Abstract
Recent work suggests that the Stagonospora nodorum-wheat pathosystem is controlled by host-selective toxins (HSTs; SnToxA, SnTox1, and SnTox2) that interact directly or indirectly with dominant host genes (Tsn1, Snn1, and Snn2) to induce disease. Here we describe and characterize a novel HST designated SnTox3, and the corresponding wheat sensitivity/susceptibility gene identified on chromosome arm 5BS, which we designated as Snn3. SnTox3 is a proteinaceous necrosis-inducing toxin between 10 and 30 kD in size. The S. nodorum isolates Sn1501 (SnToxA−, SnTox2+, and SnTox3+), SN15 (SnToxA+, SnTox2+, and SnTox3+), and SN15KO18, a strain of SN15 with a disrupted form of SnToxA, were evaluated on a population of wheat recombinant inbred lines. A compatible Snn3-SnTox3 interaction played a significant role in the development of disease caused by isolates Sn1501 and SN15KO18, with Snn2 being epistatic to Snn3. Snn3 was not significantly associated with disease caused by SN15 presumably due to the major effects observed for Snn2 and Tsn1, which were largely additive. This work introduces a fourth HST produced by S. nodorum and builds on the notion that the S. nodorum-wheat pathosystem is largely based on multiple host-toxin interactions that follow an inverse gene-for-gene scenario.
Stagonospora nodorum blotch (SNB) caused by the necrotrophic fungal pathogen Phaeosphaeria nodorum (anamorph; Stagonospora nodorum) is a devastating foliar and glume disease of common wheat (Triticum aestivum; 2n = 6x = 42, AABBDD genomes) and durum wheat (Triticum turgidum; 2n = 4x = 28, AABB genomes). The disease occurs in all major wheat growing areas of the world and has the ability to cause significant yield losses (Eyal, 1981; King et al., 1983) and negatively impact grain quality (Eyal et al., 1987). A high level of genetic resistance, when available, is the most economical and environment-friendly means of controlling losses to SNB.
Reports of the inheritance of resistance to SNB have ranged from qualitative to quantitative (for review, see Xu et al., 2004). Numerous studies conducted to identify genomic regions associated with Stagonospora nodorum glume blotch and Stagonospora nodorum leaf blotch (SNLB) have been conducted and have revealed the presence of multiple quantitative trait loci (QTLs) on various chromosomes. However, only a few of the QTLs have been found to be associated with both leaf and glume blotch resistance (Czembor et al., 2003; Aguilar et al., 2005).
Host-selective toxins (HSTs) are important in many host-pathogen systems as determinants of disease specificity (Wolpert et al., 2002). Unlike classical gene-for-gene interactions, a compatible host-toxin interaction usually relies on the recognition of the toxin by a dominant host gene, which leads to toxin sensitivity and enhanced disease susceptibility. Absence of either the toxin or the dominant host gene precludes recognition and results in an incompatible, or resistant, response. Therefore, host-toxin interactions are mirror images of classical gene-for-gene interactions and may be referred to as inverse gene-for-gene systems.
To date, three proteinaceous necrosis-inducing HSTs produced by S. nodorum have been reported. Liu et al. (2004a) described the characterization and partial purification of the first HST from S. nodorum, which was designated SnTox1. Host sensitivity to SnTox1 was conferred by a single dominant gene designated Snn1, which was physically and genetically mapped to the distal end of the short arm of chromosome 1B using the ‘Chinese Spring’ wheat chromosome deletion lines (Endo and Gill, 1996) and the International Triticeae Mapping Initiative (ITMI) mapping population, respectively. Inoculation of the ITMI population with spores of the SnTox1-producing S. nodorum isolate Sn2000 and subsequent QTL analysis indicated that the Snn1 locus accounted for as much as 58% of the phenotypic variation in susceptibility to SNLB, demonstrating that SnTox1 played an important role in causing disease (Liu et al., 2004b).
The second S. nodorum toxin to be identified was SnToxA (Friesen et al., 2006). The toxin was first identified in the tan spot fungus (Pyrenophora tritici-repentis; Tomás and Bockus, 1987; Lamari and Bernier, 1989), which is another important necrotrophic pathogen of wheat and durum, and designated Ptr ToxA. The purification of Ptr ToxA indicated that it was a protein of about 13 kD (for review, see Ciuffetti and Tuori, 1999), and was encoded by a single gene (Ciuffetti et al., 1997). Friesen et al. (2006) identified a gene with 99.7% similarity to the Ptr ToxA gene in the S. nodorum genome and showed that the S. nodorum toxin, designated SnToxA, was a major factor in the development of SNLB on wheat genotypes that carry the Tsn1 gene. Tsn1, which maps to the long arm of chromosome 5B, was previously known to confer sensitivity to Ptr ToxA (Faris et al., 1996; Haen et al., 2004; Lu et al., 2006). More recently, it was demonstrated that Tsn1 confers sensitivity to both Ptr ToxA and SnToxA and accounted for as much as 62% of the phenotypic variation in susceptibility to SNLB (Friesen et al., 2006; Liu et al., 2006).
The third S. nodorum HST identified was SnTox2 (Friesen et al., 2007), which was described as a proteinaceous HST that caused extensive necrosis on wheat genotypes harboring the Snn2 gene on the short arm of chromosome 2D. Spore inoculation experiments of a wheat recombinant inbred (RI) population derived from BR34 × ‘Grandin’ segregating for Tsn1 and Snn2 were done with a S. nodorum isolate that produced both SnToxA and SnTox2. Results indicated that the Tsn1 and Snn2 loci accounted for 47% and 20% of the variation in SNLB susceptibility, respectively. The effects of these two loci were mainly additive, and together they accounted for 66% of the phenotypic variation indicating that SnToxA and SnTox2 are highly important factors in conferring disease.
The three HSTs identified in the S. nodorum system so far have been shown to be major determinants of disease susceptibility. Here, we report the identification of a fourth HST produced by S. nodorum and the host gene that confers sensitivity to the toxin. We also report the chromosomal location of the host gene and evaluation of the role of the toxin-host gene interaction in conferring susceptibility to SNLB.
RESULTS
Identification of a New HST
Culture filtrates were prepared from the isolate Sn1501 and used to infiltrate leaves of BR34, ‘Grandin’, and individual RI lines of the BG population derived from the cross of BR34 × ‘Grandin’ (BG). Three days after infiltration, leaves were scored as either sensitive or insensitive based on the presence or absence of cell death (Fig. 1). BR34 was insensitive to the culture filtrate and ‘Grandin’ was sensitive. The population segregated in a ratio of 84:34 sensitive to insensitive, which fit a 3:1 ratio (χ2 = 0.915, P = 0.339) for the segregation of two genes controlling toxin sensitivity.
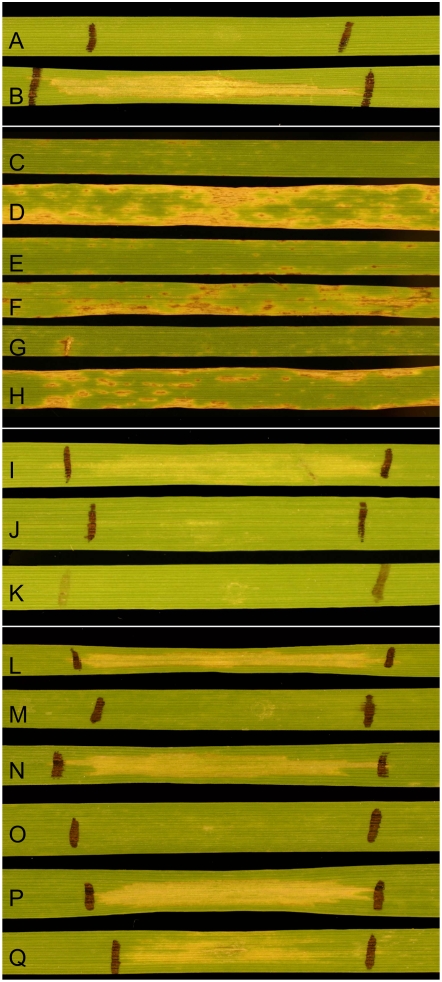
Figure 1.
