Abstract
Protein dephosphorylation by the serine/threonine protein phosphatase 2A (PP2A) modulates a broad array of cellular functions. PP2A normally acts as a heterotrimeric holoenzyme complex comprising a catalytic subunit bound by regulatory A and B subunits. Characterization of the regulatory A subunit isoforms (ROOTS CURL IN NAPHTHYLPHTHALAMIC ACID1 [RCN1], PP2AA2, and PP2AA3) of Arabidopsis thaliana PP2A has shown that RCN1 plays a primary role in controlling root and hypocotyl PP2A activity in seedlings. Here we show that hypocotyl and root growth exhibit different requirements for RCN1-mediated regulation of PP2A activity. Roots of rcn1 mutant seedlings exhibit characteristic abnormalities in cell division patterns at the root apical meristem, as well as reduced growth under ionic, osmotic, and oxidative stress conditions. We constructed chimeric A subunit genes and found that restoration of normal root tip development in rcn1 plants requires both regulatory and coding sequences of RCN1, whereas the hypocotyl elongation defect of rcn1 plants can be complemented by either RCN1 or PP2AA3 transgenes. Furthermore, the RCN1 and PP2AA3 proteins exhibit ubiquitous subcellular localization patterns in seedlings and both associate with membrane compartments. Together, these results show that RCN1-containing PP2A has unique functions that cannot be attributed to isoform-specific expression and localization patterns. Postembryonic RCN1 function is required to maintain normal auxin distribution and stem cell function at the root apex. Our data show that RCN1-regulated phosphatase activity plays a unique role in regulating postembryonic root development and stress response.
Regulated dephosphorylation by protein phosphatases (PPs) has emerged as a universal control mechanism in physiology and development. Important roles have been identified for a variety of PP species in plants. For instance, several PP2C enzymes negatively regulate abscisic acid (ABA) response (for review, see Schweighofer et al., 2004; Yoshida et al., 2006), whereas other PP2Cs modulate wound signaling, stress signaling, and meristem development (Stone et al., 1998; Song and Clark, 2005; Schweighofer et al., 2007). Similarly, distinct dual-specificity phosphatases regulate carbohydrate metabolism (Kerk et al., 2006; Niittyla et al., 2006; Sokolov et al., 2006) and oxidative, saline, and genotoxic stress tolerance (Ulm et al., 2002; Lee and Ellis, 2007). PP2A, which constitutes an abundant population of oligomeric enzymes, plays crucial roles in the regulation of growth and development. Altered PP2A activity in plants has been linked to defects in hormone homeostasis and signaling, defense responses, cell division, morphogenesis, and reproduction (for review, see DeLong, 2006). Analysis of Arabidopsis (Arabidopsis thaliana) PP2A mutants suggests that important substrates for PP2A include proteins that control microtubule dynamics (Camilleri et al., 2002) and components of the auxin transport apparatus (Rashotte et al., 2001; Shin et al., 2005; Michniewicz et al., 2007).
The predominant form of PP2A is a heterotrimeric complex containing a catalytic (C) subunit, a scaffolding/regulatory (A) subunit, and a regulatory (B) subunit (Janssens and Goris, 2001). Combinatorial diversity of these heterotrimers enhances the versatility of the enzyme complex. The C and A subunits are abundant, ubiquitous, and highly conserved, whereas B subunits exhibit more specific expression patterns and are encoded by several unrelated gene families. Localization and substrate specificity of PP2A action are controlled largely through the effects of bound A and B subunits. The regulatory A subunit comprises 15 imperfect repeats of the α-helical HEAT (huntingtin, elongation factor 3, A subunit, and TOR proteins; Andrade and Bork, 1995). The carboxy-terminal repeats bind the catalytic subunit, and the amino-terminal repeats bind the B subunit. Both binding interactions employ a hydrophobic binding interface formed by short and variable loops located in the center of each HEAT repeat (see Supplemental Fig. S1A; Ruediger et al., 1994; Xing et al., 2006). The A subunit performs at least three crucial regulatory functions. First, binding of the A subunit alters the kinetic properties of the C subunit (Price and Mumby, 2000). Second, A subunit binding also allows interaction of C subunits with the diverse B subunits involved in targeting PP2A function to its physiological targets (Ruediger et al., 1994). Third, recent work indicates that A subunit binding is required for acquisition of the fully activated C subunit conformation (Hombauer et al., 2007). A single regulatory A subunit isoform appears to suffice in rice (Oryza sativa) and maize (Zea mays), as well as in several fungi (van Zyl et al., 1992; Kinoshita et al., 1996; Yu et al., 2001), whereas mammalian systems rely on two differentially expressed and functionally distinct isoforms (Zhou et al., 2003; Sablina et al., 2007).
The Arabidopsis genome encodes three functionally distinct A subunit isoforms, ROOTS CURL IN NAPHTHYLPHTHALAMIC ACID1 (RCN1), PP2AA2, and PP2AA3 (Slabas et al., 1994; Zhou et al., 2004), with RCN1 alone acting as a key positive regulator of PP2A activity in seedlings. Biochemical and physiological analyses show reduced PP2A enzymatic activity in rcn1 plants, and rcn1 mutant phenotypes result from loss of PP2A activity in vivo (Deruère et al., 1999; Rashotte et al., 2001; Kwak et al., 2002; Larsen and Cancel, 2003). Basipetal auxin transport is increased in hypocotyls and roots of rcn1 seedlings, resulting in altered gravitropic response in both organs (Rashotte et al., 2001; Shin et al., 2005; Muday et al., 2006). RCN1 also functions as a transducer of ABA signals, acting upstream of ABA-induced increases in cytosolic Ca2+, but downstream of the PP2C ABI1 (Kwak et al., 2002), and as a negative regulator of ethylene synthesis (Larsen and Chang, 2001; Muday et al., 2006). Additional data suggest a negative regulatory role for RCN1 in ethylene signaling in shoots (Larsen and Chang, 2001; Larsen and Cancel, 2003).
The abnormal phenotypes of rcn1 mutant plants show that RCN1-containing PP2A species perform regulatory functions that are not mediated by complexes containing the other regulatory A subunit isoforms. Despite gene expression patterns that overlap with that of RCN1, loss of PP2AA2 and/or PP2AA3 function does not significantly alter phosphatase inhibitor sensitivity or produce dramatic mutant phenotypes (Zhou et al., 2004). However, plants carrying a pp2aa2 or pp2aa3 mutation in combination with rcn1 exhibit severe morphological and developmental abnormalities including arrested primary root growth (Zhou et al., 2004; Michniewicz et al., 2007). Degeneration of the primary root apical meristem in rcn1 pp2aa2 and rcn1 pp2aa3 seedlings appears to be caused by loss of auxin signaling, and is associated with relaxed or reversed localization of PIN-FORMED (PIN) proteins in embryos and seedling roots (Michniewicz et al., 2007). The radial cell and organ expansion phenotypes exhibited by these seedlings are similar to those of seedlings grown in the presence of high doses of phosphatase inhibitors, and therefore demonstrate the effect of drastic loss of PP2A activity (Rashotte et al., 2001; Shin et al., 2005; Muday et al., 2006).
We asked whether we could distinguish between functions that specifically require the RCN1 protein sequence and functions that are sensitive to overall A subunit dosage but insensitive to isoform specificity. To address this question, we undertook functional analyses in vivo, using constructs that carry regulatory and coding sequences from different A subunit-encoding genes to rescue rcn1 defects in seedling hypocotyls and in roots. The reduced hypocotyl elongation phenotype of rcn1 can be rescued by PP2AA3 constructs; however, restoration of wild-type root tip organization requires both promoter and coding sequences of RCN1. Thus hypocotyl growth is sensitive to A subunit gene dosage but relatively insensitive to isoform specificity, whereas regulation of root growth requires PP2A complexes containing the RCN1 regulatory subunit. We show that loss of rcn1 alone causes increased sensitivity to a broad panel of stress treatments and compromises the maintenance of organized stem cell populations. Inhibition of phosphatase activity in seedlings is sufficient to recapitulate the rcn1 phenotype at the root apex, indicating that RCN1-mediated phosphatase regulation is required postembryonically for normal meristem function. Our data suggest that the RCN1 protein specifically mediates interactions targeting PP2A to substrates required for root stress response and meristem function.
RESULTS
Arabidopsis RCN1-YFP Fusions Retain Biological Activity
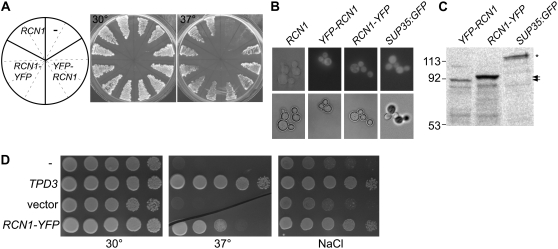
To facilitate our investigation of the biological specificity determinants for RCN1 function in plants, we constructed amino- and carboxy-terminal fusions of RCN1 with the yellow fluorescent protein (YFP) reporter (see “Materials and Methods”). We first tested cDNA fusions for biological activity using a complementation assay in the yeast PP2A regulatory A subunit mutant, tpd3-1. The tpd3-1 mutation confers temperature sensitivity and slow growth phenotypes that are complemented by the RCN1 cDNA (Garbers et al., 1996), as well as sensitivity to stress conditions such as nitrogen starvation and osmotic stress (Santhanam et al., 2004). RCN1-YFP amino- and carboxy-terminal fusion proteins were expressed under control of the constitutive alcohol dehydrogenase promoter (Ammerer, 1983). Both constructs rescued growth of tpd3 mutant at high temperature, indicating that the fusion proteins are competent to regulate the yeast PP2A complex (Fig. 1A). YFP fluorescence was detected in yeast cells carrying RCN-YFP and YFP-RCN (Fig. 1B) and full-length fusion proteins were detected by anti-GFP (Fig. 1C) and anti-RCN1 (see Supplemental Fig. S1B) antibodies. Although the predicted molecular masses of the two fusion proteins were nearly identical (94 kD), the RCN1-YFP fusion consistently exhibited a slightly slower SDS-PAGE migration when extracted from both yeast and plant cells (see below). The RCN1-YFP fusion also complemented the tpd3 stress sensitivity phenotype and rescued growth in the presence of salt (Fig. 1D) and sorbitol (data not shown). Thus fusion of YFP to either terminus does not impair the regulatory A subunit function of RCN1 in yeast.
