Abstract
High-performance liquid chromatography coupled to ultraviolet photodiode array detection and ion-trap mass spectrometry was used to analyze the intra- and extracellular secondary product metabolome of Medicago truncatula cell suspension cultures responding to yeast elicitor (YE) or methyl jasmonate (MeJA). Data analysis revealed three phases of intracellular response to YE: a transient response in mainly (iso)flavonoid metabolites such as formononetin and biochanin-A that peaked at 12 to 18 h following elicitation and then declined; a sustained response through 48 h for compounds such as medicarpin and daidzin; and a lesser delayed and protracted response starting at 24 h postelicitation, e.g. genistein diglucoside. In contrast, most compounds excreted to the culture medium reached maximum levels at 6 to 12 h postelicitation and returned to basal levels by 24 h. The response to MeJA differed significantly from that to YE. Although both resulted in accumulation of the phytoalexin medicarpin, coordinated increases in isoflavonoid precursors were only observed for YE and not MeJA-treated cells. However, MeJA treatment resulted in a correlated decline in isoflavone glucosides, and did not induce the secretion of metabolites into the culture medium. Three novel methylated isoflavones, 7-hydroxy-6,4′-dimethoxyisoflavone (afrormosin), 6-hydroxy-7,4′-dimethoxyisoflavone (alfalone), and 5,7-dihydroxy-4′,6-dimethoxy isoflavone (irisolidone), were induced by YE, and labeling studies indicated that the first two were derived from formononetin. Our results highlight the metabolic flexibility within the isoflavonoid pathway, suggest new pathways for complex isoflavonoid metabolism, and indicate differential mechanisms for medicarpin biosynthesis depending on the nature of elicitation.
Medicago truncatula is a rapidly developing model organism for the study of legume biology and a close relative of alfalfa (Medicago sativa), a premium and globally grown forage legume. As a legume, M. truncatula establishes symbiotic relationships with nitrogen-fixing rhizobia (Oldroyd, 2001) as well as beneficial arbuscular mycorrhizae (Harrison, 1999). Legumes produce an array of natural products that have a substantial impact upon mutualism as well as plant disease/defense (Dixon and Sumner, 2003). Particularly important are the phenylpropanoid-derived isoflavonoids that serve as key signaling molecules in plant-microbe interactions and as primary defense compounds. Isoflavonoids have also been ascribed a large number of pharmacological and nutraceutical properties, including chemoprevention of osteoporosis (Alekel et al., 2000; Uesugi et al., 2001) and other postmenopausal disorders (MerzDemlow et al., 2000), antioxidants related to improved cardiovascular health (Lichtenstein, 1998; Setchell and Cassidy, 1999; Heim et al., 2002), and reduced risk of breast and prostate cancers in humans (Adlercreutz, 1998; Lamartiniere, 2000). However, a more comprehensive understanding of isoflavonoid biosynthesis is still needed for the efficient engineering and exploitation of these compounds for plant, animal, and human benefit.
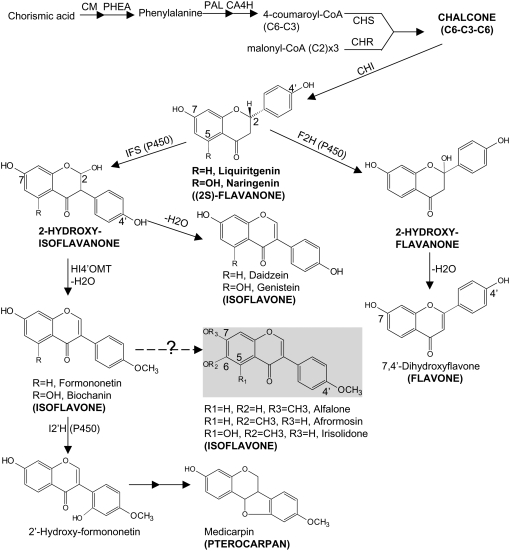
Flavanones are ubiquitous intermediates leading to the biosynthesis of all other flavonoid subclasses (Fig. 1). Isoflavones are synthesized from the flavanones naringenin and liquiritigenin via migration of the B-ring from the 2- to the 3-position, followed by hydroxylation at the 2-position. This complex reaction is catalyzed by isoflavone synthase (IFS), a cytochrome P450 enzyme, and yields the immediate product 2-hydroxyisoflavanone that is subsequently dehydrated, either spontaneously or enzymatically, to the corresponding isoflavone (Dixon, 1999; Akashi et al., 2005). In this way, IFS converts liquiritigenin to daidzein and naringenin to genistein. 4′-O-Methylation of the 2-hydroxyisoflavanone formed by the action of IFS on liquiritigenin yields an intermediate that is dehydrated to yield formononetin, a precursor of the phytoalexin medicarpin. Figure 1 illustrates the core pathways related to isoflavonoid biosynthesis, as well as the primary metabolic pathways that provide precursors for isoflavonoid production. Although the basic features of these pathways have been known for more than a decade, several areas of isoflavonoid metabolism, including specific dehydration and substitution reactions (e.g. glycosylation, O-methylation, and prenylation), as well as the steps involved in the biosynthesis of related compounds such as coumestans, have yet to be elucidated at the molecular level. Furthermore, little is known concerning sites of flux control, cross talk between isoflavonoid biosynthesis and competing pathways, the physical basis for association of biosynthetic enzymes in metabolic channels, or the regulatory mechanisms involved in isoflavonoid biosynthesis in response to different stress responses (Dakora and Phillips, 1996; Dixon and Sumner, 2003).
Figure 1.
Proposed biosynthetic pathways leading to the major classes of flavonoids in M. truncatula: chalcones, flavanones, isoflavones, and pterocarpans. Solid arrows indicate established biochemical reactions, whereas dashed arrows indicate possible steps not yet described. Isoflavones highlighted in yellow are identified for the first time in M. truncatula. The carbon numbering schema for flavone, flavanone, and isoflavone is marked. Enzymes are as follows: CA4H, cinnamate 4-hydroxylase; CHI, chalcone isomerase; CHR; CHS; CM, chorismate mutase; F2H, flavanone 2-hydroxylase; HI4′OMT, 2,7,4′-trihydroxyisoflavanone 4′-O-methyltransferase; I2′H, isoflavone 2′-hydroxylase; IFS; PAL; PHEA, prephenate deyhdratase.
M. truncatula is an ideal model for addressing legume natural product biosynthesis at the molecular genetic level due to the availability of nearly 227,000 Medicago EST sequences (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=medicago), a soon-to-be-completed genome sequence (Young et al., 2005; Sato et al., 2007), and the availability of both a custom 16,000 unigene oligonucleotide set (Suzuki et al., 2005) and a 61,200 probe set Affymetrix GeneChip for DNA microarray analyses (http://www.affymetrix.com/products/arrays/specific/medicago.affx). Unfortunately, the application of such molecular tools to the study of plant secondary metabolism is currently limited by deficient and/or inaccurate annotation of secondary metabolic enzymes and by the incomplete knowledge of the full secondary metabolic composition of plants. The union of global scale, nontargeted metabolite profiling, i.e. metabolomics (Fiehn, 2002; Sumner et al., 2003) with parallel analysis of the transcriptome and proteome, provides a powerful integrated platform for the assessment of metabolic networks and the in vivo functions of biosynthetic genes. We are currently using such an integrated functional genomics approach to study stress responses and secondary metabolism in M. truncatula.
Cell suspension cultures of M. truncatula undergo massive genetic reprogramming in response to elicitation with yeast elicitor (YE) or the wound signal methyl jasmonate (MeJA). Previous studies used targeted metabolite profiling to demonstrate that MeJA elicits the accumulation of triterpene saponins (Suzuki et al., 2002, 2005; Achnine et al., 2005), whereas YE, a pathogen mimic, induces the accumulation of both free and glycosylated isoflavonoids (Kessmann et al., 1990; Suzuki et al., 2005). The changes in primary metabolism in these elicited cell cultures have been described earlier (Broeckling et al., 2005), and a detailed analysis of transcript profiles will appear in a parallel article (Naoumkina et al., 2007). The goals of this work were to obtain a more holistic metabolomics data set for constructing a global image of the Medicago phenylpropanoid biosynthetic network. Here we describe the characterization of a number of previously unreported isoflavonoids, including afrormosin, alfalone, irisolidone, and related gluco-conjugates, in elicited M. truncatula cell cultures. Use of stable isotope labeling provided evidence for the biosynthetic origins of medicarpin and several of the other isoflavonoids described here. The results also revealed differential mechanistic and elicitor-specific responses to YE and MeJA. YE induced de novo biosynthesis of the isoflavonoids, whereas MeJA induced the accumulation of the phytoalexin medicarpin through the remobilization of preformed stores of isoflavone glycosides.
RESULTS
Experimental Design and Analytical Parameters
Liquid suspension cell cultures generated from root (Broeckling et al., 2005) were exposed independently to two elicitors (YE and MeJA) and then harvested at 21 time points following elicitation. Biological samples were harvested in triplicate from independent culture flasks for both the control and elicited cell cultures. Cells and cell culture medium were extracted, and analyzed using reverse-phase HPLC coupled to UV photodiode array and electrospray ionization ion-trap mass spectrometry detection (HPLC-PDA-ESI-ITMS) to examine the nature and extent of induced phenylpropanoid biosynthesis.