Infiltration and inoculation reactions. A and B, BR34 (A) and ‘Grandin’ (B) infiltrated with culture filtrates of Sn1501 containing both SnTox2 and SnTox3. C and D, BR34 (C) and ‘Grandin’ (D) inoculated with SN15 (SnToxA+, SnTox2+, and SnTox3+). E and F, BR34 (E) and ‘Grandin’ (F) inoculated with SN15KO18 (SnToxA−, SnTox2+, and SnTox3+). G and H, BR34 (G) and ‘Grandin’ (H) inoculated with Sn1501 (SnToxA−, SnTox2+, and SnTox3+). The SnTox3-sensitive differential line BG220 was infiltrated with fractions containing SnTox3 treated with 1× MOPS buffer alone (I) and pronase in 1× MOPS buffer (J) with pronase alone in 1× MOPS buffer (K) being used as a control. Size estimation was done using Centricon MWCO filters and infiltrating the filtrate and concentrate into the SnTox3-sensitive differential line BG220. Line BG220 was sensitive to the filtrate (P) and concentrate (Q) of the 30,000 MWCO filter as well as the concentrate of the 10,000 (N) and the 3,000 (L) MWCO filter; however, the filtrate of the 10,000 (O) and 3,000 (M) MWCO filter showed no necrosis when infiltrated into the SnTox3-sensitive BG220 differential line.
The sensitive/insensitive reactions of the RI lines were given numerical values and used to perform simple linear regression (SLR) analysis on the BG population marker data set to identify genomic regions significantly associated with toxin sensitivity. The results indicated that the genomic region on chromosome arm 2DS harboring the Snn2 gene and a region on the distal end of the short arm of chromosome 5B were significantly associated with sensitivity to the Sn1501 culture filtrate (Fig. 2). Because Snn2 confers sensitivity to SnTox2 (Friesen et al., 2007), it was presumed that one of the toxins in the Sn1501 culture filtrate was SnTox2. The second toxin was presumed to be a novel toxin because no host toxin sensitivity loci have been mapped to chromosome arm 5BS. Therefore, the Sn1501 culture filtrate contained at least two HSTs, one of which is most likely SnTox2 (Friesen et al., 2007), and a second previously unidentified toxin, which we propose to designate SnTox3.
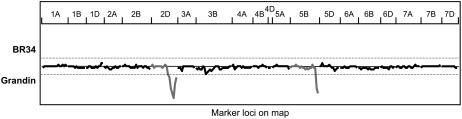
Figure 2.
SLR analysis of reactions to culture filtrates of S. nodorum isolate Sn1501 and 354 markers mapped in the BG population. Marker loci are represented on the x axis and chromosomes are indicated in horizontal positions across the top. The dotted lines represent the significance threshold of P < 0.001. Peaks below the x axis indicate toxin sensitivity is contributed by ‘Grandin’. Regression lines for chromosomes 2D and 5B, which harbor Snn2 and Snn3, respectively, are shown in gray.
Because the reactions of the BG RI lines to SnToxA and SnTox2 were previously obtained (Friesen et al., 2006, 2007; Liu et al., 2006), we were able to identify those BG lines that were sensitive to only SnTox3. The RI line BG220 was sensitive to SnTox3 but insensitive to both SnTox2 and SnToxA and was therefore used as a differential line for additional SnTox3 bioassays. The S. nodorum isolate SN15, which was used for the genomic sequencing of S. nodorum (Hane et al., 2007), was previously reported to produce SnToxA (Friesen et al., 2006). Using RI lines BG223 and BG220, the differential lines for SnTox2 (Friesen et al., 2007) and SnTox3 sensitivity, respectively, we determined that SN15 also produces both SnTox2 and SnTox3 in culture in addition to SnToxA (data not shown).
Partial Purification and Characterization of SnTox3
Dialyzed culture filtrates of isolate Sn1501 containing SnTox2 and SnTox3 were loaded onto a 1-mL HiTrap SP XL cation exchange column (GE Healthcare) followed by a gradient elution of 0 to 300 mm sodium chloride. SnTox2 activity peaked at approximately 205 mm NaCl, whereas SnTox3 activity peaked at approximately 160 mm NaCl, giving baseline separation between the two toxins.
Ion exchange fractions containing active SnTox3 were subjected to size exclusion chromatography using a HiLoad 16/60 Superdex 30 prep-grade gel filtration column (GE Healthcare). The strongest SnTox3 activity eluted after blue dextran (2,000 kD), in the same fraction as cytochrome c (12.4 kD) and significantly before aprotinin (6.5 kD), indicating that it is in a size range close to that of cytochrome c. This is in contrast to SnTox2, which eluted in the same fraction as aprotinin (6.5 kD). Culture filtrates containing active fractions of SnTox3 were subjected to ultrafiltration using 30-kD and 10-kD Centricon molecular mass cutoff filters (Millipore). Both the concentrate and the filtrate of active fractions of SnTox3 subjected to a 30-kD filter contained necrosis-inducing activity on BG line 220 (Fig. 1). Activity was also present in the 10-kD concentrate (Fig. 1), but no SnTox3 activity was present in the 10-kD filtrate (Fig. 1). Collectively, the results of ultrafiltration and size exclusion chromatography indicate that SnTox3 is between 10 and 30 kD (Fig. 1) and is likely similar in size to cytochrome c (12.4 kD).
BG220 plants subjected to SnTox3 treated with pronase showed no sensitivity, whereas fractions subjected to buffer alone started to developed necrosis after 24 h and were completely necrotic at 72 h (Fig. 1), indicating that SnTox3, like SnToxA, SnTox1, and SnTox2, is a protein. Pronase alone had no visible effect on BG line 220.
Genetic Analyses of Host Sensitivity to SnTox3
The partial purification of SnTox3 allowed us to evaluate the genetics of host sensitivity to SnTox2 and SnTox3 independently. Semipurified preparations containing SnTox3 were used to infiltrate BR34, ‘Grandin’, and the BG RI lines. As with the Sn1501 culture filtrate, ‘Grandin’ and sensitive RI lines developed necrosis within the site of infiltration, whereas BR34 and insensitive RI lines showed no reaction (Fig. 1). The BG population segregated in a ratio of 53:65 sensitive to insensitive for reaction to the partially purified SnTox3 culture, which fit the expected ratio for a single gene conferring sensitivity (χ2 = 1.22, P = 0.269). This indicates that the partially purified SnTox3 preparation contained only SnTox3 and not SnTox2. We propose the symbol Snn3 to designate the gene conferring sensitivity to SnTox3 in wheat.
Leaves of F1 individuals derived from the cross between BR34 and ‘Grandin’ were infiltrated with partially purified SnTox3 to determine whether host sensitivity was dominant or recessive in nature. Three F1 plants were infiltrated and all showed necrosis development 3 d after infiltration. A total of 137 F2 plants of the BR34 × ‘Grandin’ cross were also infiltrated. F2 plants segregated in a ratio of 98:39 sensitive to insensitive for reaction to partially purified SnTox3 fractions. This fit the expected 3:1 ratio for a single gene conferring sensitivity (χ2 = 0.878, P = 0.349). Therefore, host sensitivity to SnTox3 is conferred by the dominant allele of Snn3.