Figure 1.
RCN-YFP fusions supply regulatory A subunit function in yeast. RCN1-YFP and YFP-RCN1 fusions were expressed under control of the constitutive alcohol dehydrogenase promoter (Ammerer, 1983) in tpd3-1 yeast cells (van Zyl et al., 1992). A, Transformants carrying RCN-YFP fusions, an RCN1 construct, or the empty vector were streaked on duplicate YPD plates and incubated at 30°C or 37°C. The diagram at left indicates the construct carried by cells in the corresponding sectors on both plates. B, Cells carrying a native RCN1 construct, YFP-RCN1, RCN1-YFP, or a SUP35:GFP fusion (Satpute-Krishnan and Serio, 2005) were grown to early log phase and mounted for differential interference contrast (bottom panels) or fluorescence (top panels) microscopy. C, Protein extracts of cells carrying the constructs indicated were subjected to SDS-PAGE and immunoblotting, using anti-GFP antibodies to detect the fusion proteins. The positions of the RCN1-YFP (black arrows) and SUP35:GFP (asterisk) proteins are indicated at right. D, Cells carrying the constructs indicated were grown in liquid culture and 10-fold serial dilutions were spotted on plates containing YPD medium or YPD plus 600 mm NaCl. YPD plates were incubated at 30°C or 37°C and YPD NaCl plates were incubated at 30°C.
YFP-RCN1 and YFP-PP2AA3 Fusions Rescue the rcn1 Hypocotyl Elongation Defect
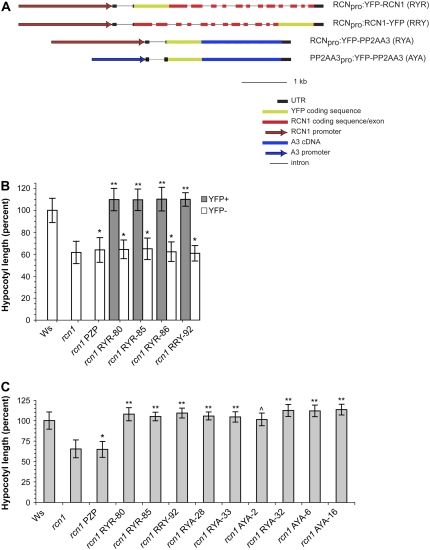
To assay the biological activities of RCN1-YFP fusions in planta, we used a triple-template PCR (TT-PCR) template overlap strategy (Tian et al., 2004) to generate translational fusions in the context of the full-length genomic RCN1 sequence (Fig. 2A). The resulting constructs carried 2.1 kb of genomic sequence upstream from the RCN1 transcript, the transcribed region (including introns), and 550 bp of downstream sequence. Both amino- and carboxy-terminal fusions were generated in the plant transformation vector pPZP221 (Hajdukiewicz et al., 1994) and transformed into rcn1-1 mutant plants. To test for biological function, we assayed for complementation of the rcn1 hypocotyl elongation defect (Fig. 2B). In families segregating for an RCNpro:YFP-RCN1 (RYR) or RCNpro:RCN1-YFP (RRY) fusion, YFP fluorescence segregated with rescued hypocotyl elongation, whereas hypocotyl lengths of YFP-negative siblings matched those of the rcn1 parent and the empty vector control. These data indicate that the YFP fusion proteins provide A subunit function in plants, as well as in yeast.
Figure 2.
RCN1 and PP2AA3 fusions to YFP rescue hypocotyl elongation in the rcn1 mutant. A, The YFP coding sequence was fused at the N- or C-terminal end of the RCN1 gene (see “Materials and Methods”) to generate in-frame fusions within the genomic RCN1 sequence. In the RCNpro:YFP-PP2AA3 (RYA) construct, the RCN1 coding sequence was replaced with the PP2AA3 cDNA. The PP2AA3pro:YFP-PP2AA3 (AYA) construct was derived from RYA by substituting upstream sequences from PP2AA3 for the RCN1 regulatory region. All constructs were transformed into the rcn1-1 mutant. B, Transformant families segregating for a single YFP-RCN1 or RCN1-YFP transgene were scored for hypocotyl elongation and for YFP fluorescence after 5 d of growth in the dark. Values shown represent the average hypocotyl lengths for each class (YFP+ or YFP−) relative to the wild-type control (Ws). Error bars indicate sd; n > 25 for all controls; n > 45 for all segregating families. C, Homozygous transformant families carrying the transgene constructs indicated were scored for hypocotyl elongation after 5 d growth in the dark. Values shown represent the average hypocotyl lengths for each family relative to the wild-type control (Ws). Error bars indicate sd; n > 25 for all controls; n > 45 for all families carrying YFP fusion constructs. rcn1 PZP is an rcn1 transgenic line carrying the empty vector. Lines rcn1 RYA-32 and rcn1 AYA-6 each carry two copies of the fusion T-DNA, whereas line rcn1 AYA16 carries three or more copies. All other lines carry the transgene construct in single copy. Levels of statistical significance as determined by Student's t test: *, P > 0.15 versus rcn1 and P < 10−6 versus Ws; **, P < 10−30 versus rcn1 and P < 0.05 versus Ws; ^, P < 10−30 versus rcn1 and P > 0.8 versus Ws.
To allow direct comparison of the localization and biological activities of the RCN1 and PP2AA3 isoforms, we constructed equivalent YFP fusions to the PP2AA3 cDNA (Fig. 2A). In one derivative the YFP-PP2AA3 fusion remained under control of the RCN1 promoter and 5′ untranslated region (RYA), while in the second construct the RCN1 upstream sequences were replaced with a 1.2-kb fragment containing the PP2AA3 leader (including a 312-bp intron), the intragenic region, and the predicted 5′ end of the next gene upstream (AYA). These constructs were designed to drive expression of the YFP-PP2AA3 fusion in the RCN1 and the PP2AA3 domains, respectively. To avoid overexpression artifacts, we focused primarily on transformants carrying single-copy T-DNAs in the rcn1 background. We assayed the ability of these fusions to complement the hypocotyl elongation defect of rcn1 seedlings (Fig. 2C). Hypocotyl growth was fully restored in lines carrying the YFP-PP2AA3 fusion under control of either the RCN1 or PP2AA3 promoter. Only a slight difference was observed between lines expressing YFP-RCN versus YFP-PP2AA3 fusions, and this difference was eliminated in lines carrying YFP-PP2AA3 in two or more copies (Fig. 2C; lines rcn1 RYA-32, rcn1 AYA-6, and rcn1 AYA-16). These data indicate that A subunit dosage is critical for supporting normal hypocotyl elongation, but RCN1-specific protein sequences are not strictly required for this activity.
RCN1-Specific PP2A Regulation Maintains Root Tip Organization
We used confocal imaging to determine whether rcn1 seedlings exhibit root tip disorganization phenotypes consistent with our hypothesis that increased basipetal auxin transport alters auxin distribution in rcn1 roots. The normal root apical meristem exhibits a stereotypical arrangement of initial cells organized around a small population of quiescent center (QC) cells (for review, see Benfey and Scheres, 2000), and root tip architecture is disrupted by factors that alter the position or magnitude of an auxin concentration maximum in the root apex (Sabatini et al., 1999). Although wild-type roots maintained highly regular cell numbers and arrangement in the QC, columella initials, and root cap columella, rcn1 seedlings showed disorganization indicating aberrant cell division patterns (Fig. 3). QC and cortical/endodermal initial cells were difficult to distinguish in rcn1 seedlings and frequently failed to form a well-defined layer at the bottom of the stele. Columella cell files were abnormal in 68% of rcn1 roots examined (versus 12% of wild-type roots), whereas columellar tiers were disrupted in 28% (versus 0 in the wild type; n = 25 for each genotype). In rcn1 roots, cells formed irregular layers and cell numbers varied between different tiers. In several roots with three files of proper columella cells, a flanking file of lateral root cap cells appeared to have been recruited into a “shoulder” of the columella. All wild-type roots scored as abnormal exhibited regular files and tiers but had an abnormal number of cell files. As shown below, an independent rcn1 allele in the Columbia (Col) genetic background showed similar defects in root tip organization.
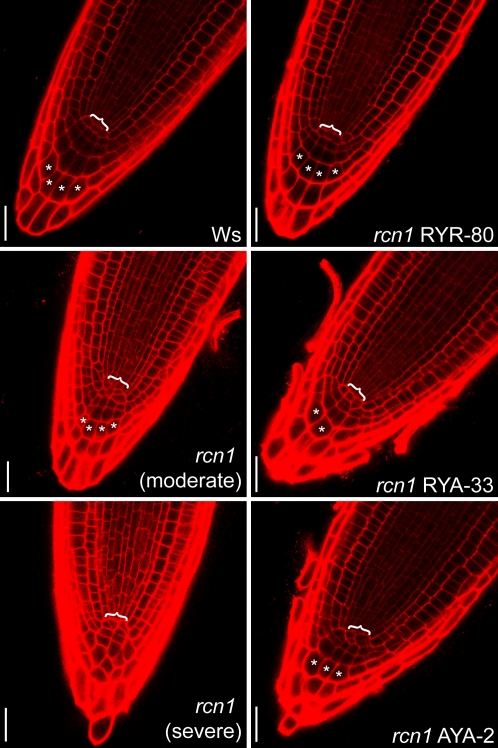
Figure 3.
