Abstract
Allantoate amidohydrolases (AAHs) hydrolize the ureide allantoate to ureidoglycolate, CO2, and two molecules of ammonium. Allantoate degradation is required to recycle purine-ring nitrogen in all plants. Tropical legumes additionally transport fixed nitrogen via allantoin and allantoate into the shoot, where it serves as a general nitrogen source. AAHs from Arabidopsis (Arabidopsis thaliana; AtAAH) and from soybean (Glycine max; GmAAH) were cloned, expressed in planta as StrepII-tagged variants, and highly purified from leaf extracts. Both proteins form homodimers and release 2 mol ammonium/mol allantoate. Therefore, they can truly be classified as AAHs. The kinetic constants determined and the half-maximal activation by 2 to 3 μm manganese are consistent with allantoate being the in vivo substrate of manganese-loaded AAHs. The enzymes were strongly inhibited by micromolar concentrations of fluoride as well as by borate, and by millimolar concentrations of l-asparagine and l-aspartate but not d-asparagine. l-Asparagine likely functions as competitive inhibitor. An Ataah T-DNA mutant, unable to grow on allantoin as sole nitrogen source, is rescued by the expression of StrepII-tagged variants of AtAAH and GmAAH, demonstrating that both proteins are functional in vivo. Similarly, an allantoinase (aln) mutant is rescued by a tagged AtAln variant. Fluorescent fusion proteins of allantoinase and both AAHs localize to the endoplasmic reticulum after transient expression and in transgenic plants. These findings demonstrate that after the generation of allantoin in the peroxisome, plant purine degradation continues in the endoplasmic reticulum.
Plant growth is often limited by nitrogen availability in the soil. While plants depend on efficient nitrogen uptake, they also require effective means to internally redistribute nitrogen during every stage of development (Rentsch et al., 2007). Most prominent are redistribution processes during germination or during senescence (Masclaux et al., 2000) when the great majority of nitrogen stored in the leaves is exported to sustain flowering and seed or storage organ development. However, “understanding the mechanisms of nitrogen remobilisation during leaf senescence is still at a preliminary stage” (p. 603) but will be required to increase nitrogen use efficiency and crop yields (Good et al., 2004).
Although proteins, mainly Rubisco, account for most of the reduced nitrogen in leaves, 8% to 19% of reduced nitrogen may be present as nucleic acids (data from wheat leaves; Peoples and Dalling, 1988). Nucleic acids undergo hydrolysis and nucleobase oxidation for nitrogen export. This results in the release of one of the two ring nitrogens of the pyrimidines and all four ring nitrogens of the purines as ammonium, which in turn is re-assimilated into amino acids (Zrenner et al., 2006).
Purine catabolism can be divided into three phases. First, xanthine is generated from the purine nucleotides by deamination and oxidation of the nucleobases and by releasing the phosphate and Rib moieties (Zrenner et al., 2006). Second, xanthine is converted via S-allantoin to allantoate in five enzymatic steps. Two of the contributing enzymes have only recently been described and the genes cloned in soybean (Glycine max) and mouse (Raychaudhuri and Tipton, 2002; Ramazzina et al., 2006). Third, four molecules of ammonium are released from allantoate and its breakdown products, making the nitrogen stored in the ring of the purine bases accessible for anabolic reactions. In this third phase, allantoate hydrolysis first yields ureidoglycolate. If catalyzed by an allantoate amidinohydrolase (EC 3.5.3.4), this conversion releases one molecule of urea from allantoate, which in turn is hydrolyzed by urease (EC 3.5.1.5) to produce two molecules of ammonium and CO2. Alternatively, if catalyzed by an allantoate amidohydrolase (AAH; EC 3.5.1.5), two molecules of ammonium and CO2 are released directly without a urea intermediate. In a second reaction, the allantoate hydrolysis product, ureidoglycolate, is converted to glyoxylate, again either releasing urea (catalyzed by an ureidoglycolate amidinohydrolase/urea lyase) or directly two molecules of ammonium and CO2 (catalyzed by an ureidoglycolate amidohydrolase, EC3.5.3.19). For a graphical overview of allantoate catalysis, see Todd and Polacco (2004).
In many warm-climate legumes like French bean (Phaseolus vulgaris) or soybean, the reactions releasing ammonium from allantoate are not only required for nitrogen recycling from nucleic acids but also are of central importance to the plants' nitrogen supply under nitrogen-fixing conditions. Most of the symbiotically fixed nitrogen is used for de novo purine synthesis in the nodules. The purines are then degraded to the ureides allantoin and allantoate, which serve as nitrogen carriers and are exported to the xylem (Schubert, 1986). In the shoot, ammonium is released from the ureides and is re-assimilated into amino acids. The enzymes involved in allantoate degradation in leguminous species have received special attention (Winkler et al., 1985, 1988; Wells and Lees, 1991; Lukaszewski et al., 1992; Munoz et al., 2001, 2006; Todd and Polacco, 2004; Raso et al., 2007). Despite efforts for over two decades, an allantoate-degrading enzyme has never been fully purified from a plant, probably because the enzymatic activity was unstable (Lukaszewski et al., 1992; Raso et al., 2007). The only AAH ever purified to homogeneity was from the soil bacterium Bacillus fastidiosus (Xu et al., 1995). However, using soybean extracts or intact leaf tissues amended with the urease inhibitor phenylphosphorodiamidate, allantoate degradation was characterized and shown to yield glyoxylate, ammonium, and CO2 without a urea intermediate (Winkler et al., 1987, 1988). The authors concluded that the reactions are carried out by an AAH and ureidoglycolate amidohydrolase that were postulated to work in a complex (Winkler et al., 1988). Ureidoglycolate amidohydrolase activity was also described in French bean (Wells and Lees, 1991). In contrast, a later study using soybean extracts indicated that one of two enzymes involved in allantoate breakdown does release urea (Todd and Polacco, 2004). In agreement with this, an enzyme releasing urea from ureidoglycolate (ureidoglycolate amidinohydrolase/urea lyase) was purified and biochemically characterized from chickpea (Cicer arietinum; Munoz et al., 2001) and French bean (Munoz et al., 2006). The yeast (Saccharomyces cerevisiae) dal2 mutant, lacking the fungal urea-releasing allantoate amidinohydrolase, could be complemented by the expression of an AAH candidate protein from Arabidopsis (Arabidopsis thaliana), called AtAAH (Todd and Polacco, 2006). An Arabidopsis T-DNA mutant in AtAAH was unable to grow on allantoin as nitrogen source and accumulated allantoate. This work described the first cloning of an allantoate-degrading enzyme from plants but did not show biochemically that it is an amidohydrolase.
AAH activity in soybean extracts was activated by the addition of manganese, whereas the presence of manganese, calcium, iron, cobalt, and nickel did not activate the enzyme. The addition of metal chelators like EDTA and acetohydroxamate abolished the activity (Winkler et al., 1985; Raso et al., 2007). Borate (Winkler et al., 1985) and l-Asn (Lukaszewski et al., 1992) were shown to inhibit allantoate-degrading activity in soybean extracts, and both inhibitors were postulated to function by chelating manganese (Lukaszewski et al., 1992). The inhibition of allantoate-degrading activity by l-Asn may be of physiological relevance. Upon application of mineral nitrogen (Bacanamwo and Harper, 1997) or during drought stress (Serraj et al., 1999), ureide accumulation in leaves is observed and coincides with the shutdown of nitrogen fixation in the nodules. Also, under these conditions, the l-Asn concentration in the shoot rises drastically, and it was postulated that either l-Asn or the accumulating ureides serve as feedback signal for the reduction of nitrogen fixation (summarized in Todd et al., 2006). However, this is still under debate, and it was recently reported that changes in the amount of shoot amino acids or ureides have no influence on nitrogen fixation (King and Purcell, 2005; Ladrera et al., 2007).
The subcellular localization of the enzymes participating in plant purine degradation has been almost exclusively studied in nodules (Schubert, 1986). Using Suc gradient centrifugation, xanthine dehydrogenase was found in the cytosol, uricase in the peroxisome, and allantoinase was assigned to the microsomal endoplasmic reticulum (ER) fraction, although a peroxisomal origin of the microsomes could not be excluded (Hanks et al., 1981). Immunocytochemical methods localized uricase (Webb and Newcomb, 1987) as well as hydroxyisourate hydrolase (Raychaudhuri and Tipton, 2002) exclusively to the peroxisomes of uninfected nodule cells. In animals, uricase is also localized to the peroxisome, whereas allantoinase was found in the peroxisome or the cytosol in fish but in the mitochondria in amphibians (Hayashi et al., 2000). The subcellular localization of enzymes involved in allantoate degradation have so far not been determined in plants. Only a ureidoglycolate amidohydrolase from French bean was associated with the peroxisome (Wells and Lees, 1991), consistent with the possibility that the entire purine catabolic pathway from urate to glyoxylate is present in this organelle.