Genomic Mapping of Snn3
Reactions of BG RI lines to the partially purified SnTox3 cultures were converted to genotypic values and used to assess linkage of Snn3 with markers that were mapped in the BG population. The analysis indicated that Snn3 mapped 1.4 cM distal to the microsatellite marker Xcfd20 on the distal end of the short arm of chromosome 5B (Fig. 3).
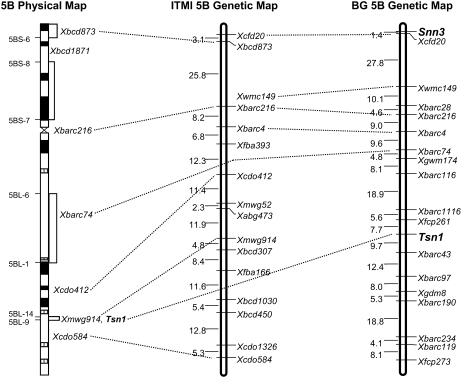
Figure 3.
Genetic and physical maps of wheat chromosome 5B. On the deletion-based physical map (left; Gill et al., 1996; Sourdille et al., 2004), positions of deletion breakpoint are shown along the left of the chromosome and bin-mapped markers are shown along the right. Black and hatched regions indicate locations of chromosome c bands. Genetic maps of chromosome 5B generated in the ITMI population (middle; http://wheat.pw.usda.gov/ggpages/map_summary.html) and the BG population (right; Liu et al., 2005) have centiMorgan distances indicated along the left and markers along the right.
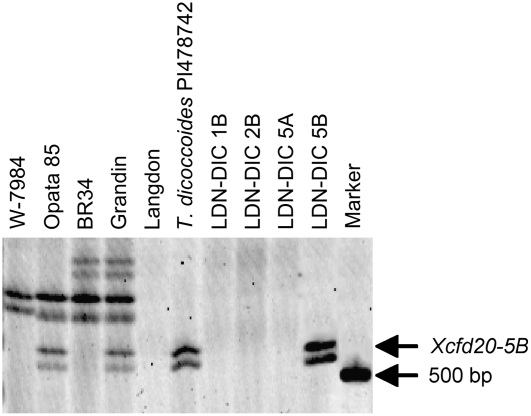
Because Xcfd20 and Snn3 were linked at nearly 30 cM distal to the next closest marker (Xwmc149), we performed experiments to verify that the assignment of Xcfd20 and Snn3 to chromosome 5B was correct and that this was not a false observation due to spurious linkage. Xcfd20 was evaluated on the durum cultivar ‘Langdon’ (LDN) along with the Triticum dicoccoides (2n = 4x = 28, AABB genomes) accession PI478742 (478), which have been used in the development of disomic chromosome substitution lines (Joppa, 1993). The LDN-T. dicoccoides (LDN-DIC) chromosome substitution lines have a pair of T. dicoccoides chromosomes substituted for a homologous pair of LDN chromosomes in a homozygous LDN background. In this experiment, LDN-DIC lines involving substitutions for chromosomes 1B, 2B, 5A, and 5B were evaluated to determine the chromosome assignment of Xcfd20 (Fig. 3). A fragment detected by Xcfd20 and linked to Snn3 was present in the T. dicoccoides accession 478, but absent in LDN. Among the LDN-DIC chromosome substitution lines, only LDN-DIC 5B(IsA) and LDN-DIC 5B(478) had the fragment indicating that the marker resides on chromosome 5B (Fig. 4).
Figure 4.
Polyacrylamide gel electrophoresis of fragments amplified with the SSR primer set CFD20, which correspond to the marker Xcfd20 on chromosome arm 5BS. Lanes are annotated across the top, and the Xcfd20-5B fragment and a 500-bp marker are indicated by arrows along the right. The presence of the Xcfd20 fragment in T. dicoccoides accession PI478742 and the LDN-DIC 5B chromosome substitution line indicates that the fragment resides on chromosome 5B. The same fragment was polymorphic between W-7984 and Opata 85, and between BR34 and ‘Grandin’, and therefore was mapped in the ITMI and BG populations, respectively, to the short arm of 5B (Fig. 3).
The same fragment detected by Xcfd20 was also polymorphic between W-7984 and Opata 85 (Fig. 4), which are the parents of the ITMI mapping population, and subsequently assigned to the short arm of chromosome 5B by linkage analysis (Fig. 3). In the ITMI population, Xcfd20 mapped 3.1 cM distal to the RFLP marker Xbcd873, which was previously shown by Gill et al. (1996) to map to the most distal bin on the deletion-based physical map of wheat chromosome 5B (Fig. 3). Therefore, we demonstrated conclusively that Xcfd20 and Snn3 reside at the distal end of chromosome 5BS.
QTL Analysis of SNLB Susceptibility
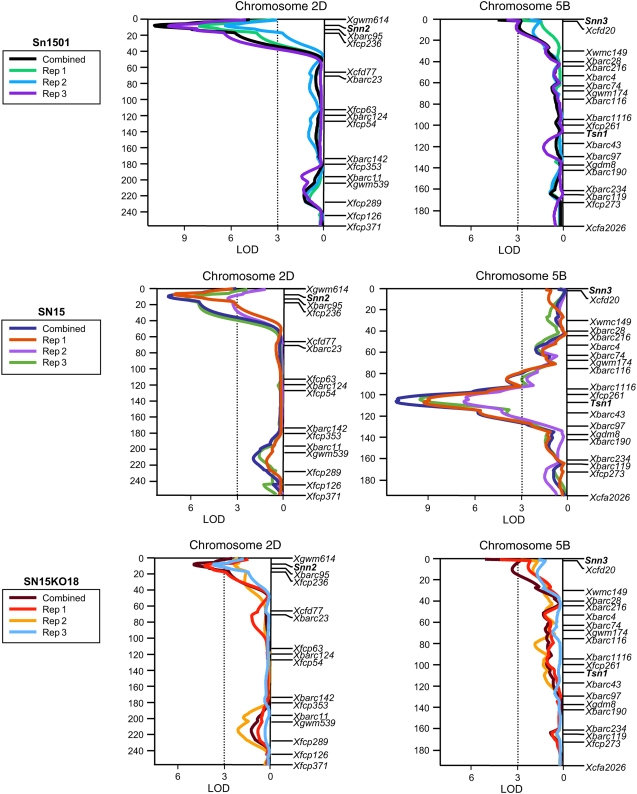
QTL analysis was conducted to quantify the effects of compatible host-toxin interactions in disease susceptibility caused by isolates Sn1501, SN15, and SN15KO18, which is identical to SN15 except that it contains a disrupted SnToxA gene and therefore does not produce SnToxA. The BG population and parents were evaluated for reaction to SNLB using conidial inoculations as described in “Materials and Methods.” Disease reaction types were rated using the 0 to 5 scale described by Liu et al. (2004b) with 0 being highly resistant and 5 being highly susceptible. For each isolate, disease reaction types of three replicates as well as the combined means were regressed on the genotypic marker data available in the BG population using SLR, simple interval mapping (SIM), and composite interval mapping (CIM). For disease reaction to isolate Sn1501, significant QTLs were detected in genomic regions of chromosome arms 2DS and 5BS that corresponded to the Snn2 and Snn3 loci, respectively (Tables I and II; Fig. 5). The 2DS QTL associated with the Snn2 locus, designated QSnb.fcu-2DS (Friesen et al., 2007), was significantly associated with susceptibility to Sn1501 in all three replicates as well as the combined means using interval regression, and explained 37% of the phenotypic variation (Table I). The QTL on the short arm of chromosome 5B associated with the Snn3 locus, hereafter referred to as QSnb.fcu-5BS, was significantly associated with susceptibility to Sn1501 in replicate 3 and the combined means using interval regression (Table I; Fig. 5), and explained 8% and 13% of the phenotypic variation, respectively (Table I). QSnb.fcu-5BS was not significant in replicates 1 and 2 using SIM and CIM, but SLR indicated that the Snn3 locus was significant in all replicates and the combined means at the 0.001 level of probability (Supplemental Table S1). The SLR analysis also indicated that the marker Xgdm125 on chromosome arm 1BS was significantly associated with susceptibility to Sn1501 in replicate 1 (Supplemental Table S1), but this region was not significant using interval regression analysis. Using the combined Sn1501 means, a significant interaction between the Snn2 and Snn3 marker loci was detected at the 0.001 level of probability. Together, Snn2, Snn3, and the interaction between Snn2 and Snn3 explained 48% of the phenotypic variation for susceptibility to Sn1501 (Supplemental Table S2).