Isoform specificity of RCN1 function in root tip organization. Median longitudinal sections of propidium iodide-stained 4-dpg root tips were captured using confocal laser microscopy, revealing normal meristem organization in wild-type (Ws) roots and highlighting the disorganization of columella and initial cells in rcn1 root tips. Examples of moderate (rcn1 moderate) and severe (rcn1 severe) rcn1 disorganization phenotypes are shown. The YFP-RCN1 fusion restores wild-type morphology in rcn1 plants carrying the RYR construct, whereas the YFP-PP2AA3 fusions carried in RYA and AYA transformants fail to fully rescue the rcn1 phenotype. Brackets indicate the region of the QC; asterisks indicate cells representative of clearly defined columellar cell files. Scale bars, 25 μm.
We asked whether YFP-RCN and YFP-PP2AA3 could rescue the abnormal morphology of rcn1 root tips (Fig. 3). RRY and RYR lines exhibited identifiable QCs and clear restoration of columellar cell organization. In a representative RYR line, only 7% of roots exhibited an abnormal number of columellar files (n = 15). In contrast, columellar file number remained abnormal in roots of a representative RYA line (35%; n = 23) and a representative AYA line (26%; n = 31). Similarly, a regular columella initial cell layer was present in all wild-type and RYR root tips, with only 7% exhibiting an abnormal columella initial cell number, whereas 53% of rcn1-1 roots, 30% of RYA roots, and 35% of AYA roots exhibited abnormal cell numbers in a columella initial cell layer that frequently was poorly defined. These data suggest that normal root tip organization specifically requires RCN1 function; YFP-PP2AA3 constructs that support normal hypocotyl elongation do not fully restore normal root growth. To ensure that fusion to YFP did not impair function of PP2AA3 protein in roots, we also assayed for rescue by a native PP2AA3 construct (PP2AA3pro:PP2AA3). The native PP2AA3 construct also provided only weak complementation of rcn1 root tip defects (data not shown). These data show that the requirements for A subunit function are more stringent in the root tip than in the hypocotyl, and suggest that increased PP2AA3 dosage does not completely compensate for loss of RCN1 function in the root tip.
Our earlier work on A subunit double mutants revealed severe root growth defects in rcn1 pp2aa2 and rcn1 pp2aa3 double mutants, but not in pp2aa2 pp2aa3 double mutant seedlings (Zhou et al., 2004). We asked whether root growth in the pp2aa2 pp2aa3 double mutant background was sensitive to decreased RCN1 dosage. We assessed root tip morphology by visualizing amyloplasts in cleared root tips after staining for starch accumulation. As expected, starch staining revealed a highly regular arrangement of columellar cells in wild-type and pp2aa2 pp2aa3 root tips, with clearly defined tiers and files of cells (see Supplemental Fig. S2). In rcn1 root tips, columellar cell files and tiers often were irregular, indicating aberrant patterns of columella initial divisions. Overall columellar morphology in the pp2aa2 pp2aa3 rcn1/+ mutant was similar to that observed in the rcn1 background, with most roots exhibiting four poorly defined tiers of cells, and many lacking part or all of one columellar cell file. Consistent with a recent report (Michniewicz et al., 2007), columellar cell numbers were severely reduced and columellar files and tiers were difficult to identify in rcn1 pp2aa2 and rcn1 pp2aa3 root tips even at 4 d postgermination (dpg; see Supplemental Fig. S2). We conclude that RCN1 function is required for normal root tip organization, and root growth is sensitive to RCN1 dosage. Although RCN1 becomes haploinsufficient in the pp2aa2 pp2aa3 background, the heterozygous RCN1 dose in pp2aa2 pp2aa3 rcn1/+ mutants supports more normal development than a homozygous PP2AA3 dose in the rcn1 pp2aa2 double mutant.
Normal Root Development Requires Postembryonic RCN1 Function
Abnormal embryogenesis in rcn1 pp2aa2 and rcn1 pp2aa3 plants (Zhou et al., 2004; Michniewicz et al., 2007) as well as enhancement of pin1 and pid embryogenesis defects by rcn1 (Zhou et al., 2004) indicate that RCN1 is required for normal embryo development. To determine whether the root tip disorganization phenotype reflects an embryonic or a postembryonic requirement for RCN1 action, we asked whether chemical inhibition of PP activity in seedling roots was sufficient to produce a phenocopy of rcn1 root tip defects. We and others have previously used cantharidin to produce a phenocopy of rcn1 in organ elongation, root curling, and basipetal auxin transport assays (Deruère et al., 1999; Rashotte et al., 2001; Shin et al., 2005), and other phosphatase inhibitors have been used to mimic the ethylene and ABA response phenotypes of rcn1 (Kwak et al., 2002; Larsen and Cancel, 2003). We compared the root apex phenotypes of wild-type (Col) seedlings grown in the absence or presence of cantharidin with those of seedlings carrying rcn1-6, a T-DNA insertion allele (see “Materials and Methods”). RCN1 protein is undetectable in extracts of rcn1-6 seedlings, and the gross hypocotyl and root phenotypes of rcn1-6 seedlings are indistinguishable from those of the rcn1-1 allele (data not shown). Poorly defined QCs and irregular columellar cell arrangements were observed in 86% of cantharidin-treated wild-type roots (n = 24), closely matching the defects exhibited by 94% of rcn1-6 seedlings grown in the absence of inhibitor (n = 18; Fig. 4, A–D). Similar results were obtained with cantharidin-treated wild-type roots of another accession (Ws; data not shown). Thus the rcn1 phenotype reflects a postembryonic requirement for normal PP2A regulation, as PP inhibition in seedlings is sufficient to disrupt normal root tip development.
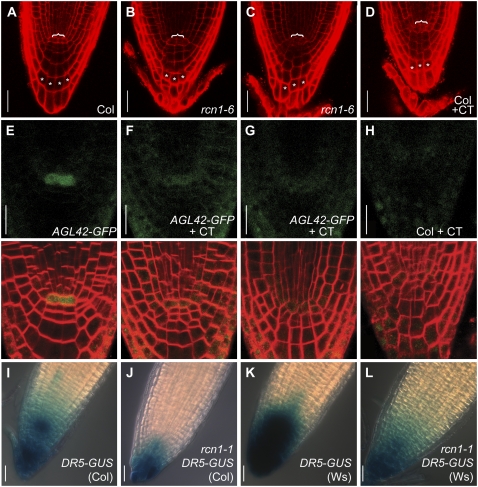
Figure 4.
Postembryonic PP2A function maintains normal root tip development. Median longitudinal sections of propidium iodide-stained 4-dpg root tips were captured using confocal microscopy. One wild-type (A) and two representative rcn1-6 mutant (B and C) root tips grown in the absence of cantharidin plus one wild-type root tip grown in the presence of 10 μm cantharidin (D) are shown. Cantharidin-treated wild-type roots show abnormalities around the QC (brackets) and reduced columellar cell file numbers matching those of rcn1-6 mutant roots. Asterisks indicate cells representative of clearly defined columellar cell files. The AGL42-GFP reporter is expressed in QC cells in 4-dpg seedling roots (E), but its expression is severely reduced in seedlings grown in the presence of 10 μm cantharidin (F and G). Each FM4-64-stained seedling was scanned sequentially for GFP fluorescence (upper row) and FM4-64 fluorescence (overlay shown in lower row). Under the imaging conditions used, a low level of background fluorescence is detected in wild-type Col root tips grown in the presence of cantharidin (H). DR5-GUS reporter activity is reduced in roots of seedlings carrying the rcn1-1 mutation in both the Col (I versus J) and Ws (K versus L) genetic backgrounds (see “Materials and Methods”). Scale bars, 25 μm (A–D and I–L) and 20 μm (E–H).
The disrupted columella organization observed in rcn1 roots suggests abnormal function of the stem cell populations, particularly the columella initials and/or QC. We asked whether loss of phosphatase activity altered the expression of a molecular marker for QC identity. In roots the expression of a GFP reporter fused to the AGL42 MADS-box gene is tightly restricted to the QC at 4 dpg (Nawy et al., 2005). Normal root tip organization and clear QC expression of AGL42-GFP were observed in 87% of untreated roots (n = 23; Fig. 4E). Strongly reduced expression of AGL42-GFP and abnormal root tip architecture were observed in 91% of cantharidin-treated seedling roots (n = 23; Fig. 4, F and G). In most cantharidin-treated roots, AGL42-GFP expression was reduced to background levels (Fig. 4, F–H). These observations are consistent with the hypothesis that loss of RCN1-regulated phosphatase activity compromises QC function. Reduced AGL42-GFP expression might be a consequence of decreased auxin accumulation at the root apex; however, we do not observe altered AGL42-GFP expression in auxin-treated root tips (data not shown). Furthermore, publicly available microarray data indicate that AGL42 expression changes less than 1.5-fold in response to indole-3-acetic acid or naphthylacetic acid treatment (AtGenExpress Hormone Response and Genvestigator Stimulus data sets).