The selected chromatographic parameters described in “Materials and Methods” resulted in the separation and differentiation of a large number of secondary metabolites within 60 min. The elution order of phenolic compounds correlated with decreasing polarity, whereby phenolic acids and flavonoid diglucosides eluted first, followed by monoglucosides, acylated monoglucosides, and finally free aglycones. To obtain a more comprehensive profile of the cell culture metabolome, and to minimize competitive ionization effects commonly observed in electrospray ionization (ESI), samples were analyzed in both positive- and negative-ion ESI modes. Approximately 178 cellular and 113 media components were detected in negative-ion HPLC-PDA-ESI-ITMS compared to 140 cellular and 75 media components in positive-ion HPLC-PDA-ESI-ITMS mode. Positive-ion ESI mass spectra provided a greater number of fragment ions for each component that aided in structural identification, whereas negative-ion ESI yielded better sensitivity and higher signal to noise ratios, as previously noted (Huhman and Sumner, 2002). A total of 480 HPLC-PDA-ESI-MS chromatographic analyses were acquired with each profile containing approximately 180 components. Data tables containing relative quantitative values for all detected components, including both identified and unidentified components, are provided as supplemental data.
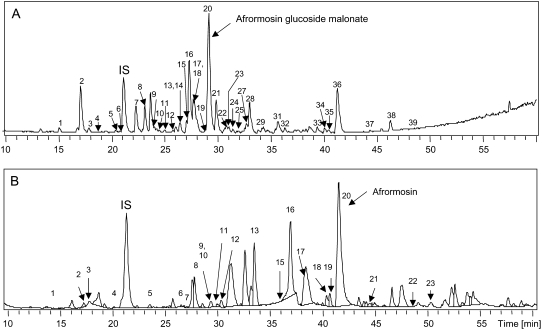
Metabolites were identified based upon their UV absorption spectra (200–600 nm), HPLC coupled to high-resolution quadrupole time-of-flight mass spectrometry (HPLC-QTofMS) analysis for improved mass accuracy measurements, comprehensive analysis of fragmentation patterns obtained by tandem ion-trap mass spectrometry, and enzymatic hydrolysis followed by gas chromatography-mass spectrometry analysis for the differentiation and determination of isomeric sugar moieties. In addition, extensive custom-made UV and mass spectral libraries of authentic flavonoids allowed for the structural confirmation of compounds identified in both cells and medium without ambiguity. A detailed description of the cell culture peak identifications and strategy has been published elsewhere (Farag et al., 2007). Approximately 23% (40 of 178 components) and 20% (23 of 113 components) of differentiated and quantified HPLC peaks were chemically identified in cell culture and medium extracts, respectively (Fig. 2). Complete lists of identified peaks in cells and media, along with their characteristic UV and mass spectral data, are provided in Tables I and II, respectively.
Figure 2.
Negative-ion HPLC-ESI-MS chromatograms contrasting compositional differences between unelicited M. truncatula cellular extracts (A) and medium (B). Peak numbers correspond to identified compounds listed in Tables I and II. Unidentified components are not labeled but listed in supplemental data.
Table I.
HPLC-PDA-MS peak identifications for (iso)flavonoids in extracts from cultured Medicago cells (peaks are listed in order of retention time)
| Code | Retention Time | Identificationa | λ Max | ESI-MS |
|---|---|---|---|---|
| min | nm | m/z | ||
| 1 | 15.33 | Genistein diglucoside malonate | 265, 328 | 595[M+H]+ |
| 2 | 17.36 | 4′,7-Dihydroxyflavone glucoside | ND | 415[M−H]− |
| 3 | 18.26 | Daidzin | 265, 310 | 417[M+H]+ |
| 4 | 18.73 | p-Hydroxybenzaldehyde | 255 | 121[M−H]− |
| 5 | 20.45 | Liquiritigenin glucoside | 265, 312 | 417[M−H]− |
| 6 | 21.1 | Daidzein glucoside malonate | 258, 314 | 503[M+H]+ |
| 7 | 22.57 | Biochanin-A diglucoside | 259, 316 | 609[M+H]+ |
| 8 | 23.2 | Genisitin | 260, 325 | 433[M+H]+ |
| 9 | 23.94 | Biochanin-A diglucoside malonate | 260, 315 | 695[M+H]+ |
| 10 | 24.53 | 3′,5-Dimethoxyluteolin glucoside | 258, 288, 335 | 477[M+H]+ |
| 11 | 24.75 | Genistein glucoside malonate | 265, 325 | 519[M+H]+ |
| 12 | 25.38 | Genistein glucoside malonate (isomer) | ND | 519[M+H]+ |
| 13 | 26.31 | 2′-OH-formononetin glucoside | 258, 325 | 447[M+H]+ |
| 14 | 26.72 | 3′,5-Dimethoxyluteolin glucoside malonate | 257,287, 335 | 563[M+H]+ |
| 15 | 27.35 | Formononetin glucoside (ononin) | 252, 302 | 431[M+H]+ |
| 16 | 27.61 | Afrormosin glucoside | 260, 321 | 461[M+H]+ |
| 17 | 28.0 | 2′-OH-formononetin glucoside malonate | ND | 533[M+H]+ |
| 18 | 28.1 | Afrormosin glucoside malonate (isomer) | 260, 325 | 547[M+H]+ |
| 19 | 29.2 | 4′,7-Dihydroxyflavone | ND | 253[M−H]− |
| 20 | 29.45 | Afrormosin glucoside malonate | 260, 321 | 547[M+H]+ |
| 21 | 29.63 | Formononetin glucoside malonate | 250, 306 | 517[M+H]+ |
| 22 | 30.4 | Liquiritigenin | 270, 315 | 255[M−H]− |
| 23 | 30.97 | Vestitol glucoside malonate | 229, 282 | 521[M+H]+ |
| 24 | 31.3 | Irilone glucoside malonate | 260, 325 | 547[M+H]+ |
| 25 | 31.68 | Medicarpin glucoside | 233, 285 | 471[M+H+K]+ |
| 26 | 32.2 | Isoflav-3-ene-glucoside malonate | ND | 519[M+H]+ |
| 27 | 32.6 | Irisolidone glucoside | 265, 332 | 477[M+H]+ |
| 28 | 33.21 | Medicarpin glucoside malonate | 230, 285 | 557[M+H+K]+ |
| 29 | 34.03 | Irisolidone glucoside malonate | 265, 330 | 563[M+H]+ |
| 30 | 34.3 | Biochanin-A glucoside malonate | 260, 326 | 533[M+H]+ |
| 31 | 35.2 | Naringenin | 290, 324 | 271[M−H]− |
| 32 | 36.7 | Tricin | 270, 342 | 329[M−H]− |
| 33 | 39.64 | Isoliquiritigenin | 250, 300, 370 | 255[M−H]− |
| 34 | 40.26 | Alfalone | 250, 321 | 282[M−CH3−H]− |
| 35 | 40.6 | Formononetin | 252, 302 | 269[M−H]− |
| 36 | 41.4 | Afrormosin | 260, 325 | 299[M+H]+ |
| 37 | 44.5 | Medicarpin | 230, 280 | 269[M−H]− |
| 38 | 46.6 | Afrormosin isomer | 262, 325 | 297[M−H]− |
| 39 | 48.42 | Biochanin-A | 260, 327 | 283[M−H]− |
| 40 | 50.6 | Irisolidone | 265, 332 | 313[M−H]− |
Full details of the identifications are published (Farag et al., 2007). ND, not detected.
Table II.
HPLC-PDA-MS peak identifications for phenolics and (iso)flavonoids in the medium of cultured Medicago cells (peaks are listed in order of retention time)
| Code | Retention Time | Identificationa | λ Max | ESI-MS |
|---|---|---|---|---|
| min | nm | m/z | ||
| 1 | 14.0 | p-Hydroxybenzoic acid | 254 | 137[M−H]− |
| 2 | 17.3 | 7,4′-Dihydroxyflavone glucoside | 250, 329 | 415[M−H]− |
| 3 | 17.6 | p-Hydroxybenzaldehyde | 253 | 121[M−H]− |
| 4 | 21.1 | Unidentified phenolic | 253 | 121[M−H]− |
| 5 | 23.9 | genistin | 260, 325 | 433[M+H]+ |
| 6 | 26.34 | 2′-Hydroxyformononetin glucoside | 258, 325 | 447[M+H]+ |
| 7 | 27.0 | Chorismic acid | 271 | 225[M−H]− |
| 8 | 27.6 | Afrormosin glucoside | 260, 321 | 461[M+H]+ |
| 9 | 29.2 | Daidzein | 250, 305 | 255[M+H]+ |
| 10 | 29.4 | Afrormosin glucoside malonate | 260, 321 | 547[M+H]+ |
| 11 | 30.0 | Liquiritigenin | 270, 315 | 257[M+H]+ |
| 12 | 30.48 | Vestitol glucoside malonate | 229, 282 | 521[M+H]+ |
| 13 | 33.6 | Unidentified aurone | 260, 390 | 255[M+H]+ |
| 14 | 35.0 | Vestitone | ND | 287[M+H]+ |
| 15 | 35.2 | Naringenin | 285, 325 | 271[M+H]+ |
| 16 | 37.3 | Dihydroafrormosin | 265, 320 | 299[M−H]− |
| 17 | 37.8 | Daidzein dimer | ND | 507[M+H]+ |
| 18 | 40.2 | Alfalone | 250, 321 | 282[M−CH3−H]− |
| 19 | 40.6 | Formononetin | 252, 302 | 269[M−H]− |
| 20 | 41.4 | Afrormosin | 260, 325 | 299[M+H]+ |
| 21 | 44.6 | Medicarpin | 230, 280 | 269[M−H]− |
| 22 | 48.6 | Biochanin-A | 260, 327 | 283[M−H]− |
| 23 | 50.6 | Irisolidone | 265, 332 | 313[M−H]− |
Full details of the identifications are published (Farag et al., 2007). ND, not detected.
