Abstract
The NADPH-dependent thioredoxin reductase (NTR)/thioredoxin (Trx) system catalyzes disulfide bond reduction in the cytoplasm and mitochondrion. Trx h is suggested to play an important role in seed development, germination, and seedling growth. Plants have multiple isoforms of Trx h and NTR; however, little is known about the roles of the individual isoforms. Trx h isoforms from barley (Hordeum vulgare) seeds (HvTrxh1 and HvTrxh2) were characterized previously. In this study, two NTR isoforms (HvNTR1 and HvNTR2) were identified, enabling comparison of gene expression, protein appearance, and interaction between individual NTR and Trx h isoforms in barley embryo and aleurone layers. Although mRNA encoding both Trx h isoforms is present in embryo and aleurone layers, the corresponding proteins differed in spatiotemporal appearance. HvNTR2, but not HvNTR1, gene expression seems to be regulated by gibberellic acid. Recombinant HvNTR1 and HvNTR2 exhibited virtually the same affinity toward HvTrxh1 and HvTrxh2, whereas HvNTR2 has slightly higher catalytic activity than HvNTR1 with both Trx h isoforms, and HvNTR1 has slightly higher catalytic activity toward HvTrxh1 than HvTrxh2. Notably, both NTRs reduced Trx h at the acidic conditions residing in the starchy endosperm during germination. Interspecies reactions between the barley proteins and Escherichia coli Trx or Arabidopsis thaliana NTR, respectively, occurred with 20- to 90-fold weaker affinity. This first investigation of regulation and interactions between members of the NTR/Trx system in barley seed tissues suggests that different isoforms are differentially regulated but may have overlapping roles, with HvNTR2 and HvTrxh1 being the predominant isoforms in the aleurone layer.
Thioredoxins (Trxs) are small, ubiquitous proteins participating in thiol-disulfide reactions via two Cys residues found in a conserved active-site motif (CXXC; Jacquot et al., 1997). A wide range of genes encoding Trx have been identified in plants, as opposed to animals, fungi, and bacteria. Trxs are classified based on primary structures and subcellular localization. Trx f, m, x, and y are found in the chloroplast, whereas Trx o and h are localized to the cytoplasm or mitochondrion (Gelhaye et al., 2004). Trx f is reduced by ferredoxin-thioredoxin reductase and in turn reduced by ferredoxin generated during photosynthetic electron transport (Hirasawa et al., 1999). In contrast, reduction of Trx o and two large Trx h subgroups depends on NADPH and involves NADPH-dependent thioredoxin reductase (NTR; Laloi et al., 2001; Gelhaye et al., 2004). Finally, a new Trx h subgroup, first reported in poplar (Populus spp.; Gelhaye et al., 2003), is reduced via the glutathione/glutaredoxin system.
NTRs belong to a superfamily of flavoprotein disulfide oxidoreductases (Reichheld et al., 2005) and transfer electrons from NADPH to the active-site disulfide bridge of oxidized Trx h via FAD and a redox-active disulfide (Mustacich and Powis, 2000). NTRs from plants, bacteria, fungi, and archaea are homodimers of 35 kD (Dai et al., 1996), whereas mammalian NTRs are homodimers of 55 kD, usually containing an active-site seleno-Cys (Tamura and Stadtman, 1996). The NTR/Trx system in plants has a variety of functions. Trx h was suggested to act as a signal in the phloem sap (Ishiwatari et al., 1995) and it was reported to participate in the self-incompatibility reaction (Cabrillac et al., 2001). In addition, Trx h functions during seed development (Serrato and Cejudo, 2003). Many of the Trx h target proteins identified in cereal seeds suggest that cellular redox-regulated processes are necessary for germination. Thus, the NTR/Trx system may control the activity of α-amylase and trypsin inhibitors (Kobrehel et al., 1991), increase activity of pullulanase (limit dextrinase) that specifically cleaves α-1,6 linkages from starch amylopectin (Cho et al., 1999), reduce storage proteins, facilitating their mobilization (Kobrehel et al., 1992), and activate a seed-specific Ser protease, thiocalsin (Besse et al., 1996). Finally, enhancement of gibberellic acid (GA) synthesis in embryos and accelerated appearance of α-amylase in transgenic barley overexpressing Trx h in the starchy endosperm suggest that Trx h plays a role in the communication between endosperm, embryo, and aleurone layer (Wong et al., 2002).