Table I.
QTLs associated with SNB disease
QTLs for seedling resistance to SNLB caused by isolates Sn1501, SN15, and SN15KO18 in the BG RI population detected by simple/composite interval mapping for the three replicates (R1–R3) as well as the combined means. The chromosomal locations, associated loci, LOD, R2, and additive effects values are given.
| Isolate | Chromosome Arm | QTL | Locus | LOD
|
R2
|
Additive Effects
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R1 | R2 | R3 | Combined | R1 | R2 | R3 | Combined | R1 | R2 | R3 | Combined | ||||
| Sn1501 | 2DS | QSnb.fcu-2DS | Snn2 | 8.09 | 6.20 | 10.48 | 11.54 | 0.28 | 0.22 | 0.34 | 0.37 | 0.42 | 0.37 | 0.54 | 0.44 |
| Sn1501 | 5BS | QSnb.fcu-5BS | Snn3 | NSa | NS | 3.34 | 4.38 | – | – | 0.08 | 0.13 | – | – | 0.25 | 0.22 |
| SN15 | 2DS | QSnb.fcu-2DS | Snn2 | 7.59 | 3.64 | 6.03 | 7.77 | 0.19 | 0.10 | 0.15 | 0.17 | 0.45 | 0.31 | 0.44 | 0.40 |
| SN15 | 5BL | QSnb.fcu-5BL.1 | Tsn1 | 9.18 | 6.68 | 9.37 | 11.06 | 0.30 | 0.23 | 0.31 | 0.35 | 0.60 | 0.45 | 0.57 | 0.54 |
| SN15KO18 | 2DS | QSnb.fcu-2DS | Snn2 | 4.25 | NS | 3.38 | 5.36 | 0.16 | – | 0.12 | 0.19 | 0.32 | – | 0.22 | 0.24 |
| SN15KO18 | 5BS | QSnb.fcu-5BS | Snn3 | 4.60 | NS | NS | 4.71 | 0.17 | – | – | 0.17 | 0.28 | – | – | 0.20 |
NS, Nonsignificant.
Table II.
Disease reactions of Sn1501 and SN15KO18 according to genotypes
Average and range of disease reaction types of parents and RI lines of the BG population for the four allelic state combinations for Snn2 and Snn3 after inoculation with conidia of S. nodorum isolates Sn1501 and SN15KO18.
| Genotype | No. RI Lines | Sn1501
|
SN15KO18
|
||
|---|---|---|---|---|---|
| Reaction Type Range | Average Reaction Typea | Reaction Type Range | Average Reaction Typea | ||
| BR34 | – | 0–0.5 | 0.17a | 1.0–1.5 | 1.17a |
| ‘Grandin’ | – | 3.5–4.0 | 3.83e | 2.0–3.0 | 2.50d |
| Snn2/Snn3 | 27 | 1.8–3.2 | 2.57d | 1.5–3.2 | 2.12c |
| Snn2/snn3 | 31 | 1.3–3.8 | 2.46d | 1.3–3.3 | 1.92bc |
| snn2/Snn3 | 26 | 0.8–3.0 | 2.11c | 1.3–2.5 | 1.89b |
| snn2/snn3 | 34 | 0.5–2.3 | 1.46b | 0.5–2.2 | 1.37a |
Numbers in the same column followed by the same letter are not significantly different at the 0.05 level of probability.
Figure 5.
Interval regression analysis of chromosomes 2D and 5B for reaction to SNLB produced by S. nodorum isolates Sn1501, SN15, and SN15KO18. Analyses of replicates 1, 2, and 3 and the combined means for each isolate are indicated by different colored lines as depicted in the boxes to the left of the maps for each isolate. A centiMorgan scale is indicated to the left of the maps, and markers are shown in their relative positions along the right. An LOD scale is shown along the x axis, and the critical LOD threshold of 3.0 is indicated by the dotted line.
The same two QTLs (QSnb.fcu-2DS and QSnb.fcu-5BS) associated with susceptibility to Sn1501 were also associated with susceptibility to SN15KO18. Interval regression analysis indicated that QSnb.fcu-2DS was significant in replicates 1 and 3, and the combined means explaining as much as 19% of the variation in susceptibility (Table I; Fig. 5). This QTL was not significant in SIM or CIM of replicate 2, but SLR indicated that Snn2, which underlies QSnb.fcu-2DS, was significant at P < 0.005 and explained 7.5% of the variation (Supplemental Table S1). QSnb.fcu-5BS was significantly associated with susceptibility to SN15KO18 using interval regression in replicate 1 and the combined means explaining 17% of the variation (Table I; Fig. 5). QSnb.fcu-5BS was not significant in replicates 2 and 3 using interval regression, but the Snn3 locus, which underlies QSnb.fcu-5BS, was significant (P < 0.001) in SLR for replicate 2 explaining 11% of the variation (Supplemental Table S1). The marker Xgdm125 on 1BS was significant in the SLR analysis for replicate 3 and the combined means explaining 10% and 11% of the variation, respectively, but it was not significant in the interval regression analysis. As with Sn1501, analysis of SN15KO18 combined means resulted in the identification of a significant (P < 0.05) interaction between Snn2 and Snn3. A multiple regression model consisting of Snn2, Snn3, Xgdm125, and the interaction between Snn2 and Snn3 explained 38% of the variation in disease caused by SN15KO18 (Supplemental Table S3).
Analysis of disease caused by the wild-type SN15 isolate indicated that QSnb.fcu-2DS and the QTL underlaid by Tsn1, which was designated QSnb.fcu-5BL.1 in Liu et al. (2006), were both significantly associated with susceptibility in all replicates as well as the combined means (Tables I and II; Fig. 5). QSnb.fcu-2DS explained as much as 19% of the variation, whereas QSnb.fcu-5BL.1 explained as much as 35%.
Snn3 was not significantly associated with SNLB caused by SN15 using SLR in any of the replicates or the combined means. Furthermore, QSnb.fcu-5BS was not significant in any replicate or the combined means using CIM of chromosome 5B.
As with the Sn1501 and SN15KO18 data, the marker Xgdm125 on chromosome arm 1BS was associated with disease using SLR but not interval analysis. Xgdm125 was significantly associated with reaction to SN15 in replicates 2, 3, and the combined means and explained as much as 9% of the variation (Supplemental Table S1). No significant interactions were identified among loci significantly associated with susceptibility to SN15. Together, Tsn1, Snn2, and Xgdm125 explained 51% of the variation in susceptibility to SN15 (Supplemental Table S4).