We asked whether the activity of an auxin-responsive reporter construct was altered in the rcn1 mutant. As described previously (Sabatini et al., 1999), wild-type seedlings carrying the DR5-GUS reporter show most intense staining around the columella initials, with lower GUS activity levels in the QC and throughout the columella (Fig. 4I). In rcn1 DR5-GUS roots, the overall intensity of staining was reduced, and rcn1 roots frequently exhibited more intense staining in the external columella layer than in the region around the columella initials (Fig. 4J). Similar results were obtained with wild-type and rcn1 DR5-GUS lines backcrossed twice into the Ws genetic background (Fig. 4, K and L). Cantharidin-treated wild-type DR5-GUS seedlings also showed a reduction in staining intensity, as was reported previously (Shin et al., 2005; data not shown). Reduced activity of a DR5rev reporter was recently reported for seedlings carrying a pp2aa2 or pp2aa3 mutation in combination with rcn1 (Michniewicz et al., 2007). Our results indicate that even modest reductions in phosphatase activity reduce auxin concentrations around the initial cells and compromise meristem function. The ability of cantharidin treatment to mimic rcn1 defects suggests that auxin distribution and QC identity are regulated by postembryonic RCN1 function.
Abundance and Localization of YFP Fusion Proteins
We assayed the levels of transgene expression in roots via immunoblotting with anti-RCN1 and anti-GFP antibodies (Supplemental Fig. S3). Anti-RCN1 immunoblots show that accumulation of the fusion proteins in the RYR and RRY lines was comparable to that of native RCN1 protein in wild-type plants. Use of anti-GFP antibodies allowed direct comparison of fusion protein abundance, which was similar in RYA, RYR, and RRY lines, and somewhat greater than in AYA lines. These results show that intact fusion protein accumulates at levels comparable to those of the endogenous A subunits and indicate that complementation of the rcn1 defects by YFP-RCN1 does not require gross overexpression of the fusion protein.
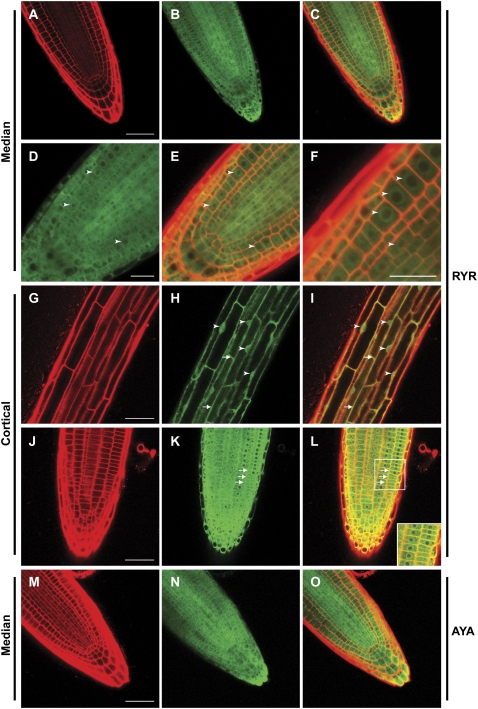
We also assayed the expression and subcellular localization of the YFP fusions using confocal microscopy. In a recent study, immunolocalization of an RCN1-YFP fusion in cells of the root apex detected cytoplasmic, perinuclear, and peripheral localization (Michniewicz et al., 2007). Consistent with this result, we observed that both RCN1-YFP fusion proteins were abundant and ubiquitous in root tips, with cytoplasmic and strong perinuclear signal accumulating in all cell types at the root apex (Fig. 5, A–F). Interestingly, although cells in the region of the root apical meristem (and distal elongation zone) exhibited very little nuclear RCN1-YFP signal, vacuolated cells in more mature regions of the root exhibited nuclear as well as cytoplasmic accumulation (Fig. 5, D–F versus G–I). Optical sectioning confirmed that the fusion protein was present inside the nucleus of these cells (data not shown). Cells in the apical meristem and in more mature regions also exhibited strong peripheral signal, indicating enrichment around the plasma membrane; membrane enrichment was especially clear in cortical cells (Fig. 5, G–L; Supplemental Fig. S4, A–C). Strong accumulation of fusion protein also was observed in root hairs and lateral root primordia, with localization in lateral root primordia recapitulating that observed at the primary root tip (data not shown). Light- and dark-grown seedlings carrying the fusion constructs exhibited YFP fluorescence in roots and hypocotyls, with YFP accumulation patterns matching the previously reported RCN1 mRNA expression pattern (Deruère et al., 1999). Like vacuolated root cells, hypocotyl cells exhibited cytoplasmic, nuclear, and peripheral signal, with little or no accumulation evident in chloroplasts (data not shown).
Figure 5.
Abundance and localization of YFP-RCN1 protein in seedling roots. Confocal microscopy reveals that the YFP-RCN1 fusion protein is abundant in all cell layers of the root tip (A–C) and shows cytoplasmic and perinuclear (arrowheads) localization (D–F) in cells of the apical meristem. In mature cortical cells (G–I), nuclear localization (arrowheads) is evident. Membrane association (arrows) is observed in both mature (G–I) and apical (J–L) cortical cells. The YFP-PP2AA3 fusion also exhibits ubiquitous accumulation in the root tip (M–O). Propidium iodide fluorescence (A, G, J, and M) and YFP fluorescence (B, D, H, K, and N) are overlaid (C, E, F, I, L, and O) in medial (A–F and M–O) and cortical (G–L) optical sections of 4-dpg roots of lines rcn1 RYR-80 (A–L) and rcn1 AYA-2 (M–O). Beam intensity was increased for imaging YFP fluorescence in line rcn1 AYA-2 (N and O; compare with Supplemental Fig. S4, E and F). Scale bars, 25 μm (A–C and G–O) and 10 μm (D–F).
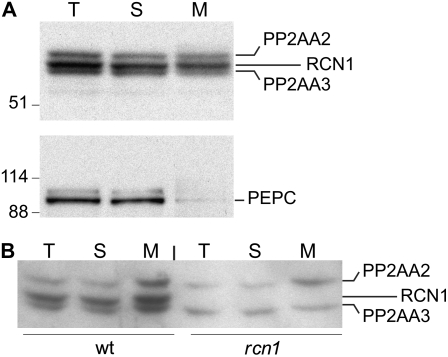
To confirm that membrane localization was characteristic of the native forms of RCN1 and PP2AA3, we assayed the distribution of endogenous A subunits in soluble and microsomal membrane fractions isolated from wild-type seedlings (Fig. 6A). All three A subunits were abundant both in soluble and microsomal fractions, consistent with the analysis of YFP-RCN1 fusion protein localization. The catalytic subunit also was detected in microsomal fractions (data not shown). In contrast to PP2A subunits, the cytosolic PEPC protein was detected in soluble but not membrane fractions, indicating minimal contamination of the membrane fraction with cytoplasmic proteins. We detected PP2AA2 and PP2AA3 in membranes extracted from rcn1 mutant roots, and all three isoforms in the membrane fraction from wild-type roots (Fig. 6B). These data indicate that PP2A complexes associate with membranes in growing seedlings. The presence of all three A subunit proteins in the microsomal fraction indicates that membrane association is not an isoform-specific characteristic. It is possible that recruitment of PP2A to a cellular membrane may allow it to interact with substrate or regulator proteins on the same membrane. Although the primary amino acid sequences of the A and C subunits do not contain motifs that predict membrane localization, several recent studies have shown that PP2A may interact with plasma membrane components, including the plasma membrane H+-ATPase and the signaling lipid phosphatidic acid (Michniewicz et al., 2007).
Figure 6.
Subcellular fractionation of native A subunit isoforms. Soluble and microsomal membrane fractions were prepared from whole wild-type seedlings (A) or roots of wild-type and rcn1 plants (B), and subjected to SDS-PAGE and immunoblotting analysis using anti-RCN1 (top) and anti-PEPC (bottom) antibodies. T, Total; S, soluble; M, microsomal membrane.
Subcellular localization of the YFP-PP2AA3 fusion protein was similar to that of YFP-RCN1. Under control of the RCN1 promoter, accumulation of the YFP-PP2AA3 fusion was similar to that of YFP-RCN1 (see Supplemental Fig. S4D). Expression of YFP-PP2AA3 under control of the PP2AA3 regulatory region resulted in decreased abundance of the fusion protein, with the strongest accumulation evident in the vascular cylinder (Fig. 5, M–O; Supplemental Fig. S4, E and F). At this lower abundance, subcellular localization patterns were difficult to assess, but perinuclear, cytoplasmic, and peripheral localization were detected in most roots. These results suggest that RCN1 is more abundant than PP2AA3 in wild-type roots.
