [PubMed] [Google Scholar]
- Obendorf RL, Dickerman AM, Pflum TM, Kacalanos MA, Smith ME (1998) Drying rate alters soluble carbohydrates, desiccation tolerance, and subsequent seedling growth of soybean (Glycine max L. Merrill) zygotic embryos during in vitro maturation. Plant Sci 132 1–12 [Google Scholar]
- Pen J, Verwoerd TC, van Paridon PA, Beudeker RF, van den Elzen PJM, Geerse K, van der Klis JD, Versteegh HAJ, van Ooyen AJJ, Hoekema A (1993) Phytase-containing transgenic seeds as a novel feed additive for improved phosphorus utilization. Biotechnology 11 811–814 [Google Scholar]
- Prattley CA, Stanley DW (1982) Protein-phytate interactions in soybeans. I. Localization of phytate in protein bodies and globoids. J Food Biochem 6 243–253 [Google Scholar]
- Raboy V, Young KA, Larson SR, Cook A (2002) Genetics of phytic acid synthesis and accumulation. In NR Reddy, SK Sathe, eds, Food Phytates. CRC Press, Boca Raton, FL, pp 63–83
- Rodriguez E, Wood Z, Karplus P, Lei X (2000) Site-directed mutagenesis improves catalytic efficiency and thermostability of Escherichia coli pH 2.5 acid phosphatase/phytase expressed in Pichia pastoris. Arch Biochem Biophys 382 105–112 [DOI] [PubMed] [Google Scholar]
- SAS (1990) SAS/STAT User's Guide, Version 6, Ed 4. SAS Institute, Cary, NC
- Sharpley AN, Charpa SC, Wedepohl R, Sims JY, Daniel TC, Reddy KR (1994) Managing agricultural phosphorus for protection of surface waters: issues and options. J Environ Qual 23 437–451 [Google Scholar]
- Shi J, Wang H, Hazebroek J, Ertl DS, Harp T (2005) The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant J 42 708–719 [DOI] [PubMed] [Google Scholar]
- Shi J, Wang H, Schellin K, Li B, Faller M, Stoop JM, Meeley RB, Ertl DS, Ranch JP, Glassman K (2007) Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat Biotechnol 25 930–937 [DOI] [PubMed] [Google Scholar]
- Shi J, Wang H, Wu Y, Hazebroek J, Meeley RB, Ertl DS (2003) The maize low-phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol 131 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear JD, Fehr WR (2007) Genetic improvement of seedling emergence of soybean lines with low phytate. Crop Sci 47 1354–1360 [Google Scholar]
- Staswick PE, Zhang Z, Clemente TE, Specht JE (2001) Efficient down-regulation of the major vegetative storage protein genes in transgenic soybean does not compromise plant productivity. Plant Physiol 127 1819–1826 [PMC free article] [PubMed] [Google Scholar]
- TeKrony DM, Egli DB (1977) Relationship between laboratory indices of soybean seed vigor and field emergence. Crop Sci 17 573–577 [Google Scholar]
- Veum TL, Bollinger DW, Buff CE, Bedford MR (2006) A genetically engineered Escherichia coli phytase improves nutrient utilization, growth performance, and bone strength of young swine fed diets deficient in available phosphorus. J Anim Sci 84 1147–1158 [DOI] [PubMed] [Google Scholar]
- Vohra P, Gray GA, Kratzer FH (1965) Phytic acid-metal complexes. Proc Soc Exp Biol Med 120 447–454 [DOI] [PubMed] [Google Scholar]
- Walker DR, Scaboo AM, Pantalone VR, Wilcox JR, Boerma HR (2006) Genetic mapping of loci associated with seed phytic acid content in CX1834-1-2 soybean. Crop Sci 46 390–397 [Google Scholar]
- Westerhaus M, Workman J Jr, Reeves JB, Mark H (2004) Quantitative Analysis. In CA Roberts, J Workman Jr, JB Reeves, eds, Near-Infrared Spectroscopy in Agriculture (#44). ASA, CSSA, and SSSA Agronomy Monographs, Madison, WI, pp 133–174
- Wilcox JR, Premachandra GS, Young KA, Raboy V (2000) Isolation of high seed inorganic P, low-phytate soybean mutants. Crop Sci 40 1601–1605 [Google Scholar]
- Zeng P, Vadnais DA, Zhang Z, Polacco JC (2004) Refined glufosinate selection in Agrobacterium-mediated transformation of soybean [Glycine max (L.) Merrill]. Plant Cell Rep 22 478–482 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Xing A, Staswick P, Clemente TE (1999) The use of glufosinate as a selective agent in Agrobacterium-mediated transformation of soybean. Plant Cell Tissue Organ Cult 56 37–46 [Google Scholar]