The presence of multiple Trx h and NTR isoforms in plants makes the NTR/Trx system particularly complex compared with other organisms. For instance, in Arabidopsis (Arabidopsis thaliana), eight genes encoding Trx h have been identified (Meyer et al., 2002, 2005) and three NTR isoforms have been characterized (Serrato et al., 2004; Reichheld et al., 2005). Five Trx h (Gautier et al., 1998; Serrato et al., 2001; Cazalis et al., 2006) and one NTR (Serrato et al., 2002) were described from wheat (Triticum aestivum). In barley (Hordeum vulgare), two seed Trx h isoforms were characterized (Maeda et al., 2003), but no barley NTR was identified until now.
The mechanisms regulating expression of Trx h and NTR in seed tissues are poorly understood; indeed, specific isoforms of Trx h and NTR may be localized in different tissues and have diverse roles during plant development. Expression of Trx h in endosperm seems to be controlled by hormones via the embryo (Lozano et al., 1996). Furthermore, an increase of Trxh1, Trxh2, and Trxh3 transcripts at the beginning of seed desiccation and a transient increase of Trxh1 after seed imbibition were observed in wheat (Cazalis et al., 2006). Others reported that Trx h (Marx et al., 2003) and NTR (Montrichard et al., 2003) increase during germination in the embryo of barley and pea (Pisum sativum), respectively.
Knowledge lags behind on the individual roles played by Trx and NTR isoforms from the same organism. In particular, insight is lacking on specificity and structural requirements for interactions between NTRs and Trxs. To address these fundamental questions, two barley NTR isoforms are cloned and characterized in this work and their gene expression and protein appearance in seed tissues are described in parallel with two Trx h isoforms. The effects of GA and abscisic acid (ABA) on the members of this NTR/Trx system were monitored in isolated aleurone layers. Production of all four proteins in recombinant form allowed the interactions between isoforms of Trx h and NTR from the same source to be characterized.
RESULTS
Isolation, Cloning, and Sequence Analysis of Two cDNAs Encoding NTR
Two cDNAs encoding NTR in barley seeds were isolated by a PCR-based cloning strategy. Each contained an open reading frame of 996 bp encoding proteins designated HvNTR1 and HvNTR2 with theoretical molecular mass (kD)/pI of 34.818/5.7 and 34.793/5.7, respectively. Nucleotide sequences of HvNTR1 and HvNTR2 are very similar to tentative consensus (TC) sequences TC142091 and TC141301 from the The Institute for Genomic Research (TIGR) barley gene index. One additional sequence in the database (TC132362) showed lower identity with HvNTR1 and HvNTR2, but was still related to NTR. TCs were assembled from few EST sequences, indicating that NTR transcripts are not highly represented in barley cDNA libraries. It therefore cannot be excluded that barley has other NTR isoforms than the three discussed here. The EST sequences originated predominantly from seed tissues.
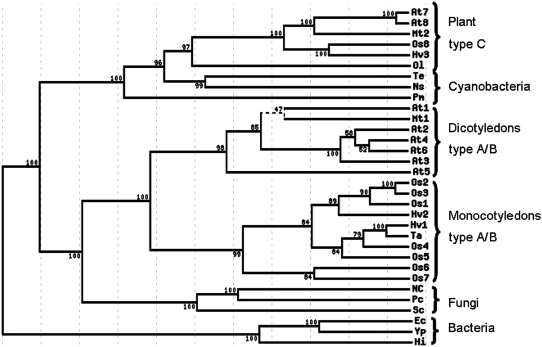
HvNTR1 and HvNTR2 proteins have 88% sequence identity. A multiple alignment, including the protein sequence deduced from TC132362 and NTR sequences from other sources (Supplemental Fig. S1), was used to generate a phylogenetic tree (Fig. 1). The tree is divided into three major clusters. One cluster contains NTR from cyanobacteria and plant chloroplast-type (C-type) NTRs that have an extra Trx active-site sequence (CGPC) in a C-terminal extension (Serrato et al., 2004; Alkhalfioui et al., 2007a). The second cluster has two main branches; one contains fungal NTRs and the other contains plant cytoplasmic or mitochondrial-type NTRs (A/B type; Reichheld et al., 2005), which are further subdivided into sequences from monocots and dicots. Bacterial NTRs constitute the third cluster. HvNTR1 and HvNTR2 belong to the monocotyledon subgroup of the plant A/B type. Despite their high sequence identity, both have even higher similarity to other cereal NTRs. HvNTR1 is thus 97% identical with wheat NTR (TaNTR) and HvNTR2 is 91% identical to a putative rice (Oryza sativa) NTR (Os1; see Fig. 1). HvNTR1 and HvNTR2 are each 75% identical to an Arabidopsis NTR (At3; Fig. 1). Barley enzymes are thus expected to share tertiary and quaternary structure with Arabidopsis NTR—the only plant NTR for which the three-dimensional (3-D) structure has been solved (Dai et al., 1996).