RCN1 Performs an Isoform-Specific Function in Root Stress Response
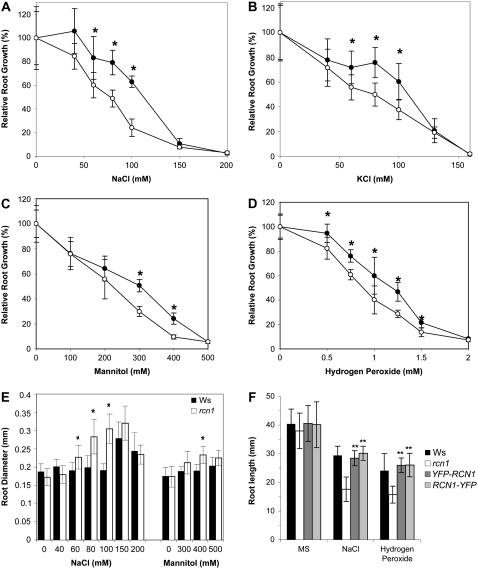
Given the unique role of RCN1 in regulating root growth and the stress sensitivity of PP2A mutants in yeast, we asked whether rcn1 mutant plants show stress sensitivity similar to that observed in tpd3 yeast cells. Roots of mutant seedlings exhibited increased sensitivity to ionic (Na+, K+), osmotic (mannitol), and oxidative (hydrogen peroxide) stress with decreased elongation across a range of concentrations (Fig. 7, A–D). Sodium and mannitol treatment also caused radial expansion, which was enhanced in the cortical cell layer of rcn1 roots (Fig. 7E; Supplemental Fig. S5). Oxidative stress inhibited elongation (Fig. 7D) but did not cause radial swelling of wild-type or mutant roots (data not shown). Both YFP-RCN1 and RCN1-YFP fusion transgenes restored wild-type stress tolerance to rcn1 mutant roots (Fig. 7F). These data indicate that RCN1 regulation of PP2A activity is required for normal stress tolerance in seedling roots. We did not detect differences in salt sensitivities of adult wild-type and rcn1 plants, suggesting that the stress sensitivity of rcn1 is limited to the seedling stage (data not shown).
Figure 7.
Stress sensitivity is increased in rcn1 seedlings. Wild-type (black symbols) and rcn1-1 mutant seedlings (white symbols) were transferred from standard medium to plates containing the indicated concentrations of NaCl (A), KCl (B), mannitol (C), and hydrogen peroxide (D). New growth (elongation from the point of transfer) was measured after 7 d of additional growth. E, Root diameter at the midpoint of the new growth segment was measured for plants grown on NaCl and mannitol. F, Overall root length was measured on wild-type, mutant, and complemented mutant seedlings (transgenic lines R1H9 and R2Q3; see “Materials and Methods”) transferred to NaCl- or hydrogen peroxide-containing plates as described above. For all panels, each value shown represents the average for 12 to 15 seedlings; error bars indicate sd. Asterisks indicate levels of statistical significance as determined by Student's t test: *, P < 0.002 for rcn1 versus wild type; **, P < 10−7 versus rcn1 and P > 0.2 versus Ws.
Unlike rcn1, the pp2aa2 and pp2aa3 single mutants and the pp2aa2 pp2aa3 double mutant exhibited stress sensitivities that very nearly matched that of the parental wild type (see Supplemental Fig. S6). Despite the presence of PP2AA2 and PP2AA3 proteins in root tissue (Zhou et al., 2004), these regulatory A subunit isoforms do not appear to play an equivalent role in stress tolerance. We asked whether the amino acid sequences for RCN1, PP2AA2, and PP2AA3 exhibit discrete differences that might confer biological specificity. The predicted RCN1 protein shares 86% identity with both PP2AA2 and PP2AA3 (see Supplemental Fig. S1A; Slabas et al., 1994), and most of the strongly conserved residues defined in mammalian A subunits are conserved in all three Arabidopsis isoforms. However, Tyr-450, one putative component of the hydrophobic interaction interface (Ruediger et al., 1994; Groves et al., 1999; Xing et al., 2006), is replaced by a basic residue (His) in PP2AA2 and PP2AA3 (see Supplemental Fig. S1A). Mutagenesis of the RCN1-YFP fusion to generate a His-450 allele does not compromise complementation of temperature, sorbitol, and sodium chloride sensitivity of tpd3 yeast cells (data not shown), suggesting that Tyr-450 does not play a required role in mediating stress tolerance. Intriguingly, database searches suggest that the His-450 A subunit may be a plant-specific variant. A Tyr residue is conserved at this position in A subunits of all vertebrates and insects, in yeast, and in many plant species (pea [Pisum sativum], tobacco [Nicotiana tabacum], Brassica spp., Medicago spp., and Lolium spp.). Rice, maize, several other grasses, many fungi, and Caenorhabditis elegans carry Phe at this position. His-450 isoforms are found only in plant genomes that also encode a Tyr-450 isoform (e.g. those of Vicia, Medicago, Brassica, and Arabidopsis) and in Thellungiella salsuginea, for which limited sequence data are available. The His-450 variant thus appears to be a plant-specific regulatory A subunit isoform, and may occur only in plants that also encode a Tyr-450 isoform.
DISCUSSION
Our analysis of Arabidopsis A subunit functions in vivo demonstrates unique functions for RCN1 in specific cell types in seedlings. The requirement for RCN1 function in maintaining normal root stem cell organization is not explained by cell- or tissue-specific mRNA expression patterns because expression of PP2AA3 under control of the RCN1 promoter does not fully rescue normal cell division patterns. In hypocotyl tissue, rescue of normal cell expansion by PP2AA3 expression demonstrates overlapping function of the A subunit isoforms. Our data support the hypothesis that root growth and stress response are regulated by PP2A substrates specifically targeted by RCN1, whereas targeting of substrates involved in hypocotyl growth is not dependent on a particular A subunit isoform. Organization and function of stem cells at the root apex, as measured by formation of normal columella tiers and expression of a marker for QC identity, is compromised by reduced PP2A function during postembryonic development. Given that the subcellular localization of RCN1 and PP2AA3 proteins appears similar, functional specificity is likely to depend on isoform-specific protein-protein interactions with substrates or regulators of the complex.
Dosage Sensitivity versus Isoform Specificity in A Subunit Function
YFP-PP2AA3 fusion constructs that provide only weak complementation of the rcn1 root tip phenotype robustly rescue hypocotyl elongation. These findings suggest that hypocotyl elongation requires a threshold level of A subunit function, with no stringent requirement for RCN1-specific amino acid sequences. Even a modest level of YFP-PP2AA3 expression in the rcn1 mutant is sufficient to promote normal hypocotyl elongation. Because increased ethylene synthesis in dark-grown rcn1 seedlings contributes significantly to reduced hypocotyl growth (Larsen and Chang, 2001; Muday et al., 2006), this result suggests that PP2A complexes containing the PP2AA3 isoform are competent for down-regulation of ethylene biosynthesis. In contrast, PP2AA3 rescues root tip organization weakly even when expression is driven by the RCN1 promoter, demonstrating a more stringent requirement for A subunit function in the root apical meristem. Although it is possible that high-level overexpression of PP2AA3 could suppress this stem cell defect, our data clearly indicate that RCN1-containing PP2A complexes effectively target the key substrates for root growth at physiological expression levels, whereas PP2AA3-containing complexes do not.
RCN1 may be the preferred interaction partner for C and B subunits most active in seedling roots. The Aα and Aβ isoforms of mammalian PP2A differ in their binding activities (Zhou et al., 2003; Sablina et al., 2007). However, the positive regulatory effect of RCN1 also is consistent with the hypothesis that RCN1 plays a role in an activation cycle for Arabidopsis C subunits analogous to that of TPD3 in yeast, promoting interaction with PTPA/RRD (Hombauer et al., 2007). The modest effects of loss of PP2AA2 and PP2AA3 function could indicate that these scaffolds do not interact efficiently with PTPA/RRD-like activators, and therefore do not have equivalent effects on overall PP2A activity, at least in the presence of functional RCN1.
RCN1 Plays an Isoform-Specific Role in Root Development
Our data provide new insight into the developmental effects of increased phosphorylation of RCN1-specific PP2A substrates, demonstrating that loss of RCN1 regulation alters stem cell function and auxin distribution without producing the meristem collapse phenotype caused by more drastic reductions in PP2A activity. Previous studies have documented ectopic expression of cell identity markers and abnormal cell division patterns caused by treatment with exogenous auxin or loss of auxin transporter function in shoots and roots (Sabatini et al., 1999; Benkova et al., 2003; Blilou et al., 2005). Our results indicate that meristem function is also sensitive to subtle changes in auxin flux, as well as to those more profound ones. Moreover, inhibition of phosphatase function during the seedling phase alone is sufficient to perturb meristem function, showing that root tip patterning is a dynamic process that responds rapidly to altered phosphorylation levels.
RCN1 plays a key role in maintaining the patterns of root meristem cell division and function. Decreased expression of the DR5-GUS reporter in rcn1 roots and of the QC marker AGL42-GFP in cantharidin-treated roots suggests that normal establishment of the local auxin concentration maximum and full expression of QC identity require RCN1-regulated PP2A activity. Because positioning of a local maximum in the auxin pool at the root apex is an important patterning determinant (Sabatini et al., 1999), increased basipetal auxin transport in rcn1 could affect root tip organization by altering the position of the auxin concentration maximum or by more generally increasing flux through the auxin transport stream in the root tip. Consistent with this hypothesis, the strongly reduced expression of DR5rev-GFP that is observed in rcn1 pp2aa3 and rcn1 pp2aa2/+ root tips is rescued by 1-N-naphthylphthalamic acid treatment (Michniewicz et al., 2007).
Previous studies have revealed complex feedback loops connecting ethylene response with auxin homeostasis in roots (Stepanova et al., 2005; Chilley et al., 2006). Furthermore, increased ethylene response was recently shown to stimulate cell divisions in the QC (Ortega-Martinez et al., 2007). Two observations argue against the hypothesis that increased ethylene response accounts for aberrant QC function in rcn1. First, we observe altered root tip morphology in light-grown seedlings, whereas increased ethylene synthesis was observed only in dark-grown rcn1 seedlings (Muday et al., 2006). Second, although rcn1 may enhance ethylene sensitivity in shoots (Larsen and Chang, 2001), ctr1 root phenotypes are suppressed in the rcn1 ctr1 double mutant (Larsen and Chang, 2001; A. DeLong, unpublished data), suggesting that loss of rcn1 reduces ethylene response in roots. Interestingly, ethylene treatment stimulates auxin accumulation in wild-type root tips, and mutations that reduce ethylene-induced auxin production confer weak ethylene insensitivity (Stepanova et al., 2005). Thus increased basipetal auxin transport also may result in decreased sensitivity to ethylene in rcn1 roots.