In this article, we characterized AAHs from legume and non-legume plants biochemically using proteins purified to homogeneity from a plant source. The enzymatic activities releasing ammonium, the activation by several metal ions, and the direct inhibition by borate, fluoride, l-Asn, l-Asp, and a range of other substances were assessed. Our data shows that previous hypotheses regarding the mechanism of borate and l-Asn inhibition need to be rejected. The functionality of the AAHs and of Arabidopsis allantoinase (AtAln) was demonstrated in vivo by generating transgenic lines expressing tagged proteins in the corresponding mutants. Finally, we investigated the subcellular localization of the AAHs and AtAln. From this, we concluded that after the generation of allantoin in the peroxisome, plant purine degradation continues in the ER.
RESULTS
Cloning, Transient Expression, and Purification
Database searches using the AAH sequences of Escherichia coli (allC, P77425) and Bacillus subtilis (allC, O32149) as query against the protein dataset of the Arabidopsis Genome Initiative identified two similar Arabidopsis AAH candidate proteins (Supplemental Fig. S1), tentatively called AtAAH (locus At4g20070) and AtAAH-2 (locus At5g43600). Amino acid identities to the enzyme from E. coli are approximately 26% to 27% (similarities 39%–43%) for AtAAH and AtAAH-2, respectively. A full-length reading frame for a soybean AAH candidate protein (GmAAH) was also identified by assembling five EST sequences. The predicted protein was 29% identical (43% similar) to the E. coli protein.
The crystal structure of the E. coli enzyme has recently been resolved (Agarwal et al., 2007), revealing a dimeric structure with two metal binding sites per monomer. Only the dimer is likely to have activity because each active center contains amino acids from both monomers. A sulfate or phosphate ion was found to be bound as cofactor in the active site, contributing to substrate binding. An alignment of the plant AAH candidate protein sequences to the sequence of the E. coli enzyme shows that all amino acids found to be important for substrate, cofactor, and metal binding are conserved in AtAAH and GmAAH but not in AtAAH-2 (Supplemental Fig. S1).
Coding sequences without a stop codon for the plant AAH candidate proteins were amplified by reverse transcription (RT)-PCR from ecotype Columbia for Arabidopsis and the cultivar ‘Williams 82’ for soybean. The products were cloned into the binary vectors pXCS-haemagglutinin-StrepII (HAStrep) and pXCS-yellow fluorescent protein (YFP; Witte et al., 2004) to allow the expression of C-terminally HAStrep- or YFP-tagged proteins in planta. Additionally, AtAln (locus At4g04955) was cloned into similar vectors (see “Materials and Methods”).
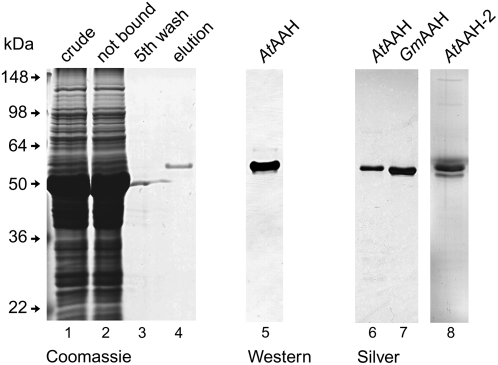
Agrobacteria carrying these binary vectors were injected into tobacco plants for transient expression. Nicotiana benthamiana was used for the transient expression of HAStrep-tagged proteins, while Nicotiana tabacum served for the transient expression of YFP-tagged proteins. Three to 4 d after infiltration, the Strep-tagged proteins were purified by StrepTactin affinity chromatography (Witte et al., 2004) in a batch format starting from soluble protein extracts of N. benthamiana leaves. Protein purification could be performed in <60 min, and highly purified enzymes were obtained (Fig. 1). The purification yield was consistently higher when fresh (not frozen) leaf material was used and severalfold higher when the extraction was performed in the presence of EDTA and absence of manganese (data not shown). About 10 to 12 μg of protein was routinely obtained from 0.75 g of leaf material. Once the transgenic lines were available (see below), the protein purification could also be performed from Arabidopsis plants. Proteins were analyzed by SDS-gel electrophoresis and matrix-assisted laser desorption mass spectrometry (coverage 47.2% for AtAAH and 57.3% for GmAAH; data not shown). No differences between the proteins obtained from N. benthamiana or from Arabidopsis were observed.
Figure 1.
Affinity purification of plant AAH candidates. Purification of C terminally StrepII-tagged AtAAH, AtAAH-2, and GmAAH from leaf extracts of N. benthamiana with StrepTactin affinity chromatography. Lanes 1 to 4, Purification of AtAAH visualized by Coomassie Blue staining, 12 μL of protein solution loaded per lane. Lane 1, Extract of soluble proteins; lane 2, proteins not bound after incubation with StrepTactin affinity matrix; lane 3, protein in fifth wash supernatant; lane 4, pool of eluted protein; lane 5 (12 μL loaded), pool of eluted AtAAH visualized by western blotting and StrepTactin-alkaline phosphatase conjugate detection; lanes 6 to 8 (4 μL loaded), purity of AtAAH, GmAAH, and AtAAH-2 visualized by silver staining.
Enzymatic Activity
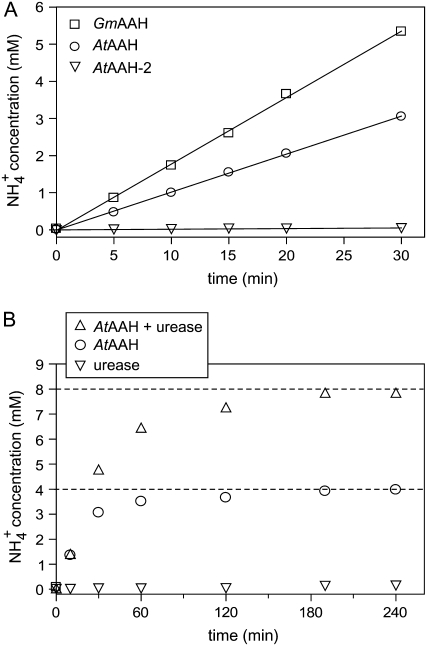
In an enzymatic assay using allantoate as substrate, a linear increase of ammonium concentration with time was observed for reactions containing AtAAH and GmAAH but not for the reaction with AtAAH-2 (Fig. 2A). We concluded that the protein encoded on locus At4g20070 is the Arabidopsis AAH and called it AtAAH in accordance with Todd and Polacco (2006). We did not further analyze AtAAH-2. GmAAH is the soybean ortholog of AtAAH. The enzymes were stable under the assay conditions for at least 30 min (Fig. 2A), and the measured activities varied proportionally with enzyme concentration (data not shown). Two mole of ammonium was released per mole allantoate hydrolyzed by AtAAH (Fig. 2B) and GmAAH (data not shown). The data does not resolve whether both ammonium molecules are released by enzymatic catalysis or if an unstable intermediate (for example, ureidoglycine) is generated by the enzyme, which subsequently decays nonenzymatically. Evidence for a reactive intermediate of allantoate breakdown has been presented for reactions performed in crude soybean extracts (Winkler et al., 1985), in yeast cells expressing AtAAH (Todd and Polacco, 2006), and in enzyme extracts from Streptococcus allantoicus (van der Drift et al., 1970). In S. allantoicus, the unstable ureidoglycine intermediate was proposed to be converted enzymatically by AAH to ureidoglycolate and ammonium.
Figure 2.
Enzymatic activity and stoichiometric conversion of allantoate. A, Test of ammonium-releasing activity from allantoate (6 mm) as substrate using purified AtAAH, AtAAH-2, and GmAAH. B, Stoichiometric conversion of 2 mm allantoate by AtAAH. The inclusion of 0.5 units of jackbean urease allowed the detection of urea using an ammonium assay.
The AtAAH reaction product, probably ureidoglycolate, is unstable under the assay conditions and decays to glyoxylate and urea, which was monitored by the inclusion of urease in the assay. Ureidoglycolate instability at different pH values and in the presence of divalent cations is well documented in the literature (Gravenmade et al., 1970). Nonenzymatic allantoate hydrolysis releasing either ammonium (Fig. 2A, AtAAH-2 curve) or urea (Fig. 2B, urease curve) is not observed under the employed assay conditions.
To maintain activity, the presence of dithiothreitol (DTT) was required at all times. Under reducing conditions, the activity of purified AtAAH was stable for a minimum of 2 h, whereas GmAAH was more labile and slowly lost activity during storage on ice. Both purified proteins could be frozen in liquid nitrogen and stored in the freezer without activity loss. Activity assays for GmAAH were therefore always carried out with freshly thawed enzyme.