Figure 1.
Phylogenetic tree of NTR sequences. Values indicate percent bootstrap support for each branch. For accession numbers and organisms, see “Materials and Methods.”
Plant NTRs are homodimers with each subunit containing an FAD- and an NADP-binding domain. HvNTR1 and HvNTR2 contain both FAD-binding motifs GXGXXA and TXXXXVFAAGD (residues 17–22 and 283–293, respectively), the NADP-binding motif GXGXXA (residues 164–169), and the two active-site Cys residues in the motif CAVC (residues 145–148). A C-terminal extension containing an additional Trx active site, characteristic for C-type NTRs, is present in the third, NTR-related sequence in barley TC132362 (Hv3; Fig. 1).
Gene Expression and Protein Profiling of NTR and Trx h in Germinating Embryos
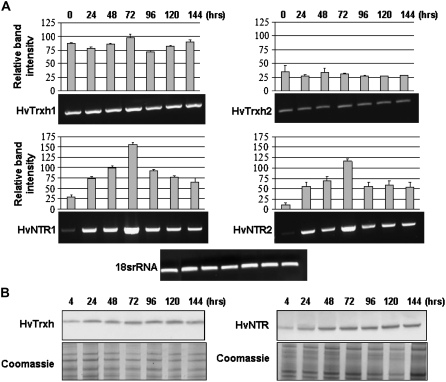
Expression of HvTrxh1, HvTrxh2, HvNTR1, and HvNTR2 genes was analyzed by semiquantitative reverse transcription (RT)-PCR using total RNA from embryos dissected from mature seeds and seeds at different time points during germination. The appearance of the corresponding proteins was monitored by western-blot analyses of soluble proteins extracted from the embryo during germination. Transcripts of both HvTrxh1 and HvTrxh2 were present in embryos from mature seeds and remained constant during germination (Fig. 2A). Trx h protein, however, increased slightly in amount from 24 h after imbibition (Fig. 2B), corresponding to the time of radicle protrusion, and then remained constant up to 144 h. HvNTR1 and HvNTR2 transcripts were both detected at low levels in embryos from mature seeds (Fig. 2A). Expression increased considerably up to 72 h after imbibition, then started to decrease. NTR protein (Fig. 2B) was detected at low levels in embryos 4 h after imbibition, increased up to 48 h, and then remained essentially constant. For both Trx h and NTR, the western-blotting profiles represent the combined appearance of all isoforms.
Figure 2.
Gene expression and protein appearance for Trx h and NTR isoforms in barley embryos during germination (0–144 h). A, RT-PCR analysis of HvTrxh1, HvTrxh2, HvNTR1, and HvNTR2. One representative gel is shown from three independent replicates. Relative band intensities were normalized to the 18S rRNA band intensity (100%). Each histogram represents the mean ± sd obtained from three independent RT-PCR reactions. B, Western blot of embryo protein extracts using antibodies against Trx h and NTR.
Gene Expression and Protein Profiling of NTR and Trx h in Aleurone Layers Responding to Hormones
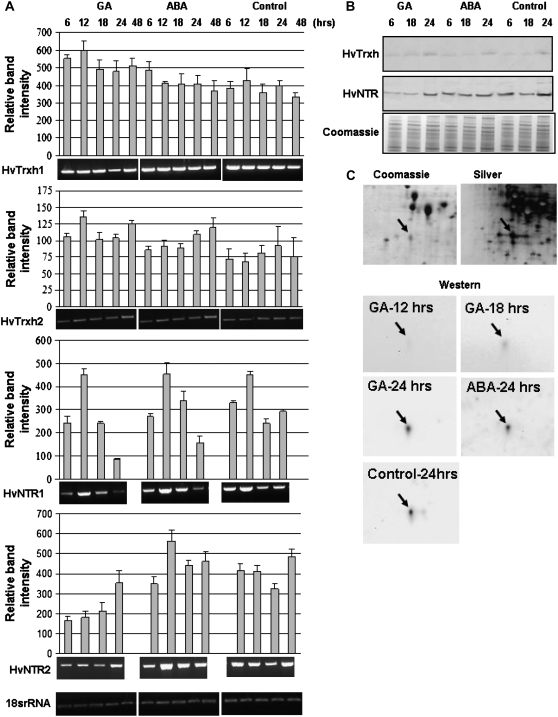
The cereal seed aleurone layer has a key role in germination, responding to hormone signals from the embryo and producing storage reserve mobilizing enzymes. Isolated aleurone layers can be maintained in buffer, providing a well-defined system for analysis of hormonal regulation of seed germination (Fath et al., 2001). Aleurone layers were incubated in buffer containing GA or ABA and harvested at time points between 6 and 48 h. HvTrxh1 and HvTrxh2 expression was detected in aleurone layers incubated with or without the addition of GA or ABA and the expression level varied only slightly (Fig. 3A). Trx h protein, however, was hardly detected in aleurone layers after 6-h incubation (Fig. 3B) and the level increased slightly during the course of the incubation. A similar increase was observed in aleurone layers treated with GA or ABA, suggesting that these hormones do not regulate gene expression or protein accumulation of HvTrxh1 and HvTrxh2.