Subcellular Localization of RCN1 and A3 Proteins
Our data suggest that nuclear localization of RCN1 is developmentally regulated in roots. YFP-RCN1 was abundant in perinuclear and cytoplasmic compartments, but was underrepresented in nuclei of meristematic and central elongation zone cells (Fig. 5). Nuclear localization was observed in more mature cells in and above the proximal elongation zone, with little or no perinuclear accumulation evident in these cells. These data are consistent with the idea that RCN1 localization is dynamic during root cell differentiation, with enrichment in the perinuclear compartment in rapidly dividing cells and nuclear enrichment in postmitotic cells. Interestingly, a GFP fusion to the OXIDATIVE SIGNAL-INDUCIBLE1 (OXI1) protein kinase, a member of the AGC kinase family that also includes the PINOID kinase, suggests that subcellular localization of OXI1 in root hairs also may be developmentally regulated, with nuclear accumulation occurring late in root hair development (Anthony et al., 2004; Rentel et al., 2004). Like RCN1, OXI1 plays a role in oxidative stress response; OXI1 activity increases under oxidative stress conditions, and promotes pathogen resistance and root hair development. Developmentally regulated localization of kinase/phosphatase pairs would provide an additional level of fine-tuning in phosphorylation-based control circuits.
We also observed peripheral localization of YFP-RCN1 and YFP-PP2AA3 in all cell types in the root tip. Membrane association appeared fairly uniform around the cell periphery, and did not exhibit obvious polarity or asymmetry in these experiments. Our cell fractionation data are consistent with the existence of a significant pool of membrane-associated PP2A in seedlings. Membrane association of PP2A complexes has been reported in several contexts previously, including early mouse development, during tight junction formation, and during associations with endothelial nitric oxide synthase at the plasma membrane (Gotz et al., 2000; Nunbhakdi-Craig et al., 2002; Wei and Xia, 2006). Arabidopsis PP2A interacts with the C terminus of the plasma membrane ATPase AHA2 in vitro and exhibits partial colocalization with the PIN1 and PIN2 proteins in roots (Fuglsang et al., 2006; Michniewicz et al., 2007). Additionally, RCN1 protein binds phosphatidic acid, a lipid signaling molecule that recruits target proteins to the plasma membrane and plays a significant role in abiotic stress response (Meijer and Munnik, 2003; Testerink et al., 2004). Recruitment of PP2A to a membrane compartment via phosphatidic acid binding could alter phosphatase activity toward membrane-associated substrates. Membrane-associated PP2A activity may be critical for regulation of auxin transport, stress response, and regulation of stem cell function in roots.
Stress Sensitivity in rcn1 Seedlings
The data presented here indicate that RCN1 function plays a unique role in mediating root stress response. Our working model states that positive regulation of PP2A activity by the RCN1 protein contributes to a response that maintains normal growth under a wide range of stress conditions. As a modulator of auxin, ethylene, and ABA levels and/or responses, RCN1 is well positioned to act as an integrator of stress signaling. Abiotic stress may alter the cellular localization and amount of PP2A activity, resulting in PP2A-induced alterations in hormone responses. Loss of RCN1 function compromises these adaptive alterations in PP2A localization and/or activity, leading to increased growth inhibition under stress conditions.
Previous studies focusing on two PP2A interactors, TAP46 and the AtCHIP E3 ubiquitin ligase, suggested that PP2A may play a role in the chilling response in Arabidopsis (Harris et al., 1999; Luo et al., 2006). Additionally, gene expression studies show that mRNAs for PP2A catalytic subunits in rice are differentially expressed in response to drought, salinity, and heat stress (Yu et al., 2003). An Arabidopsis PP2A catalytic subunit mutant was recently found to affect ABA-related stress responses through negative regulation of ABA signaling. Although loss of PP2AC-2 function confers ABA hypersensitivity (Pernas et al., 2007), loss of RCN1 function results in reduced ABA sensitivity (Kwak et al., 2002). Paradoxically, both mutants show increased sensitivity to NaCl treatment. However, the pp2ac-2 mutant exhibits a sensitivity phenotype specific for ABA-related stress (Pernas et al., 2007) unlike the general stress sensitivity phenotype reported here for rcn1. These disparities indicate that the effect of rcn1 loss of function is unlikely to be mediated by a specific effect on regulation of PP2AC-2 activity.
The parallel stress sensitivity of rcn1 plants and tpd3 yeast is striking, but it is not clear that the mechanism involved in the plant and yeast stress responses is similar. Interestingly, the transition between perinuclear enrichment and intranuclear YFP-RCN1 localization occurs in cells of the elongation zone, the same cell population that would be responsible for altered elongation under stress conditions. In yeast, PP2A is required for nuclear accumulation of the stress-responsive transcription factor Msn2p after nutrient deprivation, rapamycin treatment, and temperature and ionic stress (Santhanam et al., 2004). Regulation by PP2A has been proposed to involve dephosphorylation of a nuclear export signal in Msn2p. Although we cannot rule out a similar explanation for the stress sensitivity of rcn1 seedlings, there are no obvious orthologs of the Msn2 and Msn4 transcription factors in the Arabidopsis genome (A. DeLong, unpublished data). The Arabidopsis C2H2 zinc finger proteins that produce the best BLAST scores against Msn2 and Msn4 are more closely related to the TFIIIA family of transcription factors than to Msn2p or Msn4p. Additionally, we assayed for but did not observe an enhancement of nuclear RCN1 localization under stress conditions (see Supplemental Fig. S7). Over a range of salt treatment times from minutes to 18 h we did not detect any alteration in the nuclear versus perinuclear YFP-RCN1 localization pattern, though some experiments suggested enrichment of the membrane-associated population after short-term salt treatment (J.J. Blakeslee and A. DeLong, unpublished data). Additional biochemical experiments will be required to explore this possible membrane recruitment more rigorously.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The rcn1-1 allele (Garbers et al., 1996) and derived transgenic lines (see below) were compared with the parental Ws line. An rcn1-1 line homozygous for the DR5-GUS reporter (Rashotte et al., 2001) was backcrossed twice to the parental DR5-GUS line (Col background) or to rcn1-1 and Ws to introgress the rcn1-1 mutation or the reporter, respectively, into a more uniform genetic background. Plants homozygous for the rcn1-1 mutation were identified among the self-progeny of the DR5-GUS backcross products; homozygosity of the DR5-GUS marker was confirmed in self-progeny of these rcn1 individuals. A similar procedure was used to isolate families homozygous for the DR5 reporter from the rcn1-1 backcross. The rcn1-6 allele is the T-DNA insertion allele SALK_059903 (kind gift of X. Wang and J. Chory, Salk Institute) and is compared with the parental Col-0 line. AGL42-GFP (Nawy et al., 2005) was the kind gift of B. Kelley and P. Benfey (Duke University).
Plants were grown as described previously (Zhou et al., 2004). For stress sensitivity measurements, seedlings were grown on vertical plates for 4 d in constant light at 24°C on standard medium (0.5× Murashige and Skoog salts containing 1% Suc and 1% agar), then transferred to the same medium supplemented with the indicated salt, mannitol, and hydrogen peroxide concentrations. Root tips of five mutant and five wild-type seedlings were aligned at a marked position on each plate and plates were returned to constant light. New growth was measured 7 d later using National Institutes of Health ImageJ software, and relative new root growth was calculated as a percentage of that obtained on fresh standard medium. Each data point represents the average for 15 seedlings. Hypocotyl elongation assays were performed as described previously (Deruère et al., 1999; Zhou et al., 2004). After scanning each plate to allow measurement of hypocotyl lengths, each seedling was scored for YFP fluorescence using a Leica MZFLIII dissecting microscope equipped with a mercury arc lamp and a GFP filter set.
Construction of YFP Fusions
The ADC1pro:RCN1-YFP fusions for yeast were constructed using the TT-PCR strategy (Tian et al., 2004), with the RCN1 cDNA (Garbers et al., 1996), the ADC1 alcohol dehydrogenase promoter of pAAH5 (Ammerer, 1983), and pYFP3 (kind gift of D. Jackson, Cold Spring Harbor Laboratory) as templates for the partial PCR products. PCR primer sequences are given in Supplemental Table S1. The ADC1pro:RCN1-YFP fusions were obtained and cloned into YEplac195 (Pitluk et al., 1995) using the Gateway and TOPO-XL systems (Invitrogen). The genomic RCN1-YFP and YFP-RCN1 fusions for plant transformation also were constructed via TT-PCR using pYFP3 and wild-type Col-0 genomic DNA. To simplify cloning, the original TT-PCR products contained a short promoter region (861 bp total upstream from the RCN1 ATG). The resulting fusion was cloned into pPZP221 (Hajdukiewicz et al., 1994) using the Gateway system (Invitrogen). The insert was sequenced and coding errors were corrected by QuikChange II XL site-directed mutagenesis (Stratagene). A longer promoter fragment containing 2.1 kb of genomic sequence upstream from the start of the RCN1 transcript was amplified from Col-0 genomic DNA and substituted for the short promoter in the binary vector, yielding RCNpro:YFP-RCN (RYR). To generate the RCNpro:YFP-PP2AA3 (RYA) fusion, the PP2AA3 coding sequence was amplified from RAFL09-82-A21 (RIKEN BRC) and substituted for the RCN1 coding region in RYR. For PP2AA3pro:YFP-PP2AA3 (AYA), a 1.2-kb PP2AA3 promoter fragment was amplified from Col-0 genomic DNA and substituted for the RCN1 promoter region in RCNpro:YFP-PP2AA3. For PP2AA3pro:PP2AA3 the YFP coding sequence was deleted by oligonucleotide-mediated mutagenesis. All constructs were sequence verified and coding sequence errors were corrected by oligonucleotide-mediated mutagenesis. All clones were electroporated into Agrobacterium tumefaciens strain GV3101 for transformation into rcn1-1 plants via floral dip (Bechtold et al., 1993; Garbers et al., 1996). All transformants were selected for gentamycin resistance. T-DNA copy numbers were determined by screening gentamycin resistance segregation ratios followed by Southern-blot analysis using a probe for the pPZP221 gentamycin resistance gene. The RYR-80, RYR-85, RYR-86, RRY-90, RRY-92, RYA-28, RYA-33, and AYA-2 lines carry single-copy T-DNAs, whereas the RYR-66, RYR-74, RYA-32, and AYA-6 lines carry two T-DNA copies and AYA-16 and AYA-100 carry three or more copies. The R2Q3 line carries three or more copies of the RCN1-YFP fusion driven by the short RCN1 promoter fragment described above.