Fundamental qualitative and quantitative differences were observed between unelicited intracellular and medium phenolic profiles, with isoflavonoid conjugates being the most abundant isoflavonoid derivatives in cells (i.e. afrormosin glucoside malonate; peak 20 in Fig. 2A) and isoflavone aglycones being most abundant in media (i.e. afrormosin; peak 20 in Fig. 2B).
Metabolic Responses to YE
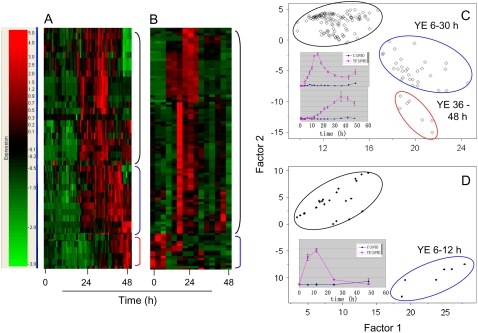
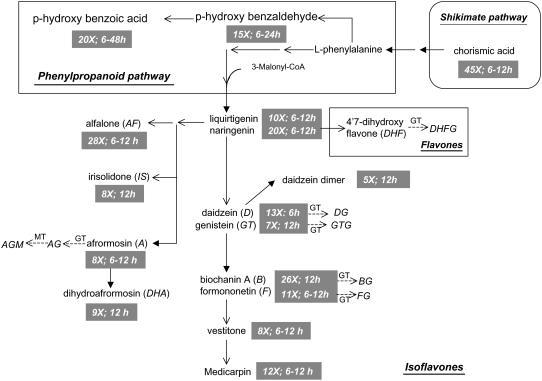
Yeast elicitation induced the shikimate, phenylpropanoid, and, most dramatically, the isoflavonoid biosynthetic pathways based upon relative quantitative changes in metabolite levels. Hierarchical cluster analysis (HCA) and principal component analysis (PCA) of cellular and medium metabolites revealed several temporal induction trends in response to YE (Fig. 3). Cellular metabolic changes included an early transient response in metabolites that peaked at 12 to 18 h and then declined, a sustained response through 48 h, and a lesser delayed and protracted response starting at 24 h postelicitation. Induction kinetics for components in the culture medium were less complex, with most compounds reaching maximum levels at 6 to 12 h postelicitation and then returning to basal levels by 24 h (Fig. 3, B and D). PCA analysis of the YE time course data showed that all the control samples and a few of the very early time points clustered together, whereas later elicited cell samples segregated into two clusters, one representing cells at 6 to 30 h postelicitation and another at 36 to 48 h postelicitation (Fig. 3C). PCA of the media extract components also showed segregation of the controls for the 12- to 24-h postelicitation samples (Fig. 3D). PCA loading plots, which define the most important components with respect to the clustering behavior, revealed that isoflavones made a larger contribution to the cluster segregation than did flavones. Many of the metabolites in the 6- to 30-h cluster had a transient accumulation pattern, whereas those in the 36- to 48-h cluster exhibited a steady and increasing accumulation pattern. ANOVA was used to assess the statistical significance of the changes observed in each metabolite. Figure 4 illustrates the temporal induction profiles for numerous metabolites overlaid upon the phenylpropanoid and (iso)flavonoid biosynthetic pathways.
Figure 3.
Metabolite induction profiles in yeast-elicited M. truncatula cell cultures. A and B, HCA of extracts from cells (A) and medium (B) from 0 to 48 h following exposure to YE. Three distinct HCA clusters were observed for cellular metabolites and are indicated by colored bars (blue, transient response that peaked at 12–18 h; red, sustained response through 48 h; green, delayed and protracted response starting at 24 h postelicitation). Metabolites included in the analyses had a minimum 2.0-fold change for at least two time points. C and D, PCA of secondary metabolites detected in cells (C) and medium (D) over 48 h following YE. The color coding for the PCA clusters is similar to that described for the HCA with the additional use of black for control and early time points containing minimal changes. Insets (C, D) depict frequent temporal trends typical of cellular and medium metabolites in each cluster. Specific metabolites implicated through the PCA loading plots as major contributors to the cluster behavior and their respective fold changes are illustrated within the metabolic pathways in Figures 4 and 6.
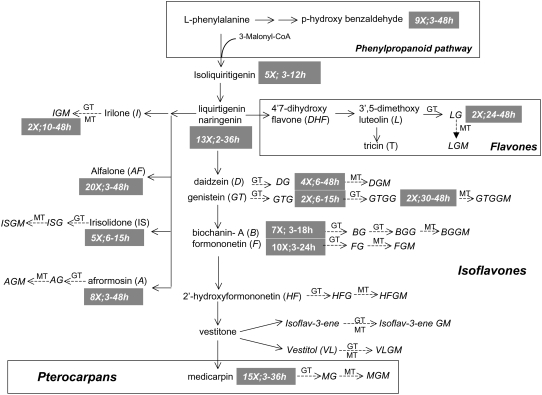
Figure 4.
Mapping of cellular phenylpropanoid metabolite responses to YE onto biosynthetic pathways. Dashed arrows after metabolite names represent conjugating enzymes responsible for the attachment of a Glc and/or a malonate moiety to form the corresponding conjugates, e.g. glucosyl transferases (GT) and malonyl transferases (MT). Metabolites that show significant differential accumulation in response to YE (P < 0.05) are followed by gray boxes illustrating the maximum fold increase detected following elicitation.
YE treatment led to transient increases in the levels of the flavanones naringenin and liquiritigenin that serve as entry points into the flavone and isoflavone biosynthetic pathways (Fig. 4). YE elicitation also led to an increase in the chalcone isoliquiritigenin, which is an immediate precursor of liquiritgenin. Increased flavanone levels were correlated with significant increases in most isoflavones, but not with subsequent increases in flavones. Marked increases in the levels of several isoflavone aglycones were detected, whereas the concentrations of the corresponding glycosidic conjugates were less affected, with the exceptions of 2- to 4-fold increases in daidzein, genistein, and irilone glucosides (Fig. 4). In Lupin (Lupinus albus and Lupinus angustifolius) seedlings, YE and fungal infection both induced marked changes in the profiles of isoflavonoid aglycones, but not of glycosidic conjugates (Bednarek et al., 2001, 2003). Medicarpin, a pterocarpan phytoalexin known to accumulate in elicited cell cultures of alfalfa (Tang and Smith, 2001), was induced up to 15-fold in response to YE. Increased levels of the medicarpin precursors formononetin and vestitone were also observed (Fig. 4).
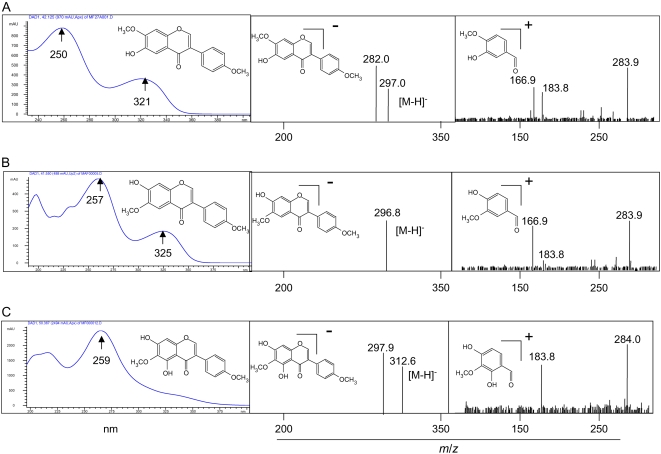
Three additional novel methylated isoflavones were also induced by YE; these were characterized as 7-hydroxy-6,4′-dimethoxyisoflavone (afrormosin), 6-hydroxy-7,4′-dimethoxyisoflavone (alfalone), and 5,7-dihydroxy-4′,6-dimethoxy isoflavone (irisolidone). Spectral data (Fig. 5) including UV, full-scan mass spectrometry (MS), and tandem MS, matched those of authentic standards for all three compounds. To the best of our knowledge, this is the first report of irisolidone in Medicago species. In the M. truncatula cell cultures, afrormosin was produced constitutively and its levels were further enhanced following exposure to YE, whereas alfalone and irisolidone were only detected following elicitation.