Figure 3.
Gene expression and protein appearance for Trx h and NTR isoforms in aleurone layers responding to GA, ABA, and buffer without hormones (control) at various time points (6–48 h). A, RT-PCR analysis of HvTrxh1, HvTrxh2, HvNTR1, and HvNTR2. One representative gel is shown from three independent replicates. Relative band intensities were normalized to18S rRNA band intensity (100%). Each data point represents the mean ± sd obtained from three independent RT-PCR reactions. B, Western blot of aleurone protein extracts using antibodies against Trx h and NTR. C, Coomassie- and silver-stained 2-D gels and 2-D western blotting using Trx h antibody in aleurone layers treated in the presence and absence of GA and ABA. The Trx h spot is marked by an arrow.
HvNTR1 transcripts were only detected in aleurone layers after 45 amplification cycles in contrast to 35 cycles in embryos, suggesting that HvNTR1 is expressed at a much higher level in embryos than aleurone layers. In contrast, HvNTR2 transcripts were detected in both tissues after 35 amplification cycles. Noticeably, expression of HvNTR1 and HvNTR2 in aleurone layers showed distinct differences. HvNTR1 expression increased considerably after 12-h incubation with or without GA or ABA, and then decreased up to 24 h (Fig. 3A). No clear trend in expression of HvNTR2 was observed; however, the expression was significantly lower in the presence of GA than in control or ABA-treated aleurone layers, in particular at early time points, suggesting that GA down-regulates HvNTR2 expression. This was supported by the western blot (Fig. 3B) showing very little NTR protein after 6- and 18-h GA treatment compared with control or ABA-treated aleurone layers. After 24 h, the NTR level was essentially the same in all three samples, supporting early-phase down-regulation by GA. Because the protein profile clearly reflected HvNTR2 gene expression, HvNTR2 might be the dominant isoform in aleurone layers.
2-D Gel Electrophoresis Pattern of Trx h in Aleurone Layers
HvTrxh1 and HvTrxh2 were identified in two and one 2-D gel spots, respectively, in mature barley embryos (Maeda et al., 2003). One HvTrxh1 spot and the HvTrxh2 spot decreased at radicle elongation (Bønsager et al., 2007). In aleurone layers dissected from mature seeds, HvTrxh2 was absent and HvTrxh1 appeared in one spot (Maeda et al., 2003). Trx h transcript levels in aleurone layers appear to be independent of GA and ABA. Therefore, 2-D western blotting was performed to examine whether the individual protein isoforms were affected by hormone treatment. A single spot with approximate 12.5-kD molecular mass and pI 5.0 was detected in all blots and on 2-D gels stained with Coomassie Blue and silver nitrate (Fig. 3C). The spot was excised from the Coomassie-stained gel for in-gel digestion with trypsin and analysis by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). Five peptide masses matched HvTrxh1 (AAP72290), giving 20% sequence coverage. The abundance of the spot was similar in aleurone layers treated with or without GA or ABA, in agreement with gene expression and the 1-D western blot, indicating that synthesis of the protein was not regulated by hormones. The HvTrxh1 spot was hardly detected in aleurone layers incubated for 12 or 18 h (Fig. 3C) and increased in intensity during incubation, in agreement with the 1-D western blot. Identification of only the HvTrxh1 protein in aleurone layers is in agreement with previous findings (Maeda et al., 2003), but is in contrast to detection of transcripts corresponding to both Trx h genes.