Microscopy and Detection of Reporter Gene Expression
For confocal imaging, seedlings were grown in constant light at 20°C on standard medium, stained lightly with propidium iodide (10 μg mL−1) or FM4-64 (5 μg mL−1), and mounted immediately for confocal analysis. For seedlings grown on NaCl, osmotic and ionic concentrations were maintained throughout the staining and mounting process. YFP fusion protein localization and seedling root tip morphology were examined by confocal microscopy using a Leica TCS SP2 AOBS spectral confocal microscope. To image YFP fusion proteins and root tip architecture, YFP fluorescence was excited at 514 nm and collected at 525 to 560 nm and propidium iodide fluorescence was excited at 593 nm and collected at 610 to 680 nm using a pinhole of 1 a.u. To image AGL42-GFP, GFP fluorescence was excited at 488 nm and collected at 495 to 550 nm and FM4-64 fluorescence was excited at 593 nm and collected at 650 to 800 nm with a pinhole setting of 2 a.u. and using sequential scanning. Images were processed using Leica confocal software (LCS Lite). To maintain comparable fluorescence signals, images were collected using constant beam intensities and settings within each experiment, unless otherwise noted. For starch staining, 4-dpg seedlings were cleared and stained with iodine-potassium iodide (Fukaki et al., 1998) before imaging on a Zeiss Axiovert 200M. To detect DR5-GUS expression, seedlings were gently vacuum infiltrated and then stained overnight in 100 mm sodium phosphatase, pH 7.0, 10 mm EDTA, 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, 0.1% Triton X-100, and 1 mm X-glucuronide, followed by clearing in 90% ethanol. Yeast cells were imaged using a Zeiss Axioplan 2 equipped with a 100× alphaPlan-FLUAR objective and Hamamatsu-ORCA camera (Hamamatsu Photonics), and images were collected with Openlabs software (Improvision).
Yeast Complementation Assays
Yeast strain Y1459 (tpd3-1; van Zyl et al., 1992) was transformed with YEpTPD3 (kind gift of J. Broach, Princeton University), YEplac195, ADHpro:RCN1, ADHpro:YFP-RCN1, ADHpro:RCN1-YFP, and ADHpro:RCN1Y450H-YFP using a standard lithium acetate transformation protocol (Ausubel et al., 1992). For serial dilution assays, cultures were grown in yeast peptone dextrose (YPD) liquid medium at 30°C, diluted to an OD600 of 2, then diluted in 10-fold steps into fresh YPD. Ten microliters of each dilution was spotted onto YPD medium or YPD supplemented with 600 mm NaCl. The SUP35:GFP strain (Satpute-Krishnan and Serio, 2005) was the kind gift of T. Serio (Brown University).
Preparation of Microsomal Membranes
Microsomal membrane fractions were prepared as described (Blakeslee et al., 2007), with the following modifications. Dark-grown whole seedlings or roots were harvested at 5 dpg, ground, and microsomal membranes were isolated by spinning at 100,000g for 1 h. Membrane fractions were washed twice, flash-frozen, and stored at −80°C. For immunoblot analysis, membrane fractions were solubilized with 1% Triton X-100.
Immunoblot Analysis
For immunoblot analysis, yeast cells were grown to log phase in YPD at 30°C, harvested by centrifugation, resuspended on ice in 100 mm Tris, pH 7.5, 100 mm EDTA, 5 mm dithiothreitol, 2 mm phenylmethylsulfonyl fluoride, 5 μg/mL pepstatin, and 100 μg/mL aprotinin and leupeptin, and lysed by vortexing with glass beads. The resulting extracts were cleared by low-speed centrifugation and immediately boiled with Laemmli buffer. Plant extract preparation, SDS-PAGE, and immunoblotting techniques were as described previously (Deruère et al., 1999), except that immunoblots were treated with 0.2 m NaOH at 37°C for 20 min immediately after transfer, followed by five PBST washes before blocking. Antisera used were anti-GFP (JL-8; CLONTECH), anti-PEP carboxylase (Rockland), anti-RCN1 (Deruère et al., 1999), and anti-C antibodies raised against the C-terminal peptide (CEPDTTRKTPDYFL) of Arabidopsis (Arabidopsis thaliana) PP2A-C1.
The Arabidopsis Genome Initiative locus identifiers for genes described in this article are as follows: RCN1 (also known as RegA, EER1, and PP2AA1; At1g25490), PP2AA3 (also known as PDF2; At1g13320), PP2AA2 (also known as PDF1; At3g25800), and AGL42 (At5g62165).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Structural model of sequence differences between Arabidopsis A subunit isoforms.
Supplemental Figure S2. RCN1 is haploinsufficient in the pp2aa2 pp2aa3 background.
Supplemental Figure S3. Regulatory A subunit transgene products accumulate to native levels.
Supplemental Figure S4. Accumulation and localization of YFP-PP2AA3 fusion proteins.
Supplemental Figure S5. Expansion of cortical cells in salt-stressed rcn1 seedlings.
Supplemental Figure S6. Normal stress sensitivity in pp2aa2 and pp2aa3 mutants.
Supplemental Figure S7. Localization of a YFP-RCN1 fusion protein after NaCl treatment.
Supplemental Table S1. PCR primers.
Supplementary Material
Acknowledgments
We gratefully acknowledge D. Jackson for TT-PCR reagents and advice, T. Serio for advice on yeast culture and imaging, A. Dunaevsky for FM4-64, and P. Benfey and T. Nawy for seed stocks and advice on QC markers. We thank J. Bender, M. Johnson, G. Muday, and F. Tax for critical reading of the manuscript and for helpful discussions. We thank G. Williams, R. Creton, and J. Nathanson for expert advice on imaging. We thank M. Clarke-Pearson, T. Simolari, R. Luo, and S. Cho for assistance with transgenic plant lines, and F. Jackson and B. Leib for greenhouse care.