Figure 5.
Identification of three novel isoflavonoids from YE-treated cultures. A, Identification of the HPLC peak at retention time (Rt) 40.26 min as alfalone was based upon UV, photodiode array, negative-ion HPLC-PDA-ITMS, and positive-ion HPLC-QTofMS/MS of the [M+H]+ ion observed at m/z 299. B, Identification of the HPLC peak at Rt 41.4 min as afrormosin was based upon UV, photodiode array, negative-ion HPLC-PDA-ITMS, and positive-ion HPLC-QTofMS/MS of the [M+H]+ ion observed at m/z 299. C, Identification of the HPLC peak at Rt 50.6 min as irisolidone was based upon UV, photodiode array, negative-ion HPLC-PDA-ITMS, and positive-ion HPLC-QTofMS/MS of the [M+H]+ ion observed at m/z 315. All mass spectra are reported as relative abundance with a scale to 100%. [See online article for color version of this figure.]
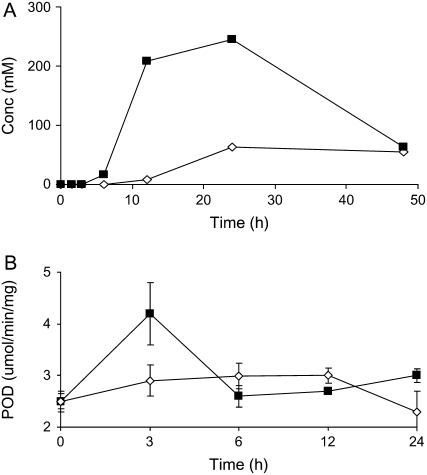
Metabolite accumulation in the medium was also evaluated and many previous studies on elicited cell cultures have overlooked this important source of information. Metabolite profiles of the medium of YE-treated M. truncatula cells broadly mirrored those observed from the cells, but with different induction kinetics for the various compounds. Significant increases in the media levels of most isoflavone aglycones were observed with maximum accumulations restricted to a period of 6 to 12 h, followed by rapid disappearance from the medium (Fig. 3D). Generally, the levels of excreted glucoside conjugates did not increase. Interestingly, four compounds were identified that accumulated exclusively in the medium (Fig. 6), namely, dihydroafrormosin, daidzein dimer, chorismic acid, and p-hydroxybenzoic acid. Peroxidase-catalyzed dimerization is an alternative to catabolic removal of isoflavonoids and can also enhance the antimicrobial activity of phenolics (Sakasai et al., 2000). Thus, the media were analyzed for hydrogen peroxide and peroxidase activity and the resultant data revealed increases in both (Fig. 7).
Figure 6.
Mapping of the extracellular phenylpropanoid metabolite responses to YE onto biosynthetic pathways. Dashed arrows after metabolite names represent conjugating enzymes responsible for the attachment of a Glc and/or a malonate moiety to form the corresponding conjugates, e.g. glucosyl transferases (GT) and malonyl transferases (MT). Metabolites that show significant differential accumulation in response to YE (P < 0.05) are followed by gray boxes showing the maximum fold increase detected over the time period at which elicitation occurs (in bold).
Figure 7.
The oxidative burst observed in the medium of yeast-elicited M. truncatula cell cultures. A, Accumulation of H2O2 in the medium of yeast-elicited (black squares) M. truncatula cell cultures relative to control (white diamonds). Results represent the average of duplicate experiments. B, Peroxidase activity measured in the medium of yeast-elicited (black squares) and control (white diamonds) M. truncatula cell cultures. Error bars represent se (n = 3).
Metabolic Responses to MeJA
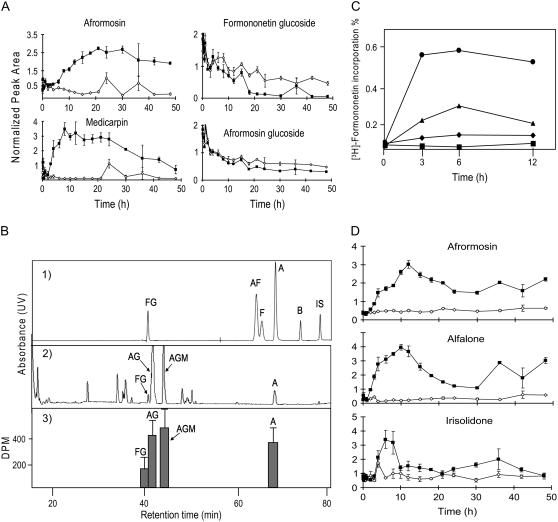
Elicitation of M. truncatula cell cultures with MeJA resulted in fewer changes in isoflavonoid profiles relative to those observed in response to YE; however, large increases in medicarpin and afrormosin (35- and 17-fold at 8 h postelicitation, respectively; Fig. 8), were observed. Interestingly, these massive increases were not preceded by a comparable increase in any of the precursors of medicarpin or afrormosin. Instead, marked decreases were observed in the levels of formononetin glucoside and 2′-hydroxyformononetin glucoside conjugates during 8 to 48 h postelicitation, and to a lesser extent in afrormosin glucoside starting at 18 h postelicitation (Fig. 8). Interestingly, no significant changes in extracellular metabolite levels were detected in response to MeJA.
Figure 8.
Elucidating the biosynthetic origins of afrormosin, alfalone, and irisolidone. A, Two isoflavones (afrormosin and medicarpin) accumulate following MeJA elicitation, whereas a decline is observed in formononetin and afrormosin glucosides. y axis value represents relative peak areas after normalization to the mean peak area for that compound. Black squares represent elicited sample means (with se bars) and white diamonds represent control sample means. B, Incorporation of will be [3H1]formononetin into afrormosin, its glycosidic conjugates, and ononin in unelicited M. truncatula cell suspension cultures. B1, HPLC/UV trace showing separation of isoflavonoid standards. Chromatography was performed as described in “Materials and Methods” (labeling experiment). B2, HPLC/UV trace of extract from M. truncatula cell cultures that had been incubated with [3H1]-formononetin for 6 h. B3, Radioactivity associated with individual HPLC fractions (n = 3) corresponding to individual isoflavonoids derived from M. truncatula cell culture incubated with will be [3H1]formononetin for 6 h. Peaks, with retention times, include: FG, formononetin glucoside (ononin), 39.2 min; AF, alfalone, 67.3 min; F, formononetin, 68.3 min; A, afrormosin, 70.1 min; B, biochanin-A, 75.6 min; IS, irisolidone, 79.5 min; AG, afrormosin glucoside, 41.1 min; and AGM, afrormosin glucoside malonate, 43.9 min. C, Accumulation of the radiolabeled isoflavonoids afrormosin (•), alfalone (▴), irisolidone (♦), and biochanin-A (▪) following treatment of yeast-elicited M. truncatula cells with will be [3H1]formononetin. Results represent average of duplicate experiments. D, Induction kinetics for afrormosin and alfalone does not mimic that of irisolidone in response to YE. Black squares represent elicited sample means with (se bars) and white diamonds represent control sample means.
Confirmation of Induction Profiles through Parallel Elicitation with YE and MeJA
The YE and MeJA elicitation experiments described above were performed at high temporal resolution with 21 sampling points between 0 and 48 h. These experiments were performed independently and sequentially over time using different passages of the same culture lines because the logistics of harvesting such large numbers of samples in a timely manner prohibited parallel analyses. To confirm the earlier findings, and to eliminate possible epigenetic effects related to the growth stage and/or passage number of the cell cultures (Kombrink and Hahlbrock, 1985), an additional experiment involving parallel YE and MeJA elicitations was performed, with sampling at a lower temporal resolution (i.e. 11 sampling points at 0, 0.25, 0.5, 1, 2, 4, 8, 12, 18, 24, and 36 h). Table III compares the fold increases in metabolites in the previous consecutive elicitations and in the additional parallel time course. For the majority of metabolites, a similar response was observed in both experiments. Importantly, the differential mechanistic responses of YE and MeJA were confirmed through the consistent observation of increased medicarpin and afrormosin accumulation at the expense of decreased glycoconjugates.
Table III.