Expression, Purification, and Biochemical Characterization of HvNTR1 and HvNTR2
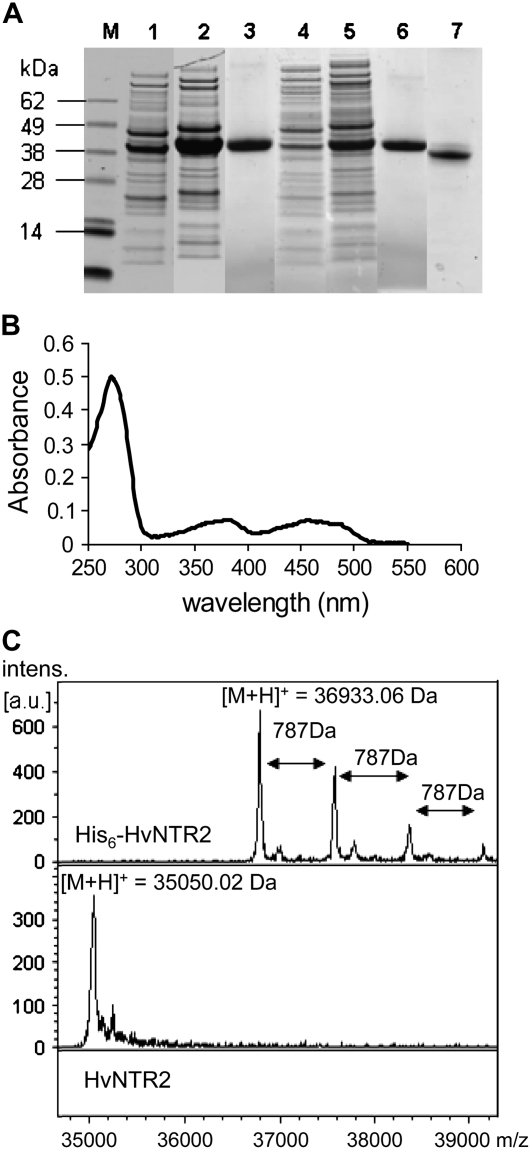
To confirm that the cDNA sequences encoded active NTR and to compare the kinetic properties of the two isoforms, recombinant proteins carrying an amino-terminal His6-tag were produced in Escherichia coli. HvNTR1 and HvNTR2 were found in the soluble fraction of the E. coli transformant culture after induction with isopropyl-β-d-thiogalactopyranoside (IPTG) and were recognized both by an antibody recognizing the His-tag and an antibody raised against wheat NTR (data not shown). SDS-PAGE of cell extracts showed a prominent polypeptide band of the expected molecular mass (Fig. 4A, lanes 2 and 5), and the recombinant His6-HvNTR1 and His6-HvNTR2 were purified from the crude extracts by nickel affinity chromatography (Fig. 4A, lanes 3 and 6) in yields of 30 and 10 mg/L, respectively. Proteins were yellow with absorption maxima at 270, 378, and 454 nm (Fig. 4B) typical for flavoproteins (Jacquot et al., 1994; Serrato et al., 2002).
Figure 4.
Expression, purification, and chemical properties of NTR. A, SDS-PAGE of NTR overexpression in E. coli, purification, and thrombin cleavage. Total protein from E. coli harboring pET15b-HvNTR1 (lanes 1 and 2) and pET15b-HvNTR2 (lanes 4 and 5), respectively, before (lanes 1 and 4) and 3.5 h after (lanes 2 and 5) induction with IPTG; purified His-HvNTR1 and His-HvNTR2, respectively (lanes 3 and 6); and HvNTR2 after thrombin cleavage to remove the His-tag (lane 7). B, Absorption spectrum of recombinant HvNTR2. C, MALDI MS spectrum of intact His-HvNTR2 and after cleavage with thrombin.
One FAD-binding site is predicted per NTR subunit. Remarkably, the MALDI-TOF mass spectrum for His6-HvNTR2 showed a series of peaks differing by 787 D corresponding to the molecular mass of FADH2 (Fig. 4C). Peaks were assigned to [M + H]+ of His6-HvNTR2 lacking FAD, and His6-HvNTR2 with one or more associated FAD molecules, respectively. The N-terminal His6-tag was subsequently removed from the recombinant proteins by thrombin cleavage (Fig. 4A, lane 7). Three amino acids (Gly, Ser, His) originating from the His-tag remain at the N terminus of NTR after thrombin cleavage. The MALDI-TOF mass spectrum for cleaved HvNTR2 exhibited a prominent peak at m/z 35050.02 D in agreement with the theoretical mass of cleaved HvNTR2 lacking FAD (35052.54 D). No additional series of peaks was observed (Fig. 4C), and it is concluded that FAD is noncovalently bound to HvNTR, allowing dissociation from the FAD-binding site and formation of adducts associated with the N-terminal His6-tag under MALDI conditions.