Determination of Kinetic Constants and Metal Requirement
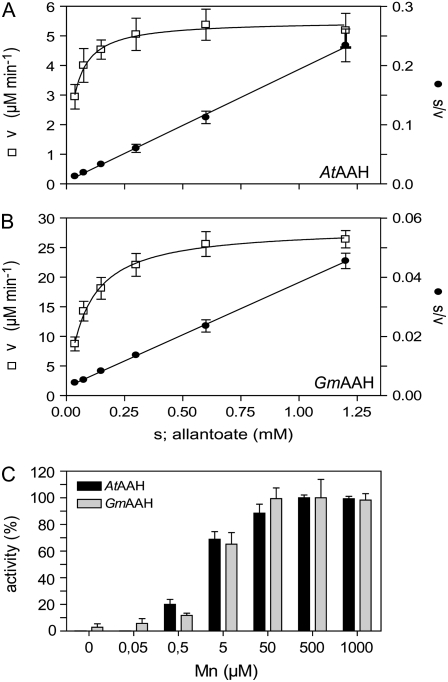
Using short 2-min assays, the initial activity at varying allantoate concentrations was measured. These assays required the optimization of the ammonium detection sensitivity (see “Materials and Methods”). The activity of both enzymes could be well described by Michaelis-Menten kinetics (Fig. 3, A and B, s/v against s graph). By fitting the data of the v against the s hyperbola, Km values of 30.1 ± 7.9 μm and 80.9 ± 12.3 μm (confidence interval [ci]; P = 95%) were determined for AtAAH and GmAAH, respectively. Specific activities of 30.7 units mg−1 and 58.1 units mg−1 corresponding to turnover numbers (kcat) of 27.2 s−1 and 50.2 s−1 were determined for AtAAH and GmAAH, respectively, using the data shown in Figure 3, A and B. In some extractions, up to 8-fold higher specific activities were found, but the basis for this variation could not be determined. The presence of Triton X-100 in the purified enzyme solution had a stimulating effect on enzyme activity (tested for AtAAH). A 10-fold reduction of the routinely used Triton X-100 concentration of 0.005% resulted reproducibly in >80% activity loss after a 30-min incubation on ice. It was not possible to recover the original activity by re-adding Triton X-100, even after incubation for several hours. Maximum activity was obtained by purifying the enzyme in the presence of 0.005 to 0.01% Triton X-100.
Figure 3.
Enzyme kinetics and activation by manganese. A, Kinetic data for AtAAH. Left axis, Enzymatic velocity (v) in terms of ammonium production against substrate concentration(s) fitted using the Michaelis-Menten equation (R2 = 0.75). Right axis, Hanes plot: ratio of substrate concentration and enzymatic velocity (s:v) against substrate concentration (s) fitted by linear regression (r2 = 0.98). Error bars are sds (n = 8). B, Kinetic data for GmAAH as in A (R2 = 0.93; r2 = 0.99). C, Activation of AtAAH (black bars) and GmAAH (gray bars) by manganese in the presence of 3 mm allantoate. Error bars are sd (n = 3).
AAHs from bacterial and plant sources are activated by manganese (Vogels, 1966; Winkler et al., 1985). For AtAAH and GmAAH, manganese concentrations of 2.1 ± 0.6 μm and 2.9 ± 1.1 μm (ci, P = 95%) were required for one-half maximal activation, respectively, while approximately 50 μm was sufficient for full activation (Fig. 3C). The crystal structure of AAH from E. coli contained two metal ions per active center that were tentatively identified as zinc (Agarwal et al., 2007). We have used zinc, cobalt, and nickel in activation tests. In comparison to the activity with manganese, which was set to 100%, we found only marginal activity with zinc but up to 20% activity with cobalt or nickel in the concentration range of 5 to 50 μm. Higher concentrations inhibited the enzyme. In general, plants contain on average 100 μg manganese, 44 μg zinc, and 2 μg nickel per kilogram dry weight (dw; Epstein, 1999). Because manganese in plants is more abundant than nickel and cobalt, it appears likely that plant AAHs are activated by manganese in vivo.
Inhibitors
The inhibition of allantoate-degrading activity by l-Asn and borate in soybean extracts has been reported by Lukaszewski et al. (1992). Experiments varying the manganese and inhibitor concentrations indicated that both inhibitors function by chelating manganese.
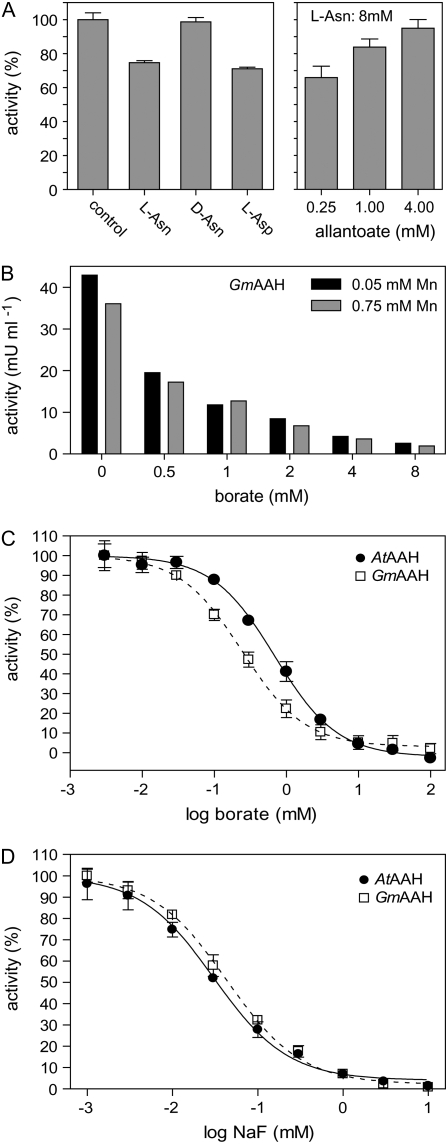
Purified GmAAH (Fig. 4A) and AtAAH (data not shown) were inhibited by l-Asn and l-Asp but barely by d-Asn, demonstrating that a chelating effect cannot be responsible for l-Asn-mediated inhibition. l-Gln had no effect on activity. Structurally, l-Asn resembles allantoate or ureidoglycolate and may rather function as a competitive inhibitor. The inhibitory effect of l-Asn can be suppressed by increasing substrate concentrations (Fig. 4A), consistent with a competitive or mixed mechanism of inhibition. None of the tested amino acids were substrates for the enzymes.
Figure 4.
Inhibition studies. A, Inhibition of GmAAH by l-Asn, d-Asn, and l-Asp (8 mm each) and suppression of l-Asn inhibition by increasing substrate concentrations. Error bars are sd (n = 3). B, Inhibition of GmAAH by increasing concentrations of borate in the presence of limiting (50 μm) or abundant (750 μm) manganese concentrations. C, Borate inhibition curve for AtAAH and GmAAH fitted using a sigmoidal dose response equation (R2 = 0.99 for both). Error bars are sd (n = 3). D, Fluoride inhibition curve of AtAAH and GmAAH fitted using a sigmoidal dose response equation (R2 = 0.99 for both). Error bars are sd (n = 3).
Borate also inhibited AtAAH and GmAAH, but not by chelating manganese. At a limiting (50 μm) or abundant (750 μm) manganese concentration, GmAAH was inhibited to the same extent by borate concentrations from 0.5 to 8.0 mm (Fig. 4B). The same was observed for AtAAH (data not shown). Using a substrate concentration of 500 μm allantoate, 233 μm (ci, 194–279 μm; P = 95%) and 708 μm (ci, 611–820 μm; P = 95%) borate inhibited 50% of AtAAH and GmAAH activity, respectively (Fig. 4C).
Fluoride at a concentration of 1 mm was reported to inhibit the activity of AAH from B. fastidiosus by 20% (Xu et al., 1995). For AtAAH and GmAAH, fluoride concentrations of 31.6 μm (ci, 26.3–38.1 μm; P = 95%) and 42.5 μm (ci, 36.4–49.7 μm; P = 95%) led to a 50% reduction of activity (Fig. 4D). These results place the plant AAHs into a small group of three enzymes (enolase, urease, and P-ATPase) that are directly inhibited by F−/HF in the micromolar concentration range (Marquis et al., 2003).
For a range of substances tested at 8 mm concentration in the presence of 500 μm allantoate, no inhibitory effect on AtAAH activity was observed. These included β-mercaptoethanol, allantoin, urea, Nα-acetyl-l-Asn and Nα-acetyl-d-Asn, and nitrate. The potent urease inhibitor phenylphosphorodiamidate did not inhibit AtAAH at a concentration of 100 μm in accordance with results for GmAAH activity in soybean extracts (Todd and Polacco, 2004).
We assessed the substrate specificity of the AAHs using several substances with similarity to allantoate at 8 mm concentration: Nα-carbamoyl-l-Asp, Nα-carbamoyl-l-Ala, and Nα-carbamoyl-Gly. None of these compounds was utilized as substrate.