The Role of a Compatible Snn3-SnTox3 Interaction in Susceptibility to SNLB
BR34 and ‘Grandin’ had average disease reaction types of 0.17 and 3.83, respectively, in response to conidial inoculations with isolate Sn1501 (Table II; Fig. 1). Average reaction types among the BG RI lines ranged from 0.5 to 3.8 (Supplemental Fig. S1) with an average of 2.11. RI lines sensitive and insensitive to SnTox2 had an average disease reaction type of 2.52 and 1.79, respectively, whereas RI lines sensitive and insensitive to SnTox3 had average disease reaction types of 2.34 and 1.96, respectively. Lines that were insensitive to both toxins were significantly more resistant than lines with sensitivity to either or both toxins (Table II; Supplemental Fig. S2). However, lines with sensitivity to both toxins were not significantly different in disease susceptibility compared to lines having sensitivity to only SnTox2. Therefore, the effects of SnTox3 produced by Sn1501 in causing SNLB were significant only among lines that were insensitive to SnTox2, i.e. harboring the recessive allele at the snn2 locus. The effects of a compatible Snn2-SnTox2 interaction were significant regardless of the allelic state of Snn3, and lines with sensitivity to only SnTox2 had significantly more disease than lines having sensitivity to only SnTox3. Together these results demonstrate that a compatible Snn2-SnTox2 interaction has a greater effect on the development of SNLB than a compatible Snn3-SnTox3 interaction and that the Snn2-SnTox2 interaction is epistatic to the Snn3-SnTox3 interaction.
When inoculated with conidia produced by SN15KO18, BR34 and ‘Grandin’ showed average disease reaction types of 1.17 and 2.50, respectively (Table II; Fig. 1). The BG population had average disease reaction types ranging from 0.5 to 3.3 (Supplemental Fig. S1) with an average of 1.81. Analysis of disease reaction types caused by SN15KO18 among the four allelic classes involving Snn2 and Snn3 revealed many similarities with the results of Sn1501. As with Sn1501, disease reaction types caused by SN15KO18 indicated that lines insensitive to both toxins were significantly more resistant to SNLB than lines with sensitivity to either or both toxins (Table II; Supplemental Fig. S3). Also, lines with sensitivity to both toxins were not more susceptible to SNLB than lines with sensitivity to only SnTox2, but lines sensitive to only SnTox3 were significantly more susceptible than lines insensitive to both toxins, indicating that, as with Sn1501, the effects of a compatible Snn3-SnTox3 interaction are significant only when there is an incompatible Snn2-SnTox2 interaction, i.e. presence of the recessive snn2 allele in the host (Table II). This result agrees with those for Sn1501, and indicates that a compatible Snn2-SnTox2 interaction is epistatic to a compatible Snn3-SnTox3 interaction. However, contrary to the Sn1501 results, the effects of a compatible Snn2-SnTox2 interaction did not have a significantly greater effect on disease susceptibility than a compatible Snn3-SnTox3 interaction at the 0.05 level of probability, indicating that, when alone, each interaction contributed equally to disease development (Table II).
The BG population was also inoculated with the SN15 wild-type isolate, which produces SnToxA, SnTox2, and SnTox3, to compare the disease reaction types with SN15KO18. The only known difference between the two strains is that SN15 produces SnToxA whereas SN15KO18 does not (Friesen et al., 2006). BR34 and ‘Grandin’ had average reaction types of 1.50 and 3.67 (Table III; Fig. 1), respectively. The BG population showed average disease reaction types ranging from 0.8 to 4.3 with an average of 2.25 (Table III; Supplemental Fig. S4). Because the Snn3 locus was not significantly associated with disease caused by SN15 in the QTL analysis (Fig. 5), we evaluated the reaction types of the four genotypic classes involving Tsn1 and Snn2, which were both significantly associated with disease susceptibility. RI lines insensitive to both SnToxA and SnTox2 were significantly more resistant to SNLB than lines with sensitivity to either or both toxins (Table III). In addition, lines with sensitivity to both toxins were more susceptible to SNLB than lines with sensitivity to only one toxin, and lines sensitive to only SnToxA were significantly more susceptible than lines sensitive to only SnTox2. Therefore, the effects of compatible Tsn1-SnToxA and Snn2-SnTox2 interactions were additive, but a compatible Tsn1-SnToxA interaction alone had a significantly greater effect on disease susceptibility than a compatible Snn2-SnTox2 interaction did by itself.
Table III.
Disease reactions of SN15 according to genotypes
Average and range of disease reaction types of parents and RI lines of the BG population for the four allelic state combinations for Tsn1 and Snn2 after inoculation with conidia of S. nodorum isolate SN15.
| Genotype | No. RI Lines | SN15
|
|
|---|---|---|---|
| Reaction Type Range | Average Reaction Typea | ||
| BR34 | – | 1.5 | 1.50a |
| ‘Grandin’ | – | 3.0–4.0 | 3.67e |
| Tsn1/Snn2 | 30 | 1.3–4.3 | 3.20d |
| Tsn1/snn2 | 29 | 1.2–3.7 | 2.28c |
| tsn1/Snn2 | 30 | 1.0–2.8 | 1.96b |
| tsn1/snn2 | 29 | 0.8–2.7 | 1.57a |
Numbers in the same column followed by the same letter are not significantly different at the 0.05 level of probability.
DISCUSSION
We previously identified three proteinaceous HSTs (SnToxA, SnTox1, and SnTox2) produced by S. nodorum, and their corresponding host sensitivity genes (Tsn1, Snn1, and Snn2; Liu et al., 2004a, 2006; Friesen et al., 2006, 2007). Here, we identified a fourth HST (SnTox3) and the corresponding host sensitivity gene (Snn3). Sensitivity to partially purified SnTox3 induces a similar necrotic reaction as that of SnToxA, SnTox1, and SnTox2. SnTox3 is also a relatively small protein, but is within the size range of previously identified proteinaceous HSTs, including those produced by S. nodorum (Liu et al., 2004a; Friesen et al., 2006, 2007).
All four toxin-host gene interactions have been shown to play significant roles in conferring disease susceptibility. The identification and characterization of the Snn3-SnTox3 interaction further strengthens the notion that the S. nodorum-wheat pathosystem provides a model inverse gene-for-gene system that involves multiple proteinaceous HSTs and corresponding host sensitivity genes that interact to cause disease (Friesen et al., 2007). In a classical gene-for-gene system (Flor, 1956), an incompatible host-pathogen interaction involving the recognition of a pathogen-produced elicitor protein by a host resistance gene is characterized by a hypersensitive response (HR) leading to localized programmed cell death (PCD) and is defined as a resistance response (Hammond-Kosack and Jones, 1996). In a classical gene-for-gene interaction, multiple race-specific resistance genes effective against a pathogen have the same effect as a single resistance gene effective against the same pathogen. Compatible S. nodorum-wheat toxin interactions involve the recognition of a pathogen-produced elicitor-like protein (toxin) by a host gene product, but the host response differs in that an interaction between gene products leads to disease susceptibility characterized by extensive cell death rather than a localized HR (Liu et al., 2004a, 2006; Friesen et al., 2006, 2007). Therefore, in the S. nodorum-wheat toxin system, successful interaction of the toxin effectors with the host leads to disease, or increased disease, whereas in a classical gene-for-gene system, interaction leads to resistance; conversely, lack of interaction in an HST system most often leads to incompatibility resulting in less disease, whereas in a gene-for-gene system the lack of interaction leads to full compatibility/susceptibility. Also in contrast to a classic gene-for-gene system, host gene-toxin interactions such as Tsn1-SnToxA and Snn2-SnTox2 have been shown to act in an additive manner (Friesen et al., 2007) where host genotypes with multiple toxin sensitivity genes will be significantly more susceptible than genotypes with sensitivity to a single toxin.