Enzyme Activity
Kinetic parameters for activity of HvNTR1 and HvNTR2 were determined using barley HvTrxh1 and HvTrxh2 and, for comparison, also with E. coli Trx as substrate (Table I). Km values indicate that HvNTR1 and HvNTR2 have similar affinities for HvTrxh1 and HvTrxh2. Activity of HvNTR1 with HvTrxh1, represented by kcat, was almost twice that with HvTrxh2. In contrast, the kcat of HvNTR2 was the same for HvTrxh1 and HvTrxh2. The kcat of HvNTR2 with E. coli Trx was similar to that of barley HvNTR1 with barley Trx h isoforms, whereas the Km for E. coli Trx was very high. The catalytic efficiency (kcat/Km) for E. coli Trx was thus 100-fold lower than for HvTrxh1 and HvTrxh2, indicating that a noncognate Trx is not a good substrate for the NTR. Similarly, thioredoxin reductases from Caenorhabditis elegans (Lacey and Hondal, 2006) and Plasmodium falciparum (Krnajski et al., 2001) also show high activity toward E. coli Trx, but with a high Km. Kinetic parameters were also determined using Arabidopsis NTR (AtNTR; Jacquot et al., 1994), which had lower affinity and lower activity for barley Trx h isoforms compared with barley NTRs (Table I). Conversely, both AtNTR and E. coli NTR showed high affinity for their own Trxs, Km being 1.1 (Jacquot et al., 1994) and 1.9 to 2.4 μm (Miranda-Vizuete et al., 1997), respectively, similar to the Km values determined here for the barley NTR/Trx h pairs. These data underline that NTR from different species prefer their cognate Trx h as substrate.
Table I.
Kinetic parameters of barley (HvNTR1, HvNTR2) and Arabidopsis (AtNTR) NTR for barley Trx h isoforms (HvTrxh1, HvTrxh2) and E. coli Trx
| Enzyme | Substrate | Km | kcat | kcat/Km |
|---|---|---|---|---|
| μm | s−1 | s−1m−1 | ||
| pH 7.4 | ||||
| HvNTR1 | HvTrxh1 | 1.18 ± 0.25 | 2.25 ± 0.08 | 1.90 × 106 |
| HvNTR1 | HvTrxh2 | 1.79 ± 0.40 | 1.31 ± 0.08 | 0.73 × 106 |
| HvNTR2 | HvTrxh1 | 1.12 ± 0.04 | 3.26 ± 0.09 | 2.91 × 106 |
| HvNTR2 | HvTrxh2 | 1.29 ± 0.25 | 2.98 ± 0.16 | 2.31 × 106 |
| HvNTR2 | E. coli Trx | 107 | 1.6 | 1.55 × 104 |
| AtNTR | HvTrxh1 | 24.70 ± 5.00 | 1.04 ± 0.09 | 0.42 × 105 |
| AtNTR | HvTrxh2 | 26.70 ± 6.50 | 0.80 ± 0.07 | 0.30 × 105 |
| pH 5.7 | ||||
| HvNTR1 | HvTrxh2 | 1.45 | 0.43 | 0.30 × 106 |
| HvNTR2 | HvTrxh2 | 0.90 | 0.8 | 0.89 × 106 |
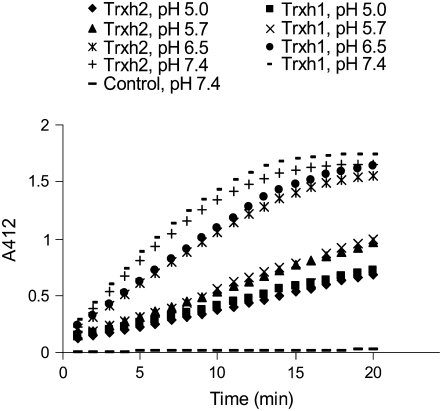
The pH used here and by others to determine NTR activity is significantly higher than that expected in some barley seed tissues because the aleurone layer acidifies the starchy endosperm to about pH 5.0 during germination (Dominguez and Cejudo, 1999). Because Trx h may have a number of roles in the starchy endosperm during germination (Buchanan and Balmer, 2005), the activity of HvNTR2 was analyzed between pH 5.0 to 7.4 using 3 μm oxidized HvTrxh1 or HvTrxh2 as substrate (Fig. 5). The velocity of 5,5′-dithio-bis (2-nitrobenzoic acid) (DTNB) reduction decreased from pH 7.4 to 5.0, but considerable reduction of DTNB was still obtained at pH 5, indicating that HvNTR2 has the capacity to reduce Trx h in the slightly acidic environment found in the starchy endosperm during germination. HvNTR1 showed similar pH dependence (data not shown). The Km of HvNTR1 and HvNTR2 determined toward HvTrxh2 at pH 5.7 was not altered (Table I); therefore, it is concluded that the decrease to pH 5.7 reduced the catalytic activity 3 to 4 times.
Figure 5.
Time course of HvTrxh1 and HvTrxh2 reduction by HvNTR2 as monitored by reduction of DTNB at various pH values (5, 5.7, 6.5, and 7.4). Control shows the time course of DTNB reduction by HvNTR2 without addition of Trx h.