This work was supported by the National Science Foundation (grant no. IOB–0446039). The work was partially supported by the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service (award no. 2007–35304–18418 to J.J.B.), and K.R.S. was partially supported by the National Institutes of Health predoctoral training program (grant no. GM007601).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Alison DeLong (alison_delong@brown.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ammerer G (1983) Expression of genes in yeast using the ADC1 promoter. Methods Enzymol 101 192–201 [DOI] [PubMed] [Google Scholar]
- Andrade MA, Bork P (1995) HEAT repeats in the Huntington's disease protein. Nat Genet 11 115–116 [DOI] [PubMed] [Google Scholar]
- Anthony RG, Henriques R, Helfer A, Meszaros T, Rios G, Testerink C, Munnik T, Deak M, Koncz C, Bogre L (2004) A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J 23 572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1992) Current Protocols in Molecular Biology. Wiley-Interscience, New York
- Bechtold N, Ellis H, Pelletier G (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Ser III Sci Vie 316 1194–1199 [Google Scholar]
- Benfey PN, Scheres B (2000) Root development. Curr Biol 10 R813–R815 [DOI] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602 [DOI] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, Makam SN, Cheng Y, Bouchard R, Adamec J, et al (2007) Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433 39–44 [DOI] [PubMed] [Google Scholar]
- Camilleri C, Azimzadeh J, Pastuglia M, Bellini C, Grandjean O, Bouchez D (2002) The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell 14 833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilley PM, Casson SA, Tarkowski P, Hawkins N, Wang KL, Hussey PJ, Beale M, Ecker JR, Sandberg GK, Lindsey K (2006) The POLARIS peptide of Arabidopsis regulates auxin transport and root growth via effects on ethylene signaling. Plant Cell 18 3058–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong A (2006) Switching the flip: protein phosphatase roles in signaling pathways. Curr Opin Plant Biol 9 470–477 [DOI] [PubMed] [Google Scholar]
- Deruère J, Jackson K, Garbers C, Söll D, DeLong A (1999) The RCN1-encoded A subunit of protein phosphatase 2A increases phosphatase activity in vivo. Plant J 20 389–399 [DOI] [PubMed] [Google Scholar]
- Fuglsang AT, Tulinius G, Cui N, Palmgren MG (2006) Protein phosphatase 2A scaffolding subunit A interacts with plasma membrane H+-ATPase C-terminus in the same region as 14-3-3 protein. Physiol Plant 128 334–340 [Google Scholar]
- Fukaki H, Wysocka-Diller J, Kato T, Fujisawa H, Benfey PN, Tasaka M (1998) Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J 14 425–430 [DOI] [PubMed] [Google Scholar]
- Garbers C, DeLong A, Deruère J, Bernasconi P, Söll D (1996) A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J 15 2115–2124 [PMC free article] [PubMed] [Google Scholar]
- Gotz J, Probst A, Mistl C, Nitsch RM, Ehler E (2000) Distinct role of protein phosphatase 2A subunit Calpha in the regulation of E-cadherin and beta-catenin during development. Mech Dev 93 83–93 [DOI] [PubMed] [Google Scholar]
- Groves MR, Hanlon N, Turowski P, Hemmings BA, Barford D (1999) The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell 96 99–110 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25 989–994 [DOI] [PubMed] [Google Scholar]
- Harris DM, Myrick TL, Rundle SJ (1999) The Arabidopsis homolog of yeast TAP42 and mammalian alpha4 binds to the catalytic subunit of protein phosphatase 2A and is induced by chilling. Plant Physiol 121 609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombauer H, Weismann D, Mudrak I, Stanzel C, Fellner T, Lackner DH, Ogris E (2007) Generation of active protein phosphatase 2A is coupled to holoenzyme assembly. PLoS Biol 5 e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V, Goris J (2001) Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J 353 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk D, Conley TR, Rodriguez FA, Tran HT, Nimick M, Muench DG, Moorhead GB (2006) A chloroplast-localized dual-specificity protein phosphatase in Arabidopsis contains a phylogenetically dispersed and ancient carbohydrate-binding domain, which binds the polysaccharide starch. Plant J 46 400–413 [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Nemoto T, Nabeshima K, Kondoh H, Niwa H, Yanagida M (1996) The regulatory subunits of fission yeast protein phosphatase 2A (PP2A) affect cell morphogenesis, cell wall synthesis and cytokinesis. Genes Cells 1 29–45 [DOI] [PubMed] [Google Scholar]
- Kwak JM, Moon JH, Murata Y, Kuchitsu K, Leonhardt N, DeLong A, Schroeder JI (2002) Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PB, Cancel JD (2003) Enhanced ethylene responsiveness in the Arabidopsis eer1 mutant results from a loss-of-function mutation in the protein phosphatase 2A A regulatory subunit, RCN1. Plant J 34 709–718 [DOI] [PubMed] [Google Scholar]
- Larsen PB, Chang C (2001) The Arabidopsis eer1 mutant has enhanced ethylene responses in the hypocotyl and stem. Plant Physiol 125 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Ellis BE (2007) Arabidopsis MAPK phosphatase 2 (MKP2) positively regulates oxidative stress tolerance and inactivates the MPK3 and MPK6 MAPKs. J Biol Chem 282 25020–25029 [DOI] [PubMed] [Google Scholar]
- Luo J, Shen G, Yan J, He C, Zhang H (2006) AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J 46 649–657 [DOI] [PubMed] [Google Scholar]
- Meijer HJ, Munnik T (2003) Phospholipid-based signaling in plants. Annu Rev Plant Biol 54 265–306 [DOI] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130 1044–1056 [DOI] [PubMed] [Google Scholar]
- Muday GK, Brady SR, Argueso C, Deruere J, Kieber JJ, DeLong A (2006) RCN1-regulated phosphatase activity and EIN2 modulate hypocotyl gravitropism by a mechanism that does not require ethylene signaling. Plant Physiol 141 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN (2005) Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17 1908–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niittyla T, Comparot-Moss S, Lue WL, Messerli G, Trevisan M, Seymour MD, Gatehouse JA, Villadsen D, Smith SM, Chen J, et al (2006) Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J Biol Chem 281 11815–11818 [DOI] [PubMed] [Google Scholar]
- Nunbhakdi-Craig V, Machleidt T, Ogris E, Bellotto D, White CL III, Sontag E (2002) Protein phosphatase 2A associates with and regulates atypical PKC and the epithelial tight junction complex. J Cell Biol 158 967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Martinez O, Pernas M, Carol RJ, Dolan L (2007) Ethylene modulates stem cell division in the Arabidopsis thaliana root. Science 317 507–510 [DOI] [PubMed] [Google Scholar]
- Pernas M, Garcia-Casado G, Rojo E, Solano R, Sanchez-Serrano JJ (2007) A protein phosphatase 2A catalytic subunit is a negative regulator of abscisic acid signalling. Plant J 51 763–778 [DOI] [PubMed] [Google Scholar]
- Pitluk ZW, McDonough M, Sangan P, Gonda DK (1995) Novel CDC34 (UBC3) ubiquitin-conjugating enzyme mutants obtained by charge-to-alanine scanning mutagenesis. Mol Cell Biol 15 1210–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NE, Mumby MC (2000) Effects of regulatory subunits on the kinetics of protein phosphatase 2A. Biochemistry 39 11312–11318 [DOI] [PubMed] [Google Scholar]
- Rashotte AM, DeLong A, Muday GK (2001) Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. Plant Cell 13 1683–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel MC, Lecourieux D, Ouaked F, Usher SL, Petersen L, Okamoto H, Knight H, Peck SC, Grierson CS, Hirt H, et al (2004) OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427 858–861 [DOI] [PubMed] [Google Scholar]
- Ruediger R, Hentz M, Fait J, Mumby M, Walter G (1994) Molecular model of the A subunit of protein phosphatase 2A: interaction with other subunits and tumor antigens. J Virol 68 123–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99 463–472 [DOI] [PubMed] [Google Scholar]
- Sablina AA, Chen W, Arroyo JD, Corral L, Hector M, Bulmer SE, DeCaprio JA, Hahn WC (2007) The tumor suppressor PP2A Abeta regulates the RalA GTPase. Cell 129 969–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhanam A, Hartley A, Duvel K, Broach JR, Garrett S (2004) PP2A phosphatase activity is required for stress and Tor kinase regulation of yeast stress response factor Msn2p. Eukaryot Cell 3 1261–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpute-Krishnan P, Serio TR (2005) Prion protein remodelling confers an immediate phenotypic switch. Nature 437 262–265 [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9 236–243 [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Kazanaviciute V, Scheikl E, Teige M, Doczi R, Hirt H, Schwanninger M, Kant M, Schuurink R, Mauch F, et al (2007) The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19 2213–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Shin HS, Guo Z, Blancaflor EB, Masson PH, Chen R (2005) Complex regulation of Arabidopsis AGR1/PIN2-mediated root gravitropic response and basipetal auxin transport by cantharidin-sensitive protein phosphatases. Plant J 42 188–200 [DOI] [PubMed] [Google Scholar]
- Slabas AR, Fordham-Skelton AP, Fletcher D, Martinez-Rivas JM, Swinhoe R, Croy RRD, Evans IM (1994) Characterisation of cDNA and genomic clones encoding homologues of the 65 kDa regulatory subunit of protein phosphatase 2A in Arabidopsis thaliana. Plant Mol Biol 26 1125–1138 [DOI] [PubMed] [Google Scholar]
- Sokolov LN, Dominguez-Solis JR, Allary AL, Buchanan BB, Luan S (2006) A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc Natl Acad Sci USA 103 9732–9737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SK, Clark SE (2005) POL and related phosphatases are dosage-sensitive regulators of meristem and organ development in Arabidopsis. Dev Biol 285 272–284 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM (2005) A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17 2230–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JM, Trotochaud AE, Walker JC, Clark SE (1998) Control of meristem development by CLAVATA1 receptor kinase and kinase-associated protein phosphatase interactions. Plant Physiol 117 1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testerink C, Dekker HL, Lim ZY, Johns MK, Holmes AB, Koster CG, Ktistakis NT, Munnik T (2004) Isolation and identification of phosphatidic acid targets from plants. Plant J 39 527–536 [DOI] [PubMed] [Google Scholar]
- Tian GW, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, Kopec CD, Li J, Ehrhardt D, Jackson D, et al (2004) High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol 135 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Ichimura K, Mizoguchi T, Peck SC, Zhu T, Wang X, Shinozaki K, Paszkowski J (2002) Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J 21 6483–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zyl W, Huang W, Sneddon AA, Stark M, Camier S, Werner M, Marck C, Sentenac A, Broach JR (1992) Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Mol Cell Biol 12 4946–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Xia Y (2006) Proteasome inhibition down-regulates endothelial nitric-oxide synthase phosphorylation and function. J Biol Chem 281 21652–21659 [DOI] [PubMed] [Google Scholar]
- Xing Y, Xu Y, Chen Y, Jeffrey PD, Chao Y, Lin Z, Li Z, Strack S, Stock JB, Shi Y (2006) Structure of protein phosphatase 2A core enzyme bound to tumor-inducing toxins. Cell 127 341–353 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T (2006) ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 140 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu RM, Zhou Y, Xu ZF, Chye ML, Kong RY (2003) Two genes encoding protein phosphatase 2A catalytic subunits are differentially expressed in rice. Plant Mol Biol 51 295–311 [DOI] [PubMed] [Google Scholar]
- Yu S, Lei H, Chang W, Söll D, Hong G (2001) Protein phosphatase 2A: identification in Oryza sativa of the gene encoding the regulatory A subunit. Plant Mol Biol 45 107–112 [DOI] [PubMed] [Google Scholar]
- Zhou HW, Nussbaumer C, Chao Y, DeLong A (2004) Disparate roles for the regulatory A subunit isoforms in Arabidopsis protein phosphatase 2A. Plant Cell 16 709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Pham HT, Ruediger R, Walter G (2003) Characterization of the Aalpha and Abeta subunit isoforms of protein phosphatase 2A: differences in expression, subunit interaction, and evolution. Biochem J 369 387–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.