Evaluation of disease caused by isolates Sn1501 and SN15KO18 allowed us to assess the effects of the Snn2-SnTox2 and Snn3-SnTox3 interactions alone and in combination. Evaluation of the four allelic state classes for Snn2 and Snn3 indicated that the effects of a compatible Snn3-SnTox3 interaction on disease susceptibility were significant only in the presence of an incompatible Snn2-SnTox2 interaction. This finding was corroborated by the identification of significant Snn2 × Snn3 interactions in both the Sn1501 and SN15KO18 reaction type data. Therefore, epistatic interactions involving toxin expression, host gene action, and/or cross talk among associated pathways likely exist, which affects the role of the Snn3-SnTox3 interaction in conferring disease susceptibility.
In the SN15 wild-type inoculation, Snn3 was not significantly associated with disease susceptibility. This lack of detection is likely due to the overwhelming effects of the Tsn1-SnToxA interaction in causing disease because the only predicted difference between SN15 and SN15KO18 is the production of SnToxA by the former. It is possible that isolates of S. nodorum contain numerous toxin-encoding genes in their arsenal, but deploy only those that may be the most efficient in causing cell death. The SN15 wild-type inoculation also showed that significantly more disease was present in a compatible Tsn1-SnToxA interaction than in a Snn2-SnTox2 interaction. This is contrary to that reported by Friesen et al. (2007) using the S. nodorum isolate Sn6. In that work, the Snn2-SnTox2 interaction contributed more to disease development than did the Tsn1-SnToxA interaction. It may be that different isolates harbor nucleotide variation within toxin-encoding genes, which allow some toxin isoforms to interact with host gene products more efficiently. Alternatively, different isolates may express toxins at different levels, which may or may not be affected by environmental or other genetic factors.
The combination of epistatic and additive results may give us some clues as to how the S. nodorum-wheat pathosystem operates. Several studies have been done on the mode of action of ToxA in P. tritici-repentis (Kwon et al., 1996, 1998; Rasmussen et al., 2004) showing that host gene expression leading to PCD is driven by the presence of ToxA. This host-induced PCD leads to susceptibility rather than resistance and indicates that the fungal-produced toxin has overtaken one or more host pathways to cause disease, aiding the fungus in vegetative proliferation and ultimately sporulation. Several pathways may be involved in the production of disease induced by toxins. Potentially, two toxins could be using the same pathway or part of a pathway and the toxin that uses that pathway most efficiently would be epistatic over the toxin using it less efficiently, whereas two toxins using different pathways or different parts of a pathway may act in an additive manner.
That the S. nodorum-wheat pathosystem may represent a toxin-based inverse gene-for-gene paradigm is intriguing. In this research, we identified four QTLs associated with disease susceptibility, and genes for three of the QTLs were identified as genes that confer sensitivity to specific toxins. Many genetic experiments to evaluate the host genetics of resistance to S. nodorum employ multiple pathogen isolates or natural infestation and result in the complex inheritance of resistance (Wicki et al., 1999) and/or the identification of multiple minor QTLs associated with host resistance (Czembor et al., 2003; Schnurbusch et al., 2003; Aguilar et al., 2005; Uphaus et al., 2007). In such experiments, multiple toxins produced by a mosaic of isolates may be present, and compatible interactions would occur if the host population harbored corresponding toxin sensitivity genes to recognize the toxins. Therefore, it is possible that the results of these experiments reflect the presence of numerous compatible host-toxin interactions. Such experiments are quite useful for the identification of germplasm resources with high levels of resistance to S. nodorum, but dissection of the S. nodorum-wheat pathosystem and its interactions requires the detailed analysis of specific isolates and host genotypes with knowledge regarding the identity of the toxins and corresponding host genes involved.
Although this and previous research (Liu et al., 2004b, 2006; Friesen et al., 2006, 2007) demonstrated the importance of compatible host-toxin interactions in conferring disease susceptibility, it is possible that the development of SNLB does not rely solely on toxins. Liu et al. (2004b, 2006) and Friesen et al. (2007) previously showed that QTLs in addition to those underlaid by toxin sensitivity genes were important to achieve complete resistance. It is possible that these additional QTLs are indicative of as yet unidentified host-toxin interactions, but they could also reflect the use of different resistance mechanisms by the host. Here, we identified a genomic region on chromosome 1BS marked by Xgdm125 that had some significant association with response to disease caused by all three isolates. Xgdm125 underlies the same QTL, QSnb.fcu-1BS, previously shown to be associated with resistance to S. nodorum isolates Sn2000 (Liu et al., 2006) and Sn6 (Friesen et al., 2007), and for race-nonspecific resistance to tan spot (Faris and Friesen, 2005) in the BG population. Therefore, it is possible that the mechanism responsible for the effects of the 1BS QTL is nonpathogen specific and may be more general in nature.
Increasingly, the S. nodorum-wheat interaction has revealed itself as a complex system of proteinaceous HSTs that interact either directly or indirectly with single dominant host gene products conferring sensitivity. This study is the first, to our knowledge, to identify the Snn3-SnTox3 interaction and to partially characterize the protein SnTox3. In this study, we have further shown a distinct interaction between SnTox2 and the other HSTs as it relates to the role of each toxin in disease. This research shows that although multiple toxins are present in this disease interaction, some toxins are more significant in disease development than others, at least under the conditions presented here. Work is ongoing to characterize additional host-toxin interactions in this important model system as well as to define the role of each toxin at other plant growth stages and under different environmental conditions.
The identification of each new toxin and its corresponding host sensitivity gene, especially as they significantly impact disease severity, is critical in understanding how to breed for the highest levels of resistance. Both semipurified toxins and molecular markers associated with toxin sensitivity are potentially valuable tools useful for the development of commercial cultivars with the highest levels of resistance.
MATERIALS AND METHODS
Plant Materials
A segregating RI population (BG population) derived from a cross between the hexaploid wheat lines BR34 (Brazilian hard red spring wheat [HRSW] breeding line) and ‘Grandin’ (North Dakota HRSW variety) was developed and provided by Dr. James A. Anderson, University of Minnesota. The population consists of 118 F7:9 RI lines used for genetic linkage mapping, phenotypic evaluations, and QTL analysis.
A subset of the ITMI population consisting of 106 RI lines was used to validate the chromosomal location of Xcfd20-5B, which was tightly linked to Snn3 in the BG population. The ITMI population was derived from crossing the synthetic hexaploid wheat W-7984 and the International Maize and Wheat Improvement Center (CIMMYT)-bred HRSW Opata 85 (PI591776) as described by Nelson et al. (1995). W-7984 was developed by A. Mujeeb-Kazi, CIMMYT. The population of RI lines was provided by M. E. Sorrells, Cornell University. In addition to mapping in the ITMI population, the chromosomal location of Xcfd20-5B was further validated using several LDN-DIC disomic chromosome substitution lines, where a pair of Triticum dicoccoides chromosomes derived from accession PI478742 were substituted for a pair of homologous LDN chromosomes.
Fungal Cultures and Culture Filtrate Production
All culture filtrates were produced as described by Liu et al. (2004a). Briefly, cultures were grown in an orbital shaker at 80 rpm at 27°C in the dark for 48 to 72 h followed by stationary growth in the dark at 21°C for 3 weeks. Culture filtrates were filter sterilized using a 0.45-μm filter followed by testing across a toxin differential set to identify which toxins are being produced by each isolate used. The differential set used in this study consisted of lines BG261 (SnToxA+, SnTox2−, SnTox3−), BG223 (SnToxA−, SnTox2+, SnTox3−), and BG220 (SnToxA−, SnTox2−, SnTox3+). These lines were shown to have single toxin sensitivities based on infiltrations of purified SnToxA (Friesen et al., 2006), and semipurified SnTox2 (Friesen et al., 2007) and SnTox3.