DISCUSSION
Production of recombinant forms of two isoforms each of barley NTR and Trx h allowed the first investigation of interactions between NTR and Trx isoforms from the same organism. The finding that both NTR isoforms have similar affinity toward the Trx h isoforms is in accordance with the high sequence identity of HvNTR1 and HvNTR2 and the conservation of residues surrounding the active site in HvTrxh1 and HvTrxh2. The higher catalytic activity of HvNTR1 toward HvTrxh1 suggests that the overall activity of an NTR/Trx system could be modulated by exploiting different isoforms under different circumstances.
Despite the different tasks proposed for the NTR/Trx system during germination (Wong et al., 2002, 2004), most experimental evidence was obtained at more basic pH values than expected to be present in vivo. Under weak acidic conditions in the starchy endosperm during germination, the Trx N-terminal active-site Cys (e.g. C46HvTrxh2), acting as a nucleophile in catalysis of the disulfide exchange reaction (Kallis and Holmgren, 1980; Maeda et al., 2006) and with pKa of 6.3 to 7.5 (in E. coli Trx; Setterdahl et al., 2003), may be protonated and hence inactive. Therefore, the central question of whether the NTR/Trx system is active at the pH of the endosperm during germination was not clearly addressed. This work demonstrates, however, that recombinant HvNTR1 and HvNTR2 are able to reduce Trx h at pH 5.0 with essentially retained affinity, but slightly decreased activity, which supports that the NTR/Trx system operates in germination.
Functional analysis was complemented by gene expression and protein profiling during germination because, for NTR/Trx pairs to interact, they must be present in the same tissue at the same time. Profiling experiments also provide information about gene expression, posttranscriptional regulation, and posttranslational regulation. Transcripts corresponding to both Trx h and NTR isoforms were detected in embryos and aleurone layers at all time points, suggesting that the NTR/Trx system is active in these tissues during germination. Levels of Trx h transcripts were relatively constant, whereas NTR transcript accumulation showed greater modulation, as characteristic for genes encoding important regulatory proteins. The comparison of Trx h gene expression and protein profiles illustrates the general problem with predicting protein expression levels from mRNA data. Differences can be due to posttranscriptional modification controlling the protein translation rate (Day and Tuite, 1998), protein modification or degradation, and protein transport to or from cytoplasm (Gygi et al., 1999; You and Yin, 2000). Distinct transcript accumulation patterns were observed for HvNTR1 and HvNTR2, indicating that the isoforms are differentially regulated and may have individual functions in seed germination. HvNTR2 perhaps has a specific role in germination because its expression in the aleurone layer was affected by GA. HvNTR1 expression level is much lower in aleurone than embryo and the NTR protein profile in the aleurone layer closely resembles the HvNTR2 gene expression pattern. In combination, gene expression and protein profiling suggest that, whereas both NTR and Trx h isoforms are present in the embryo, with HvTrxh1 protein more prominent during germination, HvNTR2 and HvTrxh1 may be the dominant isoforms in the aleurone layer during germination. Isolation of the promoter regions for the two genes would be one way to further study the regulation.
The observed increase of NTR gene expression in germinating embryos would be expected to lead to an increase in the proportion of reduced Trx h and hence reduced Trx h target proteins. These results agree with protein disulfides becoming more reduced during germination (Marx et al., 2003; Alkhalfioui et al., 2007b). The reported elevated activity during germination of oxidative pentose phosphate pathway enzymes (Lozano et al., 1996) could provide the NADPH required for activation of the NTR/Trx system in embryos. In contrast, the decrease of HvNTR2 expression in aleurone layers in response to GA at early incubation time points suggests an initial low level of reduced Trx h and target proteins in the aleurone layer. Several potential Trx target proteins (Yamazaki et al., 2004; Gelhaye et al., 2006) are involved in protection against reactive oxygen species, generated as by-products of lipid metabolism in the aleurone layer (Bethke et al., 2002) and implicated in the initiation of the cell death program (Fath et al., 2001). Down-regulation of HvNTR2 expression by GA is predicted to decrease the proportion of reduced Trx h, resulting in modulation of target protein activity. Therefore, regulation of the NTR/Trx system by hormones could affect cell death signaling in aleurone layer cells.
It still remains to be determined whether NTR isoforms are localized in the same cell compartments. In Arabidopsis, both mitochondrial and cytosolic forms (Reichheld et al., 2005) of NTR have been identified.