Molecular Structure
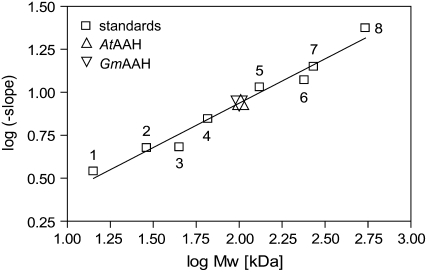
Monomeric masses of 53.1 and 51.9 kD for AtAAH and GmAAH, respectively, were calculated based on the sequence of the tagged proteins with the predicted N-terminal signal anchor cleaved off (prediction by the SignalP 3.0 Web server; cleaved signal anchor underlined in Supplemental Fig. S1). To estimate the molecular mass of the native proteins, native gel electrophoresis with eight gels of different acrylamide concentrations was resolved. Ferguson plots were generated for eight molecular mass standards and AtAAH and GmAAH. In a secondary plot, the log (−slope) determined from the Ferguson analysis was plotted against the log (molecular mass) of the standards (Bryan, 1977). The log (−slope) values of AtAAH and GmAAH were employed to estimate their native molecular masses to 103 and 100 kD, respectively (Fig. 5), indicating that both proteins form dimers. The crystal structure of AAH from E. coli indicates that the active form is dimeric (Agarwal et al., 2007). A dimeric structure was also suggested for AAH from B. fastidiosus, although based on weak experimental evidence (Xu et al., 1995). The greater instability of GmAAH compared to AtAAH was apparent in the native gels by a smear below the major band (Supplemental Fig. S2A).
Figure 5.
Native molecular mass determination by native gel electrophoresis. Secondary graph of log (negative slopes) determined from Ferguson plots using several standard proteins run on eight native gels with different acrylamide percentage plotted against log (molecular mass). Standard proteins, white squares: 1, α-lactalbumin (14.2 kD); 2, carbonic anhydrase (29 kD); 3, chicken albumin (45 kD); 4, bovine serum albumin monomer (66 kD); 5, bovine serum albumin dimer (132 kD); 6, bovine catalase (240 kD); 7, urease trimer (272 kD); 8, urease hexamer (545 kD). The log (−slope) for AtAAH (up triangle) and GmAAH (down triangle) was determined from Ferguson plots and drawn onto the calibration line.
Using gel filtration analysis, native molecular masses of 196 kD for AtAAH and 143 kD for GmAAH were determined, respectively (Supplemental Fig. S2, B and C). The enzymes may either be able to form trimers/tetramers or have an atypical hydrodynamic radius for their molecular mass in this type of analysis. In light of the structural data from the E. coli enzyme, a trimeric structure appears very unlikely. A tailing effect of the elution peak observed during gel filtration of GmAAH (Supplemental Fig. S2C) may be caused by partial dissociation into monomers consistent with the lower stability of this enzyme.
Ataah and Ataln Mutants and Complementation
A homozygous Ataah mutant (SALK 112631) was isolated from the mutant collection of the Salk Institute Genomic Analysis Laboratory (Alonso et al., 2003). A homozygous allantoinase mutant (Ataln; SAIL 810E12) was isolated from the Syngenta Arabidopsis Insertion Library (Sessions et al., 2002). For both mutants, the absence of intact mRNA was confirmed by RT-PCR (Supplemental Fig. S3). Sequence analysis mapped the insertion position to 5′ of base 2,543 (in intron 10) for Ataah and to 5′ of base 263 (in exon 2) for Ataln. Positions mark the last unaltered base in the unspliced coding sequence.
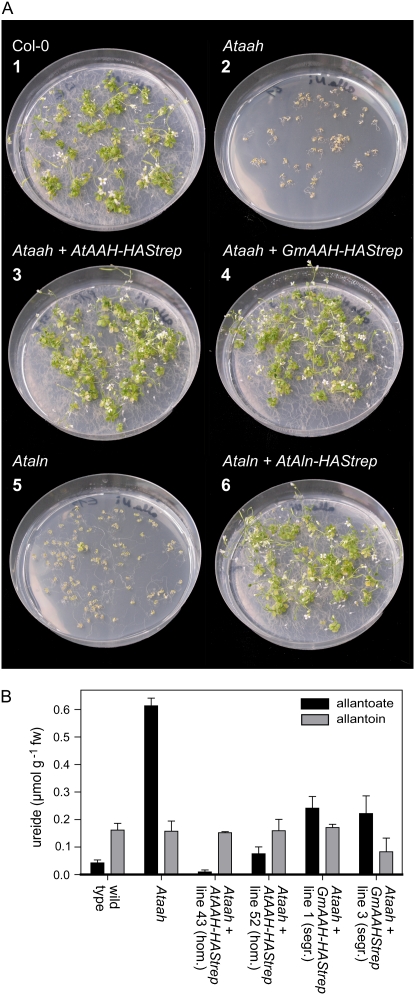
The Ataah and Ataln mutants were unable to grow with 10 mm allantoin as the sole nitrogen source (Fig. 6A, 2 and 5), while the wild type grew and finally flowered (Fig. 6A, 1). The phenotype of the Ataah mutant was more severe than that of the Ataln mutant, which still showed some root elongation with a few plants reaching the four-leaf stage. Perhaps another enzyme in Arabidopsis can partially substitute for the lacking allantoinase under these growth conditions. With nitrate as nitrogen source, growth of the mutants and the wild type was identical (data not shown). The phenotype of the Ataah mutant could be complemented in transgenic plants expressing either AtAAH-HAStrep or GmAAH-HAStrep (Fig. 6A, 3 and 4). Equally, the defect in Ataln could be complemented in transgenic plants expressing AtAln-HAStrep (Fig. 6A, 6). Also, the expression of the corresponding YFP fusions rescued the mutants (data not shown). These results demonstrate that the T-DNA insertions in AtAAH or AtAln cause the growth defect on allantoin and that C terminally tagged versions of the corresponding proteins are biologically functional.
Figure 6.
Growth assays with wild type, Ataln, and Ataah mutants and corresponding complementation lines. A, Growth on 10 mm allantoin as nitrogen source of wild-type Columbia plants (1); Ataah mutants (2); Ataah mutants complemented with an AtAAH-HAStrep construct (3); Ataah mutants complemented with an GmAAH-HAStrep construct (4); Ataln mutants (5); and Ataln mutants complemented with an AtAln-HAStrep construct (6). Pictures were taken 6 weeks after germination. B, Ureide content of 2-week-old seedlings of wild type, Ataah mutant, and complementation lines grown on KNO3. Error bars are se (n = 3).
The allantoate content of Ataah mutant seedlings grown for 2 weeks on plates with nitrate as the nitrogen source was severalfold higher than in the wild type (Fig. 6B; Todd and Polacco, 2006). In two independent homozygous complementation lines expressing tagged AtAAH, allantoate amounts were similar to those found in the wild type, confirming the successful complementation. In two segregating complementation lines expressing tagged GmAAH, the allantoate concentrations were partially reduced compared to the concentrations found in the mutants. That only partial reduction was observed is probably due to the presence of mutant plants not carrying a transgene in the segregating population.
Transcriptional Activation by Nitrogen Starvation
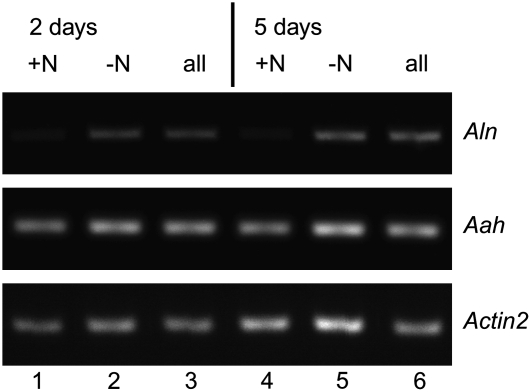
Plants were grown for 9 d on Murashige and Skoog medium with 9.35 mm KNO3 and were subsequently placed on medium containing: (1) KNO3; (2) an equimolar amount of KCl and no KNO3; or (3) KCl and no KNO3 but 10 mm of allantoin. After 2 and 5 d, plants were harvested for RNA extraction (five individuals per sample). Semiquantitative RT-PCR revealed that AtAln mRNA amounts increased due to nitrogen stress but were not further influenced by the presence of allantoin, whereas AtAAH mRNA levels did not significantly change in this experiment (Fig. 7).
Figure 7.
Transcriptional regulation by nitrogen starvation. Semiquantitative RT-PCR amplifying Arabidopsis Aln, AAH, and Actin2 transcripts 2 d (lanes 1–3) or 5 d (lanes 4–6) after exposure to nitrogen limitation in the presence or absence of allantoin. Lanes 1 and 4, Controls with KNO3; lanes 2 and 5, without nitrogen source; lanes 3 and 6, with allantoin as sole nitrogen source.
Subcellular Localization of AtAAH, GmAAH, and AtAln
Currently, it is thought that the enzymatic reactions leading to allantoin take place in the peroxisome (summarized in Todd et al., 2006). However, the allantoinases of Arabidopsis and soybean are predicted to reside in the ER (prediction by MultiLoc; Hoglund et al., 2006). A soybean allantoinase protein sequence for this prediction was obtained from the translation of a consensus full-length coding sequence constructed from the alignment of 18 GmAln ESTs. For AtAAH and GmAAH, an extracellular location is predicted. None of these proteins have a peroxisomal targeting sequence, but all contain a putative N-terminal signal peptide for the ER and secretory pathway. To clarify the subcellular localization, C-terminal YFP fusions of these proteins were co-expressed transiently in tobacco plants with markers for the ER (GFP fused to the amino acids HDEL; Batoko et al., 2000), Golgi (sialyltransferase-CFP; Brandizzi et al., 2002) and peroxisomes (CFP fused to the amino acids SKL; Sparkes et al., 2005). Additionally, transgenic Arabidopsis plants expressing fluorescent AtAln fusions and AtAAH-YFP fusions were generated. These constructs were introduced into the corresponding mutants and were able to complement the growth phenotype on allantoin as nitrogen source. The AAH-YFP fusions also maintained enzymatic activity (measured in leaf extracts; data not shown).
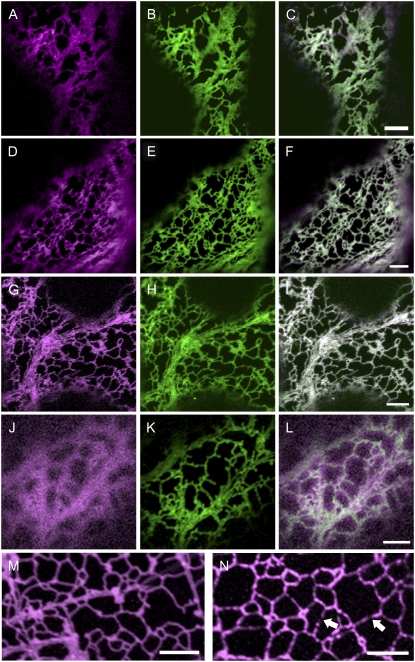
Upon transient expression in tobacco, AtAln, AtAAH, and GmAAH localized to the ER in almost all cells (Fig. 8, A–I), but were very occasionally also found in the Golgi or the peroxisomes (Supplemental Figs. S4 and S5). An N terminally truncated version of AtAAH-YFP lacking the first 71 amino acids exclusively localized to the cytosol (Fig. 8J), confirming that the N terminus is required for localization. A fraction of the full-length proteins was also present in the cytosol, maybe due to partial instability of the fusions. Western-blot analysis showed that the fluorescent fusion proteins were stable, although low amounts of free YFP were observed (data not shown).
Figure 8.
AtAln, AtAAH, and GmAAH locate to the ER. C-terminal YFP fusions to AtAln (A–C, magenta), AtAAH (D–F, magenta), GmAAH (G–I, magenta), or an N-terminal deletion of AtAAH (J–L) were transiently coexpressed in tobacco leaf epidermal cells with a marker for the ER (GFP-HDEL, in green). Merged images (C, F, I, and L) indicate AtAln, AtAAH, and GmAAH reside in the ER, while the N-terminal AtAAH deletion is present in the cytosol and not the ER. Images of stable transgenic Arabidopsis seedlings (15–18 d after imbibition) expressing AtAln (M) or AtAAH (N) in their corresponding null background display a localization pattern indicative of ER location. Note that unlike AtAln, AtAAH is not uniformly located throughout the ER in stable Arabidopsis lines but is present in discrete regions (arrows). Scale bar = 5 μm.
Several seedlings from at least two independently transformed stable transgenic lines of AtAln and AtAAH fluorescent fusions were assessed. AtAln was found exclusively in the ER in shoot and root (Fig. 8L). AtAAH was also found in the ER (Fig. 8N) but was not uniformly distributed, localizing to possible substructures within the ER. Small punctate motile structures reminiscent of Golgi or peroxisomes were also occasionally observed.
DISCUSSION
The final steps of purine degradation have long been the focus of research, especially because in tropical legumes, these reactions are central to nitrogen supply under nitrogen-fixing conditions. However, it has been difficult to isolate the enzymes involved. Native AAHs have never been purified to homogeneity from a plant source, potentially due to the instability of enzymatic activities (Lukaszewski et al., 1992). Recently, Raso et al. (2007) reported that attempts to isolate allantoate-degrading activity from developing French bean fruits using several conventional purification techniques led to complete loss of activity. We show that C terminally StrepII-tagged enzymes can be successfully isolated from a plant source. The high purity obtained (Fig. 1) and the short processing time of <1 h illustrate the potential of StrepII tag-based purification for plant biochemistry. Earlier problems with insufficient binding to the StrepTactin matrix and problematic elution under certain conditions (Witte et al., 2004) were resolved here. Despite the rapid purification, the instability of the AAH enzymes was also noted in this work. A possible explanation for the instability may be the decay of the enzymes into monomers, as indicated by results from electrophoresis and gel filtration for the less-stable GmAAH (Supplemental Fig. S2).
Expression of AtAAH was shown to complement a yeast mutant defective in allantoate and urea degradation in a growth assay on allantoin as the nitrogen source (Todd and Polacco, 2006). Our data demonstrate biochemically that AtAAH is an amidohydrolase and that an orthologous enzyme is found in legumes.
The leaf allantoate content is several times lower in Arabidopsis than in soybean. In Arabidopsis, approximately 34 μmol kg−1 fresh weight (fw) was measured in this study (Fig. 6B), and approximately 400 μmol kg−1 fw was measured by Todd and Polacco (2006). For soybean, Lukaszewski et al. (1992) measured 1,700 μmol kg−1 fw in untreated field-grown plants, and other authors report leaf ureide (allantoin and allantoate) contents between 500 and 1,500 μmol kg−1 fw (calculated by assuming 90% water content; King and Purcell, 2005; Ladrera et al., 2007). The lower KM of AtAAH compared to GmAAH may reflect the lower abundance of allantoate in Arabidopsis. The KM values for both enzymes are below the estimated substrate concentration in the respective plant, which is consistent with allantoate being the in vivo substrate. In a previous report using soybean seed coat extracts (Winkler et al., 1985), the CO2-releasing activity from allantoate was found to have a KM of 1.0 mm, over 12-fold higher than reported here. This may reflect the presence of another isoform of GmAAH in seed coats, but more likely, the KM was overestimated due to experimental difficulties of carrying out activity measurements in protein extracts.
In soybean plants grown without the addition of manganese, a leaf manganese concentration of 11.9 mg kg−1 dw was measured (Vadez et al., 2000b), and 19 to 24 mg kg−1 dw was documented by King and Purcell (2005). This corresponds approximately to a concentration of 22 to 44 μm (if manganese would be evenly distributed; 90% water content assumed) and would still be sufficient for partial GmAAH activation (full activation at over 50 μm; Fig. 2C). Consistent with only partial GmAAH activation under these conditions, supplementing these soybean plants with manganese led to increased ureide degradation (Vadez et al., 2000b; King and Purcell, 2005). However, at leaf manganese concentrations >50 mg kg−1 dw (corresponding to more than 91 μm), no further increase in ureide degradation rate was observed (Vadez and Sinclair, 2002; King and Purcell, 2005).
l-Asn inhibits AtAAH and GmAAH, likely by a competitive mechanism. The structural similarity of Asn to allantoate or ureidoglycolate also suggests that it may bind to the active site. The l-Asn concentration increases in leaf tissues of soybean during drought stress. King and Purcell (2005) measured a maximum of 7.85 μmol g−1 dw (about 0.8 mmol kg−1 fw) in leaves and Bacanamwo and Harper (1997) measured up to 3.3 mmol kg−1 fw. Considering that allantoate concentrations in soybean leaves are far higher than the KM of GmAAH, l-Asn would have to reach locally (in the ER at the site of AAH activity) very high concentrations to significantly inhibit GmAAH. It has been postulated that the rise in l-Asn inhibits allantoate breakdown in vivo, leading to the observed accumulation of ureides in leaves under drought conditions (summarized in Todd et al., 2006), which in turn may be one of the feedback signals for the shutdown of nitrogen fixation. This hypothesis has recently been challenged (King and Purcell, 2005; Ladrera et al., 2007). In light of our results, it appears questionable whether l-Asn inhibition of GmAAH is significant in vivo and is the molecular cause for the observed increase in ureide concentrations.
Borate as well as fluoride inhibit AtAAH and GmAAH (Fig. 4). No indication was found that borate functions by chelating manganese as previously postulated (Lukaszewski et al., 1992). Fluoride is a slow-binding pseudo-uncompetitive inhibitor for urease from Klebsiella aerogenes (Todd and Hausinger, 2000). It replaces the activated water molecule bridging the two nickel ions in the active center that possibly carries out the nucleophilic attack on the urea amide carbon. Fluoride may play a similar role in the bi-metal center of AAH where a water molecule also bridges the two metal ions (Agarwal et al., 2007). There may be further parallels between urease and AAHs, because urease is also inhibited by borate (Mobley and Hausinger, 1989). Borate has been employed to inhibit GmAAH in physiological studies (Lukaszewski et al., 1992; Vadez et al., 2000a). Because fluoride is a stronger inhibitor than borate, its use for in vivo inhibition studies may be preferable.