Toxin Bioassays and Preliminary Characterization of the Toxin
Culture filtrates from isolate Sn1501 were used in SnTox3 production for initial characterization of this toxin. For all toxin experiments, toxin was infiltrated into plants using a 1-mL syringe with the needle removed as described by Liu et al. (2004a). Sensitivity to protease was determined as described by Friesen et al. (2007). Active SnTox3 fractions were treated for 4 h at room temperature with pronase dissolved in MOPS buffer (pH 7.5) at a final concentration of 1 mg/mL pronase and 50 mm MOPS. Active toxin subjected to MOPS buffer (50 mm) alone and pronase (1 mg/mL) in MOPS buffer (50 mm) were also left at room temperature for 4 h and used as controls.
Preliminary size estimation of SnTox3 was done using both ultrafiltration and size exclusion chromatography. Centricon ultrafiltration devices (Millipore) were used to get a preliminary size estimation. Samples containing SnTox3 were subjected to ultrafiltration using 10,000 Mr cutoff (MWCO) and 30,000 MWCO ultrafiltration devices. Filtrates and concentrates were tested for activity by infiltration into line BG220 (SnTox3 sensitive) to identify which filters retained SnTox3 and which filters allowed flow through. Size exclusion chromatography was also used to evaluate the approximate size of SnTox3. Ion exchange fractions containing active SnTox3 were subjected to size exclusion chromatography using a HiLoad 16/60 Superdex 30 prep-grade gel filtration column (GE Healthcare). Blue dextran (approximately 2,000 kD), cytochrome c (12.4 kD), and aprotinin (6.5 kD) were used as size standards to estimate the approximate size of SnTox3. After infiltration, all plants were placed at 21°C under a 12-h photoperiod at 900 μmol m−2 s−1. Plants were evaluated at 3 d postinfiltration
Conidial Inoculations
Conidial inoculations were done as described by Friesen et al. (2006) with slight modifications. The BG population along with parental lines BR34 and ‘Grandin’ were planted in racks consisting of 98 cones (Steuwe & Sons) including a complete border of ‘Grandin’ to eliminate any edge effect. Sixty lines of a single cone each at a rate of three seeds per line were planted in each rack; therefore, an entire replicate of the BG population of 118 lines plus parents fit into two racks. Conidia were produced and harvested as described by Liu et al. (2004b) and plants were inoculated using 1 × 106 spores/mL and inoculated until runoff. After inoculations, plants were placed in 100% relative humidity at 21°C in the light for 24 h followed by 6 d at 21°C under a 12-h photoperiod at 900 μmol m−2 s−1. Plants were evaluated at 7 d postinoculation using the 0 to 5 scale developed by Liu et al. (2004b) where 0 is highly resistant and 5 is highly susceptible. Three randomized replicates were completed for each isolate tested.
Molecular Mapping
A whole-genome linkage map generated in the BG population consisting of over 700 molecular markers was published previously (Liu et al., 2005). Recently, 107 additional microsatellite markers have been placed on the BG map, which, with the exception of marker Xcfd20-5B, will be published elsewhere. Because Xcfd20-5B is tightly linked to Snn3, it was used to help validate the chromosome location of Snn3. Primer sequences for Xcfd20 were obtained from the Graingenes Web site (http://wheat.pw.usda.gov/GG2/index.shtml). Conditions for PCR were as follows: 94°C for 4 min, followed by 35 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 2 min, and a final extension at 72°C for 10 min. Amplified products were separated on 6% denaturing polyacrylamide gels, stained with Sybr Green, and visualized on a Typhoon 9410 variable mode imager (GE Healthcare) as described by Liu et al. (2005).
The chromosomal locations of the toxin sensitivity genes Tsn1 (Liu et al., 2005, 2006) and Snn2 (Friesen et al., 2007) were previously reported. Here, reactions of RI lines to SnTox3 cultures were assigned genotypic values and assessed for linkage with all other markers in the BG population data set using the Mapmaker v2.0 for Macintosh (Lander et al., 1987) “TRY” command. Using the same method, Xcfd20-5B was mapped in both the BG population and the ITMI population. Data for markers previously mapped to chromosome 5B in the ITMI population were obtained from the Graingenes database (http://wheat.pw.usda.gov/GG2/index.shtml).
QTL Analysis
QTL analysis was used to determine the effects of toxin sensitivity genes in conferring susceptibility to fungal inoculations. The methods for QTL detection in the BG population were the same as previously described for the detection of QTLs associated with resistance to tan spot (Pyrenophora tritici-repentis; Faris and Friesen, 2005), resistance to Stagonospora nodorum isolates Sn2000 (Liu et al., 2006) and Sn6 (Friesen et al., 2007), and plant height and days to heading (Liu et al., 2005). SLR was conducted to identify markers significantly associated with resistance to SNLB caused by isolates Sn1501, SN15, and SN15KO18 at the 0.001 level of probability. Simple and composite interval mapping (SIM and CIM) was performed using a subset of 354 markers to identify marker intervals associated with resistance. The critical log of the odds (LOD) threshold of 3.0 was calculated by executing 1,000 permutations. Single markers with significant main effects were tested for significant (P < 0.05) interactions with other significant main effect markers in the data set. All calculations were performed using either the Map Manager QTX software package (Manly et al., 2001) or qGene v3.06 (Nelson, 1997).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. BG population histograms showing reaction type distribution of S. nodorum isolates.
Supplemental Figure S2. Histograms using isolate Sn1501 comparing BG lines differing in Snn2 and Snn3.
Supplemental Figure S3. Histograms using isolate SN15KO18 comparing BG lines differing in Snn2 and Snn3.
Supplemental Figure S4. Histograms using isolate SN15 comparing BG lines differing in Snn2 and Snn3.
Supplemental Table S1. BG population seedling resistance QTL against S. nodorum isolates.
Supplemental Table S2. Multiple regression of loci associated with SNB caused by isolate Sn1501.
Supplemental Table S3. Multiple regression of loci associated with SNB caused by isolate SN15KO18.
Supplemental Table S4. Multiple regression of loci associated with SNB caused by isolate SN15.
Supplementary Material
Acknowledgments
The authors thank Danielle Holmes, Meghan Rolfsrud, and Erik Doehler for technical assistance, and Drs. Judith Lichtenzveig and John Weiland for critical review of the manuscript.
This work was supported by the U.S. Department of Agriculture-Agriculture Research Service Current Research Information System (projects no. 5442–22000–043–00D and no. 5442–22000–030–00D).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Timothy L. Friesen (timothy.friesen@ars.usda.gov).
The online version of this article contains Web-only data.