Comparisons in the relative fold changes in peak areas for selected identified compounds in two independent elicitation experiments
Metabolites are listed if the changes in their levels showed a similar response, at a significant value of P < 0.05, in the two experiments (large-scale experiments in which YE and MeJA elicitations were applied to consecutive passages of the culture and cultures sampled at 21 time points postelicitation, and the additional smaller experiment in which YE and MeJA elicitation were carried out in parallel on a single passage culture). Values represent the fold change in peak area at the average of the 3-, 12-, and 24-h time points (values >1.0 represent increased levels in elicited samples, values <1.0 represent decreased levels due to elicitation; empty cells, not significant in at least one of the two Student's t tests).
| Compound | YE
|
MeJA
|
||
|---|---|---|---|---|
| YE | Parallel | MeJA | Parallel | |
| p-Hydroxybenzaldehyde | 5.5 | 9.3 | – | – |
| genistin | 3.5 | 3.8 | – | – |
| 5,3′-Dimethoxyluteolin glucoside | 1.7 | 1.8 | – | – |
| Genistein glucoside malonate | – | – | −0.7 | – |
| Hispidol glucoside malonate | 4.7 | 6.2 | – | – |
| 5,3′-Dimethoxyluteolin glucoside malonate | 1.4 | 1.4 | – | – |
| Ononin | – | – | −0.5 | −0.6 |
| Afrormosin glucoside | – | – | −0.8 | −0.8 |
| 2′-Hydroxyformononetin glucoside malonate | – | – | −0.5 | – |
| Afrormosin glucoside malonate | – | – | −0.8 | – |
| Formononetin glucoside malonate | – | – | −0.6 | – |
| Naringenin | 8.3 | 10.1 | – | – |
| Isoliquiritigenin | 2.1 | 4.5 | – | – |
| Alfalone | 13 | 14.6 | – | – |
| Formononetin | 5.3 | 6.2 | – | – |
| Afrormosin | 3.9 | 5.1 | 3.2 | 2.8 |
| Medicarpin | 8 | 9.6 | 5 | 5.2 |
| Biochanin-A | 2.7 | 4.1 | 3 | – |
| Irisolidone | 3 | – | – | – |
Pulsed Labeling with Exogenous [3H1]Formononetin to Probe the Biosynthetic Origins of Afrormosin, Alfalone, and Irisolidone Isoflavones
Afrormosin is found in a wide range of legume species (Dewick, 1978; Al-Ani and Dewick, 1980); however, its biosynthetic origin has been debated. The kinetics of afrormosin formation in MeJA-treated cells was characterized by a peak at 8 h, prior to a decrease in afrormosin glucoside levels (Fig. 8). This suggests that afrormosin is unlikely to be derived solely from afrormosin glucoside in response to MeJA. To further probe the biosynthetic origin of afrormosin, unelicited M. truncatula cells were pulse labeled with exogenous [3H1]formononetin for 6 h. Isoflavonoids were then extracted, fractionated by HPLC, and the incorporation of 3H into the downstream metabolites formononetin glucoside, afrormosin, afrormosin glucoside, and afrormosin glucoside malonate was measured by liquid scintillation counting (Fig. 8B). 3H label was detected in afrormosin, afrormosin glucoside, and afrormosin glucoside malonate, with percentage incorporations of 1.2%, 1.5%, and 2%, respectively (Fig. 8B, 3). Incorporation into formononetin glucoside was only 0.3% of the applied radioactivity.
Alfalone is a structural isomer of afrormosin, whereas irisolidone has an additional hydroxyl group at the C5 position relative to afrormosin (Fig. 5). To assess the biogenetic origins of these compounds, which only accumulate following YE elicitation, [3H1]formononetin was provided to YE-treated cells. The HPLC method used yielded baseline separation of alfalone, afrormosin, formononetin, and irisolidone (Fig. 8B, 1). In elicited cells, the majority of the radiolabel accumulated in afrormosin (0.6%), and to a lesser extent in alfalone (0.35%), at 6 h postelicitation (Fig. 8C). Incorporation into irisolidone was very weak and represented less than 0.06% of the applied radioactivity (Fig. 8C). Incorporation of label into biochanin-A was likewise low at 0.02% (Fig. 8C). Correlation analyses were performed and revealed that alfalone and afrormosin had similar induction kinetics (Fig. 8D) in response to YE with a correlation of r2 = 0.76, whereas irisolidone induction correlated weakly with that of both alfalone and afrormosin (r2 = 0.2 and 0.3, respectively).
DISCUSSION
Deciphering the Isoflavonoid Biosynthetic Pathway in M. truncatula
HPLC-PDA-ESI-MS analyses indicated that the cell cultures have the requisite machinery to synthesize a range of 5-deoxyflavones (e.g. 4′,7-dihydroxyflavone), 5-hydroxyisoflavones (e.g. genistein), 5-deoxyisoflavones (e.g. formononetin), pterocarpans (e.g. medicarpin), and isoflavans (e.g. vestitol). The biogenetic origins of many of these compounds have been determined from previous studies with chickpea (Cicer arietinum) and alfalfa (Edwards and Kessman, 1992; Dixon, 1999). However, three methylated isoflavones, afrormosin, alfalone, and irisolidone, were identified here for the first time in M. truncatula, and the biosynthetic origins of these and other methylated isoflavonoids is still a matter of some uncertainty (Edwards and Dixon, 1991; Deavours et al., 2006).
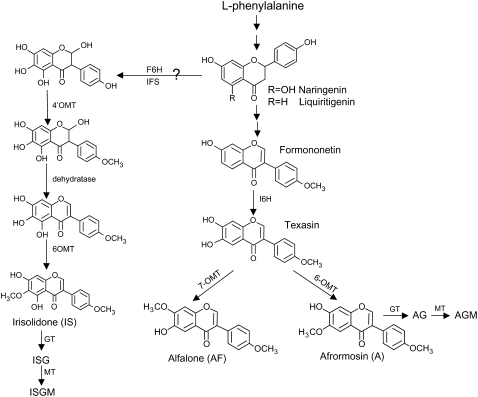
In unelicited cells, afrormosin was the major accumulated isoflavone, and alfalone was only formed in trace amounts. However, texasin, a potential intermediate in the formation of both afrormosin and alfalone, was not detected. In unelicited cells, the key enzyme directing flux into alfalone biosynthesis, presumably a texasin-7-O-methyltransferase (OMT), may be rate limiting, and flow occurs instead through a 6-OMT that favors afrormosin formation (Fig. 9). This portion of the pathway may operate as a “metabolic channel” in which hydroxylases and O-methyltransferases colocalize, as suggested to occur at the entry point into formononetin biosynthesis (Liu and Dixon, 2001).
Figure 9.
Proposed biosynthetic pathway for afrormosin, alfalone, and irisolidone in M. truncatula cell cultures. The represented enzymes are: F6H, flavanone-6-hydroxylase; I6H, isoflavone-6-hydroxylase; OMT; GT, glucosyl transferase; and MT, malonyl transferase.
Pulse labeling studies using exogenous [3H1]formononetin revealed the accumulation of 3H label in afrormosin and alfalone, which supports a biosynthetic link and evidence that formononetin is a precursor of afrormosin and alfalone. Correlation analyses provided further evidence for the close biogenetic origins of afrormosin and alfalone compared with irisolidone. Alfalone and afrormosin had very similar induction kinetics (Fig. 8D) in response to YE with a correlation relationship of r2 = 0.76, whereas irisolidone induction correlated weakly with that of both alfalone and afrormosin (r2 = 0.2 and 0.3, respectively). Correlation analyses have been used in a similar fashion to reveal relationships between primary metabolites in YE- and MeJA-elicited M. truncatula cells (Broeckling et al., 2005).
Label from exogenous [3H1]formononetin was not incorporated into irisolidone, a 5-hydroxyisoflavone. The presence of a hydroxyl group at the 5-position in irisolidone suggests that it is derived from naringenin, a precursor for 5-hydroxyisoflavones in legumes. Indeed, naringenin was detected in the cell cultures, along with several naringenin-derived isoflavones, including genistein and biochanin-A. A putative biosynthetic pathway from naringenin to irisolidone involving a flavanone-6-hydroxylase, IFS, 4′-OMT, dehydratase, and 6-OMT is shown in Figure 9. The relatively lower incorporation of 3H label into formononetin glucoside suggests that the conversion of formononetin to afrormosin is favored over the direct glucosylation of formononetin to form formononetin glucoside.
Irisolidone and other 5-hydroxyisoflavones were minor components compared with the 5-deoxyisoflavones such as afrormosin. Pronounced differences in metabolism between 5-deoxy- and 5-hydroxyisoflavones have been observed in chickpea (Jaques et al., 1985; Barz and Mackenbrock, 1994). 5-Hydroxyisoflavones are derived from naringenin chalcone, the product of chalcone synthase (CHS) acting alone, whereas 5-deoxy isoflavones are derived from isoliquiritgenin by the action of a chalcone reductase (CHR), which coacts with CHS (Strack, 1997). In M. truncatula cell cultures, CHR is likely a key regulatory step for channeling of substrates into the 5-deoxyisoflavone pathway.
Increased flavanone levels were observed in response to YE. However, this did not result in subsequent changes in flavone levels, whereas significant increases were observed in the levels of most isoflavones. Inhibition of methylation reactions involved in isoflavonoid formation diverts alfalfa cells to accumulate more flavones than isoflavones upon elicitation (Daniell et al., 1997), suggesting that this observation may reflect channeling of flavanone to isoflavone rather than a specific suppression of flavone formation.
Differential and Elicitior-Specific Induction of Isoflavonoids in Response to YE and MeJA
Increases in medicarpin and afrormosin levels were observed in response to both YE and MeJA with comparable induction kinetics; however, relatively larger fold increases were observed in response to MeJA. YE treatment resulted in marked increases in several pathway components that precede isoflavonoid biosynthesis, including chorismic acid from the shikimic acid pathway and both liquiritgenin and naringenin that serve as entry points into (iso)flavonoid biosynthesis. Moreover, the pools of most constitutively accumulated isoflavonoid glucosides remained relatively unaltered after YE treatment.
Constitutively accumulated glucosides are generally regarded as the stable, soluble storage forms of isoflavonoids and typically localized within the cell's central vacuole (Mackenbrock and Barz, 1991). The response to MeJA was in sharp contrast to that of YE, where many pathway precursors were unchanged and increases in afrormosin and medicarpin were correlated with the consumption or significant decreases in isoflavonoid conjugate levels, particularly the glucosides of formononetin and 2′-hydroxyformononetin.
Importantly, the data document that M. truncatula cell cultures are responding to YE and MeJA with two fundamentally different and elicitor specific mechanisms. Based upon the relative abundance of pathway precursors, increases in isoflavonoids and the phytoalexin end product medicarpin in response to YE are achieved from de novo biosynthesis; whereas, MeJA-induced accumulation of medicarpin occurs via hydrolysis and remobilization of vacuolar pools of formononetin glucoside. Formononetin then reenters the isoflavonoid pathway and serves as the carbon source for the synthesis of medicarpin (Liu and Dixon, 2001). The identification and characterization of glucosidase enzymes potentially involved in hydrolysis and remobilization of isoflavone glycosides are reported in a parallel publication (Naoumkina et al., 2007).
A marked decline was also observed in the levels of several early phenylpropanoid-related compounds such as p-hydroxybenzaldehyde and p-hydroxybenzoic acid in MeJA-treated cell cultures compared with control cell cultures. Decreases in these compounds suggest inhibition of flux through the early phenylpropanoid pathway in response to MeJA. This response is different from YE, which induces phenylpropanoid biosynthesis and increases in pathway precursors, but is consistent with the suggestion that endogenous MeJA suppresses the hypersensitive response and Phe ammonia-lyase (PAL) expression during bacterial elicitation (Andi et al., 2001). The differential responses observed here are also consistent with results obtained in elicited chickpea cells treated with a PAL inhibitor. Chickpea cells treated with a fungal elicitor reportedly accumulate medicarpin via de novo biosynthesis, whereas cells elicited in the presence of the PAL inhibitor l-α-aminooxy-β-phenylpropionic acid utilized vacuolar pools of formononetin glucoside conjugates as precursors for medicarpin biosynthesis (Mackenbrock and Barz, 1991). In M. truncatula cell culture, MeJA appears to be acting in a similar manner and remobilizing vacuolar stores of phenylpropanoid glycosides to serve as precursors for medicarpin and afrormosin biosynthesis. Thus, isoflavone conjugates provide a rapidly available source of isoflavonoid phytoalexin precursors under conditions of rate-limiting phenylpropanoid biosynthesis such as might occur during wounding or herbivory to mount a timely defense response.
Extracellular Secretion of Isoflavonoids
Intracellular isoflavonoid levels were induced by both YE and MeJA; however, significant increases in isoflavone levels in the culture medium were only observed in response to YE. The selective release of isoflavone aglycones as compared with isoflavone glycosides in response to YE suggests carrier-mediated transport rather than a simple diffusion mechanism. It is known that secondary metabolites are transported across membranes by specific carrier proteins (Walker et al., 2003; Yazaki, 2005). For example, an ATP-binding cassette-type transporter involved in antifungal terpenpoid secretion from tobacco (Nicotiana tabacum) cell cultures has been identified (Jasinski et al., 2001). We assume that similar transporters are present in Medicago for isoflavones; however, neither specific nor generic isoflavonoid transporters have been identified in this species to date.
Four identified compounds accumulated exclusively in the medium, including dihydroafrormosin, daidzein dimer, chorismic acid, and p-hydroxybenzoic acid. Increased chorismic acid levels in the medium are consistent with YE inducing phenylpropanoid accumulation from primary metabolism (shikimate/arogenate pathway), and gas chromatography-mass spectrometry analyses of YE-treated M. truncatula cells likewise showed an increase in shikimic acid (Broeckling et al., 2005). However, it is not clear why shikimate pools rise in the cells while chorismate is excreted.
The detection of daidzein dimers in the media suggests that oxidative oligomerization (Park et al., 1995) may account for the rapid decrease in excreted isoflavonoid levels. This hypothesis is supported by correlated increases in both hydrogen peroxide and peroxidase activity (Fig. 7). Peroxidase-catalyzed dimerization is an alternative to catabolic removal of isoflavonoids and can also enhance the antimicrobial activity of phenolics (Sakasai et al., 2000).
Functions for Induced Isoflavonoids in Medicago
MeJA is involved in signaling in wound responses, such as those that occur during insect herbivory (McConn et al., 1997; Baldwin, 1998), and medicarpin is a well-known antimicrobial phytoalexin (Lucy et al., 1988; Blount et al., 1992). With the provision that these studies were conducted using an artificial cell culture system, the marked coordinated increases in medicarpin and afrormosin levels in response to MeJA suggest that these compounds might have dual defensive roles against both insects and fungi. Antifungal activity and insect-feeding deterrence were found to be closely associated for several isoflavonoids from L. angustifolius (Lane et al., 1987). Afrormosin has been shown to be toxic to soybean loopers (Pseudoplusia includens; Caballero et al., 1986), and a large pool of afrormosin glucosides likely acts as a preformed insect deterrent (Wittstock and Gershenzon, 2002). Characterization of the enzymatic machinery involved in the synthesis and turnover of these and related isoflavone glucosides could provide a basis for manipulating defense responses in legumes by genetic engineering.
CONCLUSION
A metabolomics approach was used to investigate isoflavonoid metabolism in response to elicitation in M. truncatula. To the best of our knowledge, this study provides the most comprehensive picture of isoflavonoid biosynthesis and its regulation in the model legume M. truncatula. The results confirm and significantly extend our knowledge base concerning secondary metabolism in legume species. In addition to the identification of several novel methylated isoflavonoids and evidence for their biosynthetic routes, the reported metabolomics studies provide important evidence for the differential and elicitor-specific induction of isoflavonoids in response to YE and MeJA. The data suggest the regulation of vacuolar efflux of constitutively accumulated glycoconjugates during MeJA-induced defense responses, and for the selective activation of natural product transport mechanisms in response to YE. Studies focused on the molecular bases of these phenomena are ongoing and will pursue the exciting new hypotheses generated using metabolomics.
MATERIALS AND METHODS
Cell Cultures and Elicitation
Cell cultures derived from Medicago truncatula (‘Jemalong A17’) roots were subcultured in 40 mL of modified Schenk and Hildebrandt media (Schenk and Hildebrandt, 1971) in 125-mL Erlenmeyer flasks while shaking at 130 rpm, elicited with YE and MeJA, and harvested at 21 time points postelicitation (0, 0.07, 0.25, 0.5, 0.75, 1, 2, 3, 4, 6, 8, 10, 12, 15, 18, 21, 24, 30, 36, 42, and 48 h) as previously described (Broeckling et al., 2005). Triplicate biological replicates were collected for both control and elicited samples at each time point, with each replicate collected from a separate culture flask. Thus, each elicitation time course contained 126 independent culture flasks and biological samples. Cell culture media were also sampled, but at a lower temporal resolution that included 0, 6, 12, 24, and 48 h postelicitation (Suzuki et al., 2005).
Chemicals and Reagents
[3H1]Formononetin was purchased from SibTech. Biochanin-A, sissostrin, ononin, diadzin, diadzein, formononetin, 4′,7-dihydroxy-flavone, genistein, and genistin were purchased from Indofine, and alfalone, afrormosin, and vestitol were purchased from Apin Chemicals Ltd. Medicarpin, medicarpin glucoside, and medicarpin glucoside malonate were isolated from alfalfa (Medicago sativa). Chorismic acid, irisolidone, irilone, and tricin were kindly provided by Professor Tom Mabry (University of Texas at Austin). Other compounds were purchased from Sigma-Aldrich. Solvents used were of HPLC-grade purity.
Extraction and Liquid Chromatography-Mass Spectrometry Analysis of Phenolic Compounds
For analysis of phenolic compounds from cells, 20 ± 0.06 mg batches of lyophilized cultured cells were extracted with 1.8 mL of a solution composed of 80% methanol and 20% water containing 2 μg of umbelliferone (internal standard) for 10 h on an orbital shaker in the dark. Extracts were centrifuged at 3,000g for 60 min, and 1.4 mL of supernatant collected and evaporated under nitrogen to dryness. The residue was resuspended in 300 μL of 45% methanol and analyzed by HPLC-PDA-MS.
For analysis of phenolic compounds in the culture medium, the medium was filtered from cells and 20-mL aliquots were liquid-liquid extracted three times with 25 mL of ethyl acetate spiked with 2 μg of umbelliferone (internal standard). The extracts were pooled, evaporated to dryness under nitrogen, dissolved in 100% methanol, and analyzed using HPLC-PDA-MS.
HPLC-PDA-MS was performed using an Agilent 1100 series II LC system (Agilent Technologies) equipped with a photodiode array detector coupled to a Bruker Esquire ion-trap mass spectrometer equipped with an electrospray-ionization source. Photodiode array spectra were recorded over the range of 200 to 600 nm. A reverse-phase, C18, 5-μm, 4.6- × 250-mm column (J.T.Baker) was used for separations. The mobile phases consisted of solvent A (0.1% [v/v] CH3COOH in water) and solvent B (acetonitrile), and separations were performed using a linear gradient of 5% to 90% B (v/v) over 70 min. The flow rate was 0.8 mL/min, and the temperature of the column was kept at 28°C. Both positive- and negative-ion mass spectra were acquired. Positive-ion ESI was performed using an ion source voltage of 4.0 kV and a capillary offset voltage of 86.0 V. Nebulization was aided with a coaxial nitrogen sheath gas provided at a pressure of 60 psi. Desolvation was aided using a countercurrent nitrogen flow set at a pressure of 12 psi and a capillary temperature of 300°C. Mass spectra were recorded over the range 50 to 2,200 m/z. The Bruker ion-trap mass spectrometer was operated under an ion current control of approximately 10,000 with a maximum acquire time of 100 ms.
Data Acquisition and Statistical Analysis
Relative metabolite abundances were calculated using custom software MET-IDEA to extract peak areas of individual ions characteristic of each component (Broeckling et al., 2006). Metabolites were identified based upon UV spectral, mass spectral, and retention time matching with authentic compounds prepared and analyzed in an identical manner. Peak areas were normalized by dividing each peak area value by the mean peak area value for that compound, with each time course treated independently. PCA was performed on normalized data sets using Pirouette (InfoMetrix) software. Analysis of variance was performed using JMP5 (SAS Institute) statistical software. Means were separated using Duncan's multiple range test at P < 0.05.
Labeling Studies
M. truncatula suspension culture cells (60 mL) 5 d after subculture were incubated with 10 nmol of [3H1]formononetin (2 μCi/nmol) for 60 min. The cell cultures were then treated with YE (Broeckling et al., 2005) at a final concentration of 50 mg Glc equivalent per liter of cell medium. Cells (5-mL batches) were harvested at 0, 3, 6, and 12 h postelicitation and extracted as described above. Cell extracts (50 μL) were analyzed by HPLC with UV detection at 254 nm. Separation was performed on a reverse-phase, C18, 5-μm, 4.6- × 250-mm column (J.T.Baker) with a solvent of 0.1% (v/v) acetic acid with an increasing concentration gradient of acetonitrile (0 to 2 min, 15% [v/v]; 2 to 17 min, 15%; 17 to 37 min, 35%; 37 to 67 min, 45%; 67 to 77 min, 65%) at a flow rate of 0.3 mL/min. The HPLC eluate was monitored for radioactivity with a Beckman Coulter LS6500 multipurpose scintillation counter. Labeling experiments in unelicited cells were carried out using the same procedure, but without the addition of YE.
Quantification of H2O2
The concentration of H2O2 in the extracellular medium of cell cultures was measured using a colorimetric method (Park et al., 1995). The cells were removed from a 5.0-mL sample of cell culture by filtration through Miracloth (Calbiochem) and 2.0 mL of the resulting medium was added to 200 μL of 0.1 m sodium acetate buffer containing 80 μg of horseradish peroxidase (Sigma) and 400 μg of N,N-dimethyl-p-phenylenediamine (Sigma). The reaction mixture was incubated at room temperature for 5 min, and then the A515 was measured on a Beckman Coulter DU800 spectrophotometer. Quantification was based on an H2O2 standard calibration curve (0–50 mm) constructed following the same protocol. Results are expressed as millimolar concentration.
Assay of Peroxidase Activity
Peroxidase activity was assayed in the medium using the method of Nair and Showalter (Nair and Showalter, 1996). Cell culture samples were harvested and the medium filtered from cells using Miracloth (Calbiochem). Filtered medium (50 μg protein equivalent) was added to 2 mL of 0.1% guaiacol and 0.03% H2O2 in 50 mm potassium acetate, pH 6.0, and the increase in A470 over 3 min was measured using a Beckman Coulter DU800 spectrophotometer. Protein concentrations were determined by the Bradford assay (1976) using a commercial dye reagent (Bio-Rad) and bovine serum albumin as the standard.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Relative quantitative values for all detected components (identified and unidentified) and supporting metadata.
Supplementary Material
Acknowledgments
The authors acknowledge Drs. Pedro Mendes, Gregory D. May, Joel T. Smith, Marina Noumkina, Zhentian Lei, Satish Nagaraj, and Bharat Mehrotra for their participation and contributions to the integrated M. truncatula functional genomics project of which only a small component is described here. The authors also thank the numerous individuals that helped coordinate the logistics, large-scale culturing, elicitation, and rapid harvesting of M. truncatula suspension cultures, including: Jack W. Blount, Lahoucine Achnine, Courtney Allen, Stacy Allen, Victor Asirvatham, Naveed Aziz, Corey D. Broeckling, Fang Chen, John Cooper, Anthony Duran, Patrick Fennell, Xian Zhi He, Lisa Jackson, Parvathi Kota, Changjun Liu, Srinu Reddy, Gail Shadle, Shashi Sharma, Hideyuki Suzuki, Ivone Torres-Jerez, Bonnie Watson, and Deyu Xie (in addition to the authors). We thank Professor Tom J. Mabry (University of Texas at Austin) for his contribution of authentic chorismic acid, irisolidone, irilone, and tricin and related compounds.
This work was supported by the National Science Foundation (Plant Genome Research Program Award no. DBI–0109732). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. Additional personnel and instrumentation support was provided by The Samuel Roberts Noble Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Lloyd W. Sumner (lwsumner@noble.org).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Achnine L, Huhman DV, Farag MA, Sumner LW, Blount JW, Dixon RA (2005) Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J 41 875–887 [DOI] [PubMed] [Google Scholar]
- Adlercreutz M (1998) Epidemiology of phytoestrogens. Baillieres Clin Endocrinol Metab 12 605–623 [DOI] [PubMed] [Google Scholar]
- Akashi T, Aoki T, Ayabe S (2005) Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of carboxylesterase-like proteins in leguminous isoflavone biosynthesis. Plant Physiol 137 882–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ani HAM, Dewick PM (1980) Isoflavone biosynthesis in Onobrychis viciifolia, formononetin and texasin as precursors of afrormosin. Phytochemistry 19 2337–2339 [Google Scholar]
- Alekel DL, Germain AS, Pererson CT, Hanson KB, Stewart JW, Toda T (2000) Isoflavone-rich soy protein isolate attenuates bone loss in the lumbar spine of perimenopausal women. Am J Clin Nutr 72 844–852 [DOI] [PubMed] [Google Scholar]
- Andi S, Taguchi F, Toyoda K, Shiraishi T, Ichinose Y (2001) Effect of methyl jasmonate on harpin-induced hypersensitive cell death, generation of hydrogen peroxide and expression of PAL mRNA in tobacco suspension cultured BY-2 cells. Plant Cell Physiol 42 446–449 [DOI] [PubMed] [Google Scholar]
- Baldwin IT (1998) Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proc Natl Acad Sci USA 95 8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barz W, Mackenbrock U (1994) Constitutive and elicitatin induced metabolism of isoflavones and pterocarpans in chickpea (Cicer arietinum) cell suspension cultures. Plant Cell Tissue Organ Cult 38 199–211 [Google Scholar]
- Bednarek P, Franski R, Kerhoas L, Einhorn J, Wojtaszek P, Stobiecki M (2001) Profiling changes in metabolism of isoflavonoids and their conjugates in Lupinus albus treated with biotic elicitor. Phytochemistry 56 77–85 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Kerhoas L, Einhorn J, Franski R, Wojtaszek P, Rybus-Zajac M, Stobiecki M (2003) Profiling of flavonoid conjugates in Lupinus albus and Lupinus angustifolius responding to biotic and abiotic stimuli. J Chem Ecol 29 1127–1142 [DOI] [PubMed] [Google Scholar]
- Blount JW, Dixon RA, Paiva NL (1992) Stress responses in alfalfa (Medicago sativa L.). XVI. Antifungal activity of medicarpin and its biosynthetic precursors: implications for the genetic manipulation of stress metabolites. Physiol Mol Plant Pathol 41 333–349 [Google Scholar]
- Broeckling CD, Huhman DV, Farag MA, Smith JT, May GD, Mendes P, Dixon RA, Sumner LW (2005) Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. J Exp Bot 56 323–336 [DOI] [PubMed] [Google Scholar]
- Broeckling CD, Reddy I, Duran A, Zhao X, Sumner L (2006) MET-IDEA: data extraction tool for mass spectrometry-based metabolomics. Anal Chem 78 4334–4341 [DOI] [PubMed] [Google Scholar]
- Caballero P, Smith CM, Fronczek FR, Fischer NH (1986) Isoflavones from an insect-resistant variety of soybean and the molecular structure of afrormosin. J Nat Prod 49 1126–1129 [Google Scholar]
- Dakora FD, Phillips DA (1996) Diverse functions of isoflavonoids in legumes transcend anti-microbial definitions of phytoalexins. Physiol Mol Plant Pathol 49 1–20 [Google Scholar]
- Daniell T, O'Hagan D, Edwards R (1997) Alfalfa cell cultures treated with a fungal elicitor accumulate flavone metabolites rather than isoflavones in the presence of the methylation inhibitor tubericidin. Phytochemistry 44 285–297 [Google Scholar]
- Deavours BE, Liu CJ, Naoumkina MA, Tang Y, Farag MA, Sumner LW, Noel JP, Dixon RA (2006) Functional analysis of members of the isoflavone and isoflavanone O-methyltransferase enzyme families from the model legume Medicago truncatula. Plant Mol Biol 62 715–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewick PM (1978) Biosynthesis of the 6-oxygenated isoflavone afrormosin in Onobrychis viciifolia. Phytochemistry 17 249–250 [Google Scholar]
- Dixon RA (1999) Isoflavonoids: biochemistry, molecular biology, and biological function. In D Barton, K Nakanishi, O Meth-Cohn, eds, Comprehensive Natural Product Chemistry, Vol 1. Elsevier, New York, pp 774–821
- Dixon RA, Sumner LW (2003) Legume natural products: understanding and manipulating complex pathways for human and animal health. Plant Physiol 131 878–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R, Dixon RA (1991) Isoflavone O-methyltransferase activities in elicitor-treated cell suspension cultures of Medicago sativa. Phytochemistry 30 2597–2606 [Google Scholar]
- Edwards R, Kessman H (1992) Isoflavonoid phytoalexins and their biosynthetic enzymes. In SJ Gurr, MJ Mcpherson, DJ Bowles, eds, Molecular Plant Pathology. A Practical Approach, Vol 2. Oxford University Press, Oxford, pp 45–62
- Farag MA, Huhman DV, Lei Z, Sumner LW (2007) Metabolic profiling and systematic identification of flavonoids and isoflavonoids in roots and cell suspension cultures of Medicago truncatula using HPLC-UV-ESI-MS and GC-MS. Phytochemistry 68 342–354 [DOI] [PubMed] [Google Scholar]
- Fiehn O (2002) Metabolomics—the link between genotypes and phenotypes. Plant Mol Biol 48 155–171 [PubMed] [Google Scholar]
- Harrison MJ (1999) Molecular and cellular aspects of the arbuscular mycorrhizal symbiosis. Annu Rev Plant Physiol Plant Mol Biol 50 361–389 [DOI] [PubMed] [Google Scholar]
- Heim KE, Tagliaferro AR, Bobilya DJ (2002) Flavonoid antioxidants: chemistry, metabolism and structure-activity relationships. J Nutr Biochem 13 572–584 [DOI] [PubMed] [Google Scholar]
- Huhman D, Sumner L (2002) Metabolic profiling of saponins in Medicago sativa and Medicago truncatula using HPLC coupled to an electrospray ion-trap mass spectrometer. Phytochemistry 59 347–360 [DOI] [PubMed] [Google Scholar]
- Jaques U, Köster J, Barz W (1985) Differential turnover of isoflavone 7-O-glucoside-6″-O-malonates in Cicer arietinum roots. Phytochemistry 24 949–951 [Google Scholar]
- Jasinski M, Stukkens Y, Degand H, Purnelle B, Marchand-Brynaert J, Boutry M (2001) A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. Plant Cell 13 1095–1107 [PMC free article] [PubMed] [Google Scholar]
- Kessmann H, Edwards R, Geno PW, Dixon RA (1990) Stress responses in alfalfa (Medicago sativa L.): V. Constitutive and elicitor-induced accumulation of isoflavanoid conjugates in cell suspension cultures. Plant Physiol 94 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kombrink E, Hahlbrock K (1985) Dependence of the level of phytoalexin and enzyme induction by fungal elicitor on the growth stage of Petroselinum crispum cell cultures. Plant Cell Rep 4 277–280 [DOI] [PubMed] [Google Scholar]
- Lamartiniere CA (2000) Protection against breast cancer with genistein: a component of soy. Am J Clin Nutr 71 1705S–1707S [DOI] [PubMed] [Google Scholar]
- Lane GA, Sutherland ORW, Skipp RA (1987) Isoflavonoids as insect feeding deterrents and antifungal components from root of Lupinus angustifolius. J Chem Ecol 13 771–783 [DOI] [PubMed] [Google Scholar]
- Lichtenstein AH (1998) Soy protein, isoflavones and cardiovascular disease risk. J Nutr 128 1589–1592 [DOI] [PubMed] [Google Scholar]
- Liu CJ, Dixon RA (2001) Elicitor-induced association of isoflavone O-methyltransferase with endomembranes prevents the formation and 7-O-methylation of daidzein during isoflavonoid phytoalexin biosynthesis. Plant Cell 13 2643–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucy MC, Matthews PS, VanEtten HD (1988) Metabolic detoxification of the phytoalexins maackiain and medicarpin by Nectria haematococca field isolates: relationship to virulence on chickpea. Physiol Mol Plant Pathol 33 187–199 [Google Scholar]
- Mackenbrock U, Barz W (1991) Elicitor-induced formation of pterocarpan phytoalexins in chickpea (Cicer arietinum L.) cell suspension cultures from constitutive isoflavone conjugates upon inhibition of phenylalanine ammonia lyase. Z Naturforsch [C] 46 43–50 [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J (1997) Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA 94 5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz-Demlow BE, Duncan AM, Wangen KE, Xu X, Carr TP, Phipps WR, Kurzer MS (2000) Soy isoflavones improve plasma lipids in normocholesterolemic, premenopausal women. Am J Clin Nutr 71 1462–1469 [DOI] [PubMed] [Google Scholar]
- Nair A, Showalter A (1996) Purification and characterization of a wound-inducible cell wall cationic peroxidase from carrot roots. Biochem Biophys Res Commun 226 254–260 [DOI] [PubMed] [Google Scholar]
- Naoumkina M, Farag MA, Sumner LW, Tang Y, Liu CJ, Dixon RA (2007) Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula. Proc Natl Acad Sci USA 104 17909–17915 [DOI] [PMC free article] [PubMed]
- Oldroyd GED (2001) Dissecting symbiosis: developments in nod factor signal transduction. Ann Bot (Lond) 87 709–718 [Google Scholar]
- Park H-H, Hakamatsuka T, Sankawa U, Ebizuka Y (1995) Rapid metabolism of isoflavonoids in elicitor-treated cell suspension cultures of Pueraria lobata. Phytochemistry 38 373–380 [Google Scholar]
- Sakasai M, Fukui H, Yamane H, Kyaw A, Tahara S (2000) A new class of biflavonoids: 2′-hydroxygenistein dimers from the roots of white lupin. Z Naturforsch [C] 55 165–174 [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Asamizu E, Isobe S, Tabata S (2007) Genome sequencing and genome resources in model legumes. Plant Physiol 144 588–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk U, Hildebrandt AC (1971) Medium and techniques for induction and growth of monocotyledonous and dicotyledonous plant cell cultures. Can J Bot 50 199–204 [Google Scholar]
- Setchell KDR, Cassidy A (1999) Dietary isoflavones: biological effects and relevance to human health. J Nutr 129 758S–767S [DOI] [PubMed] [Google Scholar]
- Strack D (1997) Phenolic metabolism. In PM Dey, JB Harborne, eds, Plant Biochemistry. Academic Press, San Diego, pp 387–416
- Sumner LW, Mendes P, Dixon RA (2003) Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry 62 817–836 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Achnine L, Xu R, Matsuda SP, Dixon RA (2002) A genomics approach to the early stages of triterpene saponin biosynthesis in Medicago truncatula. Plant J 32 1033–1048 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Reddy MS, Naoumkina M, Aziz N, May GD, Huhman DV, Sumner LW, Blount JW, Mendes P, Dixon RA (2005) Methyl jasmonate and yeast elicitor induce differential transcriptional and metabolic re-programming in cell suspension cultures of the model legume Medicago truncatula. Planta 220 696–707 [DOI] [PubMed] [Google Scholar]
- Tang M, Smith CJ (2001) Elicitor induced defence responses in Medicago sativa. New Phytol 149 401–418 [DOI] [PubMed] [Google Scholar]
- Uesugi T, Toda T, Tsuji K, Ishida H (2001) Comparative study on reduction of bone loss and lipid metabolism abnormality in ovariectomized rats by soy isoflavones, daidzin, genistin, and glycitin. Biol Pharm Bull 24 368–372 [DOI] [PubMed] [Google Scholar]
- Walker TS, Bais HP, Grotewold E, Vivanco JM (2003) Root exudation and rhizosphere biology. Plant Physiol 132 44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock U, Gershenzon J (2002) Constitutive plant toxins and their role in defense against herbivores and pathogens. Curr Opin Plant Biol 5 300–307 [DOI] [PubMed] [Google Scholar]
- Yazaki K (2005) Transporters of secondary metabolites. Curr Opin Plant Biol 8 301–307 [DOI] [PubMed] [Google Scholar]
- Young ND, Cannon SB, Sato S, Kim D, Cook DR, Town CD, Roe BA, Tabata S (2005) Sequencing the genespaces of Medicago truncatula and Lotus japonicus. Plant Physiol 137 1174–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.