Plant AAHs are multimers. A dimeric structure was found by an electrophoretic method in accordance with the dimeric structure of the E. coli enzyme (Agarwal et al., 2007). Native Mr calculated from gel filtration analysis indicated that the enzymes could be tri- or tetrameric. However, it appears more likely that the hydrodynamic radius of the enzymes in gel-filtration analysis is greater than expected. Consistent with this interpretation, the E. coli enzyme possesses a rod-like overall structure (Agarwal et al., 2007).
Arabidopsis mutants carrying T-DNA insertions in AtAAH or AtAln do not grow on allantoin as sole nitrogen source. This defect can be phenotypically (Fig. 6A) and molecularly (Fig. 6B) rescued by expressing C terminally tagged versions of the corresponding proteins. These findings show that AtAln functions as allantoinase, and AtAAH and GmAAH function as AAHs in vivo. The crystal structure of the E. coli enzyme (Agarwal et al., 2007) indicates that the C terminus lies at the surface of the protein away from the active site. If the plant enzymes adopt a similar structure, the C-terminal tags could protrude freely from the enzyme.
Nitrogen starvation induces genes involved in amino acid and nucleotide breakdown, while upon re-addition of nitrate these genes are repressed. However, the changes in gene expression are generally small (Scheible et al., 2004). Induction of AtAln was observed upon nitrogen starvation (Fig. 7). The plants did not seem to perceive allantoin as a good nitrogen source, because AtAln amounts were as high in nitrogen-starved plants as in plants growing in the presence of 10 mm allantoin. This is consistent with the reduced growth of Arabidopsis observed when allantoin serves as the nitrogen source (Desimone et al., 2002). AtAAH was not induced by nitrogen starvation or allantoin (Fig. 7). Transcript profiling of plants subjected to 2 d of nitrogen starvation with subsequent addition of nitrate (Scheible et al., 2004) showed that AtAln mRNA was approximately 4-fold up- and down-regulated, while AtAAH was expressed at a lower level and the mRNA amounts changed by only approximately 2-fold, with considerable variability between repeats. This data is consistent with our results, although we could not detect significant differences in AtAAH expression.
Localization studies for enzymes involved in purine degradation were almost exclusively performed using nodules of legumes. It appears that the enzymes involved in the generation of allantoin locate to the peroxisomes (summarized in Todd et al., 2006). However, we found that plant allantoinases are predicted to reside in the ER, and our data confirmed this localization for AtAln. Soybean allantoinase in nodules was reported earlier to possibly be associated with the ER (Hanks et al., 1981). Thus, allantoin must be transported out of the peroxisomes into the ER, which may be facilitated by ureide transporters already identified in plants (Desimone et al., 2002; Pelissier et al., 2004). Although predicted to be extracellular, AtAAH and GmAAH also reside in the ER. The retention of these enzymes in the ER may be caused by interaction with other proteins that are intrinsically ER-localized. A possible candidate protein for interaction would be allantoinase. The AAHs were sporadically localized in Golgi bodies and also in peroxisomes during transient expression. All localization of AtAAH to organelles depended on the presence of the N-terminal signal sequence. Maybe the AAHs can occasionally escape from the ER into other vesicles. In transgenic plants, AtAAH did not localize uniformly to the ER but appeared often locally focused in ER substructures. These may be the sites at which the AAHs are bound and retained in the ER.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Mutants of Arabidopsis (Arabidopsis thaliana) from the SALK collection (Alonso et al., 2003) and the SAIL collection (Sessions et al., 2002) were ordered from the European Arabidopsis Stock Centre. For genetic characterization and to isolate homozygous mutants, plants were grown as described by Witte et al. (2005). Nicotiana benthamiana plants were grown as described by Witte et al. (2004). Soybean (Glycine max ‘Williams 82’) plants were grown as described by Todd and Polacco (2004).
Agar plates were prepared with half-strength Murashige and Skoog nutrients without vitamins, sugars, or amino acids. NH4NO3 was omitted and CoCl2 was replaced by 0.5 μm NiCl2. Plates contained either 9.35 mm KNO3 or 10 mm allantoin as sole nitrogen source. To allantoin plates, 9.35 mm KCl was added to maintain the same molarity of potassium ions as on the nitrate plates. Plates were incubated for up to 6 weeks in a controlled growth room in long-day conditions (16 h light of 150 μmol m−2 s−1, 20°C day, 18°C night, 60% relative humidity).
Cloning and Semiquantitative RT-PCR
RNA from plant leaves was prepared using TRI reagent (Sigma) and treated with DNaseI (Sigma) following the manufacturer's instructions. RT using 1.0 μg total RNA was performed with Moloney murine leukemia virus reverse transcriptase (Invitrogen) and a poly-T primer. AtAln cDNA was amplified by PCR using primers p1191: gaattcaaaATGGAGAGAACTTTGCTTC and p1192: cccgggAGTAGTTGCAAGTTGCAGAG introducing an EcoRI and XmaI site and a plant ribosome binding site consensus (all lowercase). The product was cloned via these restriction sites into modified versions of pXCS-HAStrep (AY457636; Witte et al., 2004) and pXCS-YFP and pXCS-CFP (Feys et al., 2005) containing kanamycin instead of Basta selectable markers. Similarly for AtAAH, the primers p1172: aagcttaaaATGGCGGTTCCTCATC and p1173: cccgggCATTTGAGACTCTAGAAACTCCA introducing a HindIII and XmaI site were used. For the generation of an N terminally truncated version of AtAAH, lacking the first 71 amino acids and adding two amino acids for Met and Ala (codons underlined), the primers p1284: aagcttaaaatggctCATATCTCTTTAGGAATCG and p1173 were used. For the gene tentatively called AtAAH-2, the primers p1167: gaattcaaaATGGAATCTTTGAAACG and p1168: cccgggGTCTAAAGAGAGCTTAGCCAG introducing an EcoRI and XmaI site were used. For cloning of GmAAH, the ESTs with the accession numbers CF806524, BM177023, BF425656, BU761060, and CX705438 were assembled and the primers p1320: gaattcaaaATGTCTTCTGCCACTGCCTC and p1321: cccgggTGATAGGTTTTCCAGAAATGAC introducing an EcoRI and XmaI site were designed at the extremes of the open reading frame. Using cDNA generated from ‘Williams 82’ leaf mRNA, the coding sequence was amplified and cloned as described above. All PCR products were confirmed by sequencing and finally cloned into pXCS-HAStrep or pXCS-YFP conferring Basta resistance.
For semiquantitative RT-PCR, 400 ng of seedling RNA was reverse transcribed in a 20-μL reaction (as described above). Five microliters of 10-fold diluted cDNA was used as template in 20-μL PCR reactions. For the amplification of AtAln cDNA, the primers p1199: GACTTAGAGATTGAAGATGGTAGTGA and p1192 (see above) were used (53°C annealing temperature; 28 cycles). AtAAH cDNA was amplified with primers p1190: GCAGGACAGACAAGACTCAAG and p1176: CGTAAATCCACAGTGAAAGTTAC (53°C annealing temperature; 35 cycles), and Actin2 (At3g18780) cDNA was amplified with primers p1034: GAGAGGTTACATGTTCACCACAAC and p1033: GTGAACGATTCCTGGACCTGCCTC (53°C annealing temperature; 28 cycles). All primer sets would yield products of increased size with genomic DNA due to the presence of introns, but only amplification from cDNA was observed.
Mutant Characterization and Transgenic Plants
Mutants from the SALK and the SAIL collections were screened with a primer binding at the left border of the T-DNA insert (p488: TGGTTCACGTAGTGGGCCATC for SALK and p1197: TTTTCAGAAATGGATAAATAGCCTTGCTTC for SAIL) together with gene-specific primers. As gene-specific primers p1198: GAAGAGATCAAGAGAGAGATATGGAG and p1193: GATGTTTGTCATTGGAAAATC were used for AtAln, and p1190 and p1173 (see above) were used for AtAAH. The PCR products from the mutants were cloned and sequenced to map the exact position of the insertions.
To determine the level of gene-specific mRNA in the mutants, cDNA from seedlings was prepared as described above. For amplification of AtAAH cDNA, the primers p1190 and p1173 were used (wild-type product size 678 bp; 53°C annealing temperature; 35 cycles). In the mutant, the T-DNA insertion is flanked by these primers. For amplification of AtAln cDNA, the primers p1198 and p1193 flanking the insertion were used (wild-type product size, 653 bp), and additionally the primers p1199 and p1192 not flanking the insertion were used (wild-type product size, 816 bp). The PCR was carried out at 53°C annealing temperature with 35 cycles. Control PCRs for Actin2 were performed as described above.
Transgenic plants were generated as described by Witte et al. (2004).
Transient Expression and Affinity Purification
Transient expression in N. benthamiana was performed as described by Witte et al. (2004) but adjusting optical densities of Agrobacteria carrying HAStrep constructs to 0.5 and of helper Agrobacterium with the silencing inhibitor construct to 0.25. After infiltration, plants were grown under short-day (8 h light) conditions for 3 to 5 d.
For enzyme purification, 0.75 g of fresh leaf material was ground in 1.5 mL of extraction buffer consisting of 100 mm HEPES, pH 8.0, 100 mm NaCl, 5 mm EDTA, pH 8.0, 15 mm DTT, 100 μg mL−1 avidin, and 0.5% (v/v) Triton X-100. The extract was centrifuged at 21,000g, 15 min, 4°C, and 40 μL of StrepTactin Macroprep (50% suspension; IBA) was added to the supernatant. After incubation for 10 min (4°C) in a rotation wheel, the affinity matrix was pelleted by centrifugation for 30 s at 700g and the supernatant discarded. The matrix was washed five times with 500 μL of wash buffer (100 mm HEPES, pH 8.0, 100 mm NaCl, 0.5 mm EDTA, 2 mm DTT, 0.005% Triton X-100), discarding the supernatant after each wash. The proteins were eluted by adding 75 μL of elution buffer (wash buffer containing 10 mm biotin) and placing the vials for 5 min in an Eppendorf thermomixer (1,000 rpm) at room temperature. The elution was repeated with further 75 μL of elution buffer and the two elutions were pooled.
Activity Assays and Total Protein Determination
For the standard enzymatic assay, 50 μL of allantoate solution (routinely 6 mm in 10 mm HEPES, pH adjusted to 8.0) and 45 μL of wash buffer (see above) containing 2 mm MnCl2 were incubated separately for 5 min at 30°C. Two minutes before the start of the reaction, 5 μL of enzyme solution was added to the wash buffer. The enzyme solution was prepared by diluting purified enzyme in wash buffer such that a linear rate for the corresponding assay was obtained. Due to the variation of specific activities, dilutions needed to be adjusted for each purification batch. Dilutions of 2- to 20-fold were routinely employed corresponding to approximately 16 to 170 ng of protein per assay. The reaction was started by the addition of the preheated allantoate. In a time course, samples of 20 μL were taken and added to 200 μL of water, followed by 50 μL of phenol nitroprusside reagent and 100 μL of hypochloride reagent prepared as described by Witte and Medina-Escobar (2001) to determine the ammonium content by a photometric measurement at 639 nm. Ammonium standard curves were generated by adding NH4Cl solution instead of enzyme solution to the wash buffer, then adding allantoate and performing the colorimetric detection. The possible interference of the inhibitors or alternative substrates with colorimetric detection was always controlled by separate standard curves in the presence of the inhibitors or alternative substrates. This ensured that partial suppression of color development as for example observed in the presence of 8 mm Asn or Asp could be taken into account.
The determination of kinetic constants required short assay times and assays with low substrate concentrations. To increase the detection sensitivity, assays were carried out in 200-μL reactions to which 50 μL of phenol nitroprusside reagent and 100 μL of hypochloride reagent were added directly. Ten-fold diluted wash buffer was used in these reactions, but the Triton X-100 concentration in this diluted buffer was maintained at 0.005% and the DTT concentration was adjusted to 1 mm. The enzyme preparations were diluted such that at the lowest substrate concentration, a linear rate could still be measured within 2 min. Substrate concentrations of 37.5, 75, 150, 300, 600, and 1,200 μm were employed and measured in eight repeats each.
Inhibitors were generally included into the allantoate solution used to start the enzymatic reactions, with the exception of fluoride, which was present at the indicated concentrations in wash buffer and allantoate solution. As a result, the enzyme was already in contact with fluoride 2 min before the addition of allantoate (during the preincubation phase). This procedure was chosen because fluoride was reported to be a slow binding inhibitor for urease (Todd and Hausinger, 2000). For the inhibitor studies, allantoate concentrations of 500 μm in the assay were generally employed, except for the measurements with amino acids, where 250 μm was used and for the assay with fluoride where 2.7 mm allantoate was present.
Purified protein was quantified using the NanoOrange kit (N6666) from Invitrogen. Twenty microliters of eluted protein was mixed with 300 μL of working solution (in three replicates) and processed according to the manufacturer's instructions. Standards were constructed with bovine serum albumin solutions in elution buffer.
Allantoin and Allantoate Measurements
Seedlings were grown on half-strength Murashige and Skoog plates containing KNO3 as nitrogen source under long-day conditions. After 15 d, three samples of 0.1 g fw were taken from the seedlings of each plant type and extracted with 1 mL of 50 mm K/Na phosphate buffer, pH 7.0. Insoluble material was removed by centrifugation, and allantoin and allantoate concentrations were determined by differential glyoxylate analysis (Vogels and Van der Drift, 1970).
Electrophoresis and Gel Filtration Analyses
SDS-gel electrophoresis, western blots for the detection of StrepII-tagged proteins, and Coomassie staining were performed according to Witte et al. (2004). To determine the native molecular mass of AtAAH and GmAAH, native gel electrophoresis with 4.5%, 5.0%, 5.5%, 6.0%, 7.0%, 8.0%, 9.0%, and 10% gels was performed using commercial native molecular mass standards (MWND500 from Sigma-Aldrich and 39064 from Serva). The analysis was performed according to Bryan (1977). Gel filtration was performed on a Superdex 200 10/300 GL column on an ÄKTA purifier HPLC system from GE Healthcare. The column was operated at room temperature with 50 mm HEPES buffer, pH 8.0, containing 150 mm NaCl, 2 mm DTT, and 0.005% Triton X-100 (flow rate 0.5 mL/min; sample volume, 25 μL). Column calibration was performed with the low molecular and high molecular mass standard protein kits from GE Healthcare according to the manufacturer's instructions.
Data Analysis
Statistical analyses were performed with the GraphPad Prism software package (www.graphpad.com). Michaelis-Menten parameters were determined by using the Michaelis-Menten equation to fit the enzymatic velocity against substrate concentration curve. Manganese activation data were fitted to a hyperbola: v = vmax ([Mn]/(Kd + [Mn])). Borate and fluoride inhibition data were fitted with a variable slope sigmoid equation (Hill equation) without constrains. The multiple alignment was generated with VectorNTI (Invitrogen) and shaded with the Boxshade program (www.ch.embnet.org/). Global pairwise alignments were performed with Needle from the EMBOSS open software suite. Subcellular targeting was predicted with the MultiLoc Web server (www-bs.informatik.uni-tuebingen.de/Services/MultiLoc/index_html). Signal anchor sequences were predicted with the SignalP 3.0 Web server (www.cbs.dtu.dk/services/SignalP). Gel filtration data was analyzed with the Unicorn software package (GE Healthcare).
Subcellular Localization
Nicotiana tabacum was grown and epidermal cells infiltrated according to Sparkes et al. (2006). Agrobacteria containing binary vectors encoding for AtAln-YFP, AtAAH-YFP, or GmAAH-YFP were infiltrated with an optical density of 0.1, whereas sialyltransferase-CFP, GFP-HDEL, and CFP-SKL were infiltrated with 0.04 density. Small sections of leaf were excised 48 to 72 h after infiltration, mounted in water, and imaged using confocal microscopy. Confocal imaging was performed using an inverted Zeiss LSM510 microscope as detailed in Sparkes et al. (2005).
The GmAAH cDNA sequence was deposited in GenBank under the accession number AM773229.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Multiple alignment of plant AAH candidate proteins with AllC from E. coli.
Supplemental Figure S2. Analysis of AtAAH and GmAAH by native electrophoresis and gel filtration.
Supplemental Figure S3. Characterization of Ataln and Ataah mutants by RT-PCR.
Supplemental Figure S4. AtAln, AtAAH, and GmAAH occasionally locate to Golgi bodies.
Supplemental Figure S5. AtAln, AtAAH, and GmAAH occasionally locate to peroxisomes.
Supplementary Material
Acknowledgments
We thank Dr. Chris Weise from the Institute for Chemistry and Biochemistry of the FU Berlin for mass spectrometry analysis, and Renate Grübnau and Janet Evins for technical assistance.
This work was supported by the Alexander von Humboldt Foundation (to A.K.W., T.R., and C.P.W.), and by the Biotechnology and Biological Sciences Research Council (to I.A.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Claus-Peter Witte (cpwitte@zedat.fu-berlin.de).
The online version of this article contains Web-only data.
References
- Agarwal R, Burley SK, Swaminathan S (2007) Structural analysis of a ternary complex of allantoate amidohydrolase from Escherichia coli reveals its mechanics. J Mol Biol 368 450–463 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Bacanamwo M, Harper JE (1997) The feedback mechanism of nitrate inhibition of nitrogenase activity in soybean may involve asparagine and/or products of its metabolism. Physiol Plant 100 371–377 [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I (2000) A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12 2201–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F, Snapp EL, Roberts AG, Lippincott-Schwartz J, Hawes C (2002) Membrane protein transport between the endoplasmic reticulum and the golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. Plant Cell 14 1293–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan JK (1977) Molecular-weights of protein multimers from polyacrylamide-gel electrophoresis. Anal Biochem 78 513–519 [DOI] [PubMed] [Google Scholar]
- Desimone M, Catoni E, Ludewig U, Hilpert M, Schneider A, Kunze R, Tegeder M, Frommer WB, Schumacher K (2002) A novel superfamily of transporters for allantoin and other oxo derivatives of nitrogen heterocyclic compounds in Arabidopsis. Plant Cell 14 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E (1999) Silicon. Annu Rev Plant Physiol Plant Mol Biol 50 641–664 [DOI] [PubMed] [Google Scholar]
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE (2005) Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good AG, Shrawat AK, Muench DG (2004) Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci 9 597–605 [DOI] [PubMed] [Google Scholar]
- Gravenmade EJ, Vogels GD, Van der Drift C (1970) Hydrolysis, racemization and absolute configuration of ureidoglycolate, a substrate of allantoicase. Biochim Biophys Acta 198 569–582 [DOI] [PubMed] [Google Scholar]
- Hanks JF, Tolbert NE, Schubert KR (1981) Localization of enzymes of ureide biosynthesis in peroxisomes and microsomes of nodules. Plant Physiol 68 65–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Fujiwara S, Noguchi T (2000) Evolution of urate-degrading enzymes in animal peroxisomes. Cell Biochem Biophys 32 123–129 [DOI] [PubMed] [Google Scholar]
- Hoglund A, Donnes P, Blum T, Adolph HW, Kohlbacher O (2006) MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics 22 1158–1165 [DOI] [PubMed] [Google Scholar]
- King CA, Purcell LC (2005) Inhibition of N2 fixation in soybean is associated with elevated ureides and amino acids. Plant Physiol 137 1389–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladrera R, Marino D, Larrainzar E, Gonzalez EM, Arrese-Igor C (2007) Reduced carbon availability to bacteroids and elevated ureides in nodules, but not in shoots, are involved in the nitrogen fixation response to early drought in soybean. Plant Physiol 145 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewski KM, Blevins DG, Randall DD (1992) Asparagine and boric acid cause allantoate accumulation in soybean leaves by inhibiting manganese-dependent allantoate amidohydrolase. Plant Physiol 99 1670–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis RE, Clock SA, Mota-Meira M (2003) Fluoride and organic weak acids as modulators of microbial physiology. FEMS Microbiol Rev 26 493–510 [DOI] [PubMed] [Google Scholar]
- Masclaux C, Valadier MH, Brugiere N, Morot-Gaudry JF, Hirel B (2000) Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211 510–518 [DOI] [PubMed] [Google Scholar]
- Mobley HLT, Hausinger RP (1989) Microbial ureases: significance, regulation, and molecular characterization. Microbiol Rev 53 85–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz A, Piedras P, Aguilar M, Pineda M (2001) Urea is a product of ureidoglycolate degradation in chickpea. Purification and characterization of the ureidoglycolate urea-lyase. Plant Physiol 125 828–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz A, Raso MJ, Pineda M, Piedras P (2006) Degradation of ureidoglycolate in French bean (Phaseolus vulgaris) is catalysed by a ubiquitous ureidoglycolate urea-lyase. Planta 224 175–184 [DOI] [PubMed] [Google Scholar]
- Pelissier HC, Frerich A, Desimone M, Schumacher K, Tegeder M (2004) PvUPS1, an allantoin transporter in nodulated roots of French bean. Plant Physiol 134 664–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples MB, Dalling JD (1988) The interplay between proteolysis and amino acid metabolism during senescence and nitrogen reallocation. In LD Noodén, AC Leopold, eds, Senescence and Aging in Plants. Academic Press, San Diego, pp 181–217
- Ramazzina I, Folli C, Secchi A, Berni R, Percudani R (2006) Completing the uric acid degradation pathway through phylogenetic comparison of whole genomes. Nat Chem Biol 2 144–148 [DOI] [PubMed] [Google Scholar]
- Raso MJ, Munoz A, Pineda M, Piedras P (2007) Biochemical characterisation of an allantoate-degrading enzyme from French bean (Phaseolus vulgaris): the requirement of phenylhydrazine. Planta 226 1333–1342 [DOI] [PubMed] [Google Scholar]
- Raychaudhuri A, Tipton PA (2002) Cloning and expression of the gene for soybean hydroxyisourate hydrolase. Localization and implications for function and mechanism. Plant Physiol 130 2061–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D, Schmidt S, Tegeder M (2007) Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett 581 2281–2289 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert KR (1986) Products of biological nitrogen-fixation in higher-plants: synthesis, transport, and metabolism. Annu Rev Plant Physiol Plant Mol Biol 37 539–574 [Google Scholar]
- Serraj R, Vadez V, Denison RF, Sinclair TR (1999) Involvement of ureides in nitrogen fixation inhibition in soybean. Plant Physiol 119 289–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Hawes C, Baker A (2005) AtPEX2 and AtPEX10 are targeted to peroxisomes independently of known endoplasmic reticulum trafficking routes. Plant Physiol 139 690–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1 2019–2025 [DOI] [PubMed] [Google Scholar]
- Todd CD, Polacco JC (2004) Soybean cultivars ‘Williams 82’ and ‘Maple Arrow’ produce both urea and ammonia during ureide degradation. J Exp Bot 55 867–877 [DOI] [PubMed] [Google Scholar]
- Todd CD, Polacco JC (2006) AtAAH encodes a protein with allantoate amidohydrolase activity from Arabidopsis thaliana. Planta 223 1108–1113 [DOI] [PubMed] [Google Scholar]
- Todd CD, Tipton PA, Blevins DG, Piedras P, Pineda M, Polacco JC (2006) Update on ureide degradation in legumes. J Exp Bot 57 5–12 [DOI] [PubMed] [Google Scholar]
- Todd MJ, Hausinger RP (2000) Fluoride inhibition of Klebsiella aerogenes urease: mechanistic implications of a pseudo-uncompetitive, slow-binding inhibitor. Biochemistry 39 5389–5396 [DOI] [PubMed] [Google Scholar]
- Vadez V, Sinclair TR (2002) Sensitivity of N-2 fixation traits in soybean cultivar Jackson to manganese. Crop Sci 42 791–796 [Google Scholar]
- Vadez V, Sinclair TR, Serraj R (2000. a) Asparagine and ureide accumulation in nodules and shoots as feedback inhibitors of N-2 fixation in soybean. Physiol Plant 110 215–223 [Google Scholar]
- Vadez V, Sinclair TR, Serraj R, Purcell LC (2000. b) Manganese application alleviates the water deficit-induced decline of N-2 fixation. Plant Cell Environ 23 497–505 [Google Scholar]
- van der Drift C, de Windt FE, Vogels GD (1970) Allantoate hydrolysis by allantoate amidohydrolase. Arch Biochem Biophys 136 273–279 [DOI] [PubMed] [Google Scholar]
- Vogels GD (1966) Reversible activation of allantoate amidohydrolase by acid-pretreatment and other properties of enzyme. Biochim Biophys Acta 113 277–291 [DOI] [PubMed] [Google Scholar]
- Vogels GD, Van der Drift C (1970) Differential analyses of glyoxylate derivatives. Anal Biochem 33 143–157 [DOI] [PubMed] [Google Scholar]
- Webb MA, Newcomb EH (1987) Cellular compartmentation of ureide biogenesis in root nodules of cowpea (Vigna unguiculata (L.) Walp.). Planta 172 162–175 [DOI] [PubMed] [Google Scholar]
- Wells XE, Lees EM (1991) Ureidoglycolate amidohydrolase from developing French bean fruits (Phaseolus-vulgaris [L]). Arch Biochem Biophys 287 151–159 [DOI] [PubMed] [Google Scholar]
- Winkler RG, Blevins DG, Polacco JC, Randall DD (1987) Ureide catabolism of soybeans. 2. Pathway of catabolism in intact leaf tissue. Plant Physiol 83 585–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler RG, Blevins DG, Randall DD (1988) Ureide catabolism in soybeans: III. Ureidoglycolate amidohydrolase and allantoate amidohydrolase are activities of an allantoate degrading enzyme complex. Plant Physiol 86 1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler RG, Polacco JC, Blevins DG, Randall DD (1985) Enzymic degradation of allantoate in developing soybeans. Plant Physiol 79 787–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte CP, Medina-Escobar N (2001) In-gel detection of urease with nitroblue tetrazolium and quantification of the enzyme from different crop plants using the indophenol reaction. Anal Biochem 290 102–107 [DOI] [PubMed] [Google Scholar]
- Witte CP, Noel LD, Gielbert J, Parker JE, Romeis T (2004) Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol Biol 55 135–147 [DOI] [PubMed] [Google Scholar]
- Witte CP, Rosso MG, Romeis T (2005) Identification of three urease accessory proteins that are required for urease activation in Arabidopsis. Plant Physiol 139 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, de Windt FE, van der Drift C (1995) Purification and characterization of allantoate amidohydrolase from Bacillus fastidiosus. Arch Biochem Biophys 324 99–104 [DOI] [PubMed] [Google Scholar]
- Zrenner R, Stitt M, Sonnewald U, Boldt R (2006) Pyrimidine and purine biosynthesis and degradation in plants. Annu Rev Plant Biol 57 805–836 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.