References
- Aguilar V, Stamp P, Winzeler M, Winzeler H, Schachermayr G, Keller B, Zanetti S, Messmer MM (2005) Inheritance of field resistance to Stagonospora nodorum leaf and glume blotch and correlations with other morphological traits in hexaploid wheat (Triticum aestivum L.). Theor Appl Genet 111 325–336 [DOI] [PubMed] [Google Scholar]
- Ciuffetti LM, Tuori RP (1999) Advances in the characterization of the Pyrenophora tritici-repentis-wheat interaction. Phytopathology 89 444–449 [DOI] [PubMed] [Google Scholar]
- Ciuffetti LM, Tuori RP, Gaventa JM (1997) A single gene encodes a selective toxin causal to the development of tan spot of wheat. Plant Cell 9 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czembor PC, Arseniuk E, Czaplicki A, Song QJ, Cregan PB, Ueng PP (2003) QTL mapping of partial resistance in winter wheat to Stagonospora nodorum blotch. Genome 46 546–554 [DOI] [PubMed] [Google Scholar]
- Endo TR, Gill BS (1996) The deletion stocks of common wheat. J Hered 87 295–307 [Google Scholar]
- Eyal Z (1981) Integrated control of Septoria diseases of wheat. Plant Dis 65 763–768 [Google Scholar]
- Eyal Z, Scharen AL, Prescott JM, van Ginkel M (1987) The Septoria Diseases of Wheat: Concepts and Methods of Disease Management. CIMMYT, Texcoco, Mexico
- Faris JD, Anderson JA, Francl LJ, Jordahl JG (1996) Chromosomal location of a gene conditioning insensitivity in wheat to a necrosis-inducing culture filtrate from Pyrenophora tritici-repentis. Phytopathology 86 459–463 [Google Scholar]
- Faris JD, Friesen TL (2005) Identification of quantitative trait loci for race-nonspecific resistance to tan spot of wheat. Theor Appl Genet 111 386–392 [DOI] [PubMed] [Google Scholar]
- Flor HH (1956) The complementary genetic systems in flax and flax rust. Adv Genet 8 29–54 [Google Scholar]
- Friesen TL, Meinhardt SW, Faris JD (2007) The Stagonospora nodorum-wheat pathosystem involves multiple proteinaceous host-selective toxins and corresponding host sensitivity genes that interact in an inverse gene-for-gene manner. Plant J 51 681–692 [DOI] [PubMed] [Google Scholar]
- Friesen TL, Stukenbrock EH, Liu ZH, Meinhardt SW, Ling H, Faris JD, Rasmussen JB, Solomon PS, McDonald BA, Oliver RP (2006) Emergence of a new disease as a result of interspecific virulence gene transfer. Nat Genet 38 953–956 [DOI] [PubMed] [Google Scholar]
- Gill KS, Gill BS, Endo TR, Boiko EV (1996) Identification and high-density mapping of gene-rich regions in chromosome group 5 of wheat. Genetics 143 1001–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haen KM, Lu H-J, Friesen TL, Faris JD (2004) Genomic targeting and high-resolution mapping of the Tsn1 gene in wheat. Crop Sci 44 951–962 [Google Scholar]
- Hammond-Kosack KE, Jones JDG (1996) Resistance gene-dependent plant defense response. Plant Cell 8 1773–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hane JK, Lowe RGT, Solomon PS, Tan K-C, Schoch CL, Spatafora JW, Crous PW, Kodira C, Birren BW, Torriani SFF, et al (2007) Dothideomycete-plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum. Plant Cell 19 3347–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joppa LR (1993) Chromosome engineering in tetraploid wheat. Crop Sci 33 908–913 [Google Scholar]
- King JE, Cook RJ, Melville SC (1983) A review of the Septoria diseases of wheat and barley. Ann Appl Biol 103 345–374 [Google Scholar]
- Kwon CY, Rasmussen JB, Francl LJ, Meinhardt SW (1996) A quantitative bioassay for necrosis toxin from Pyrenophora tritici-repentis based on electrolyte leakage. Phytopathology 86 1360–1363 [Google Scholar]
- Kwon CY, Rasmussen JB, Meinhardt SW (1998) Activity of Ptr ToxA from Pyrenophora tritici-repentis requires host metabolism. Physiol Mol Plant Pathol 52 201–212 [Google Scholar]
- Lamari L, Bernier CC (1989) Toxin of Pyrenophora tritici-repentis: host-specificity significance in disease, and inheritance of host reaction. Phytopathology 79 740–744 [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1 174–181 [DOI] [PubMed] [Google Scholar]
- Liu ZH, Anderson JA, Hu J, Friesen TL, Rasmussen JB, Faris JD (2005) A wheat intervarietal genetic linkage map based on microsatellite and target region amplified polymorphism markers and its utility for detecting quantitative trait loci. Theor Appl Genet 111 792–794 [DOI] [PubMed] [Google Scholar]
- Liu ZH, Faris JD, Meinhardt SW, Ali S, Rasmussen JB, Friesen TL (2004. a) Genetic and physical mapping of a gene conditioning sensitivity in wheat to a partially purified host-selective toxin produced by Stagonospora nodorum. Phytopathology 94 1056–1060 [DOI] [PubMed] [Google Scholar]
- Liu ZH, Friesen TL, Ling H, Meinhardt SW, Oliver RP, Rasmussen JB, Faris JD (2006) The Tsn1-ToxA interaction in the wheat-Stagonospora nodorum pathosystem parallels that of the wheat-tan spot system. Genome 49 1265–1273 [DOI] [PubMed] [Google Scholar]
- Liu ZH, Friesen TL, Meinhardt SW, Ali S, Rasmussen JB, Faris JD (2004. b) Quantitative trait loci analysis and mapping of seedling resistance to Stagonospora nodorum leaf blotch in wheat. Phytopathology 94 1061–1067 [DOI] [PubMed] [Google Scholar]
- Lu HJ, Fellers JP, Friesen TL, Meinhardt SW, Faris JD (2006) Genomic analysis and marker development for the Tsn1 locus using bin-mapped ESTs and flanking BAC contigs. Theor Appl Genet 112 1132–1142 [DOI] [PubMed] [Google Scholar]
- Manly KK Jr, Cudmore HH, Meer JM (2001) Map Manager QTX, cross platform software for genetic mapping. Mamm Genome 12 930–932 [DOI] [PubMed] [Google Scholar]
- Nelson JC (1997) QGENE: software for marker-based genomic analysis and breeding. Mol Breed 3 239–245 [Google Scholar]
- Nelson JC, Van Deynze AE, Autrique E, Sorrells ME, Lu YH, Negre S, Bernard M, Leroy P (1995) Molecular mapping in wheat. Homoeologous group 3. Genome 38 525–533 [DOI] [PubMed] [Google Scholar]
- Rasmussen JB, Kwon CY, Meinhardt SW (2004) Requirement of host signaling mechanisms for the action of Ptr ToxA in wheat. Eur J Plant Pathol 110 333–335 [Google Scholar]
- Schnurbusch T, Paillard S, Fossati D, Messmer M, Schachermayr G, Winzeler M, Keller B (2003) Detection of QTLs for Stagonospora glume blotch resistance in Swiss winter wheat. Theor Appl Genet 107 1226–1234 [DOI] [PubMed] [Google Scholar]
- Sourdille P, Singh S, Cadalen T, Brown-Guedira GL, Gay G, Qi L, Gill BS, Dufour P, Murigneux A, Bernard M (2004) Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct Integr Genomics 4 12–25 [DOI] [PubMed] [Google Scholar]
- Tomás A, Bockus WW (1987) Cultivar-specific toxicity of culture filtrate of Pyrenophora tritici-repentis. Phytopathology 77 1337–1340 [Google Scholar]
- Uphaus J, Walker E, Shankar M, Golzar H, Loughman R, Francki M, Ohm H (2007) QTL identified for resistance to Stagonospora glume blotch in wheat in the USA and Australia. Crop Sci 47 1813–1822 [Google Scholar]
- Wicki W, Winzeler M, Schmid JE, Stamp P, Messmer M (1999) Inheritance of resistance to leaf and glume blotch caused by Septoria nodorum Berk. in winter wheat. Theor Appl Genet 99 1265–1272 [Google Scholar]
- Wolpert TJ, Dunkle LD, Ciuffetti LM (2002) Host-selective toxins and avirulence determinants: What's in a name? Annu Rev Phytopathol 40 251–285 [DOI] [PubMed] [Google Scholar]
- Xu SS, Friesen TL, Cai XW (2004) Sources and genetic control of resistance to Stagonospora nodorum blotch in wheat. Recent Res Devel Genet Breeding 1 449–469 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.