CONCLUSION
This study demonstrates time interactions between two NTR and two Trx h isoforms from the same organism. The results support a functional role of the NTR/Trx system during germination and suggest that the members of the barley seed NTR/Trx system can function interchangeably. Future proteomics-based studies of individual NTR and Trx isoforms in conjunction with determination of intracellular localization and promoter structure may clarify their differential expression pattern in response to hormones and their possibly divergent in vivo functions. Finally, heterologous expression of both NTR and Trx h isoforms provides a basis for design and characterization of mutants to investigate the interaction between NTR and Trx h at the level of molecular structures.
MATERIALS AND METHODS
Plant Material
Embryos from germinated seeds were prepared from the malting barley (Hordeum vulgare ‘Barke’) provided by Sejet Plantbreeding. Mature seeds were sterilized in 5% sodium hypochlorite for 30 min and rinsed several times with water. Seeds were germinated for 0, 4, 24, 48, 72, 96, 120, and 144 h, frozen, and stored as described (Bønsager et al., 2007). Embryos were dissected from seeds prior to protein and RNA extraction.
Aleurone layers were prepared from embryoless half-grains (Hynek et al., 2006) of barley ‘Himalaya’ (purchased from Washington State University). Isolated aleurone layers (100 mg fresh weight) were incubated in 2 mL of 20 mm sodium succinate (pH 4.2), containing 20 mm CaCl2, 50 μg/mL ampicillin, and 5 μg/mL nystatin. Where appropriate, GA or ABA (5 μm) was added. Incubation was performed at room temperature with continuous gentle shaking. Aleurone layers were harvested at various time points (6–48 h), washed four times with 2 mL of the incubation medium without addition of antibiotics or hormones, frozen in liquid nitrogen, and stored at −80°C until use.
RT-PCR Analysis
Total RNA was extracted from tissues using the RNeasy plant mini kit (Qiagen) and treated with RNase-Free DNase (Qiagen). RT-PCR was performed using a PTC-200 DNA Engine Peltier thermal cycler (Bio-Rad) and a one-step RT-PCR kit (Qiagen) according to the manufacturer's recommendations. Barley 18S rRNA showing invariant expression across the samples was amplified in parallel. The optimal number of amplification cycles (between 15 and 45) for each set of primers was determined at the exponential phase range of amplification. To control for possible genomic DNA contamination, parallel reactions were carried out where reverse transcriptase activity was inactivated by incubation at 95°C. A negative control lacking template RNA was included for each set of RT-PCR reactions. Reactions were performed in triplicate. Amplification products were separated by agarose gel electrophoresis and quantified using ImageJ software (W.S. Rasband; 1997–2007; National Institutes of Health; http://rsb.info.nih.gov/ij). Signal intensities were normalized with respect to 18S rRNA from the same sample. Primers used were trxh8 and trxh9 for HvTrxh1, trxh10F and trxh10R for HvTrxh2, ntr3F and ntr3R for HvNTR1, and ntr4F and ntr4R for HvNTR2 (Supplemental Table S1).
Sequence Analysis
Multiple alignment of the region marked in Supplemental Figure S1 was performed using ClustalW (http://www.ebi.ac.uk/Tools/clustalw). A phylogenetic tree was constructed using the GeneBee TreeTop phylogenetic tree prediction server (http://www.genebee.msu.su/genebee.html). National Center for Biotechnology Information (NCBI) accessions used for the analysis were: Os, rice (Oryza sativa), Os1 (NP_001047911), Os2 (EAY87270), Os3 (EAZ24372), Os4 (BAD33510), Os5 (NP_001057531), Os6 (EAZ00754), Os7 (EAZ36842), Os8 (NP_001060515); At, Arabidopsis (Arabidopsis thaliana), At1 (Q39242), At2 (NP_195271), At3 (1VDC), At4 (AAO42318), At5 (AAO42318), At6 (CAA80656), At7 (AAL08250), At8 (NP_565954); Hv, H. vulgare, Hv1 (ABY27300), Hv2 (ABX09990), Hv3 (TC132362); Ta, wheat (Triticum aestivum; CAD19162); Mt, Medicago truncatula, Mt1 (ABH10138), Mt2 (ABH10139); Ol, Ostreococcus lucimarinus (XP_001422184); Te, Thermosynechococcus elongates (NP_682714); Ns, Nostoc sp. (NP_484780); Pm, Prochlorococcus marinus (NP_893267); Nc, Neurospora crassa (P51978); Pc, Penicillium chrysogenum (P43496); Sc, Saccharomyces cerevisiae (P29509); Ec, Escherichia coli (P09625); Yp, Yersinia pestis (NP_404967); and Hi, Haemophilus influenzae (P43788).
Protein Extraction and Western-Blot Analysis
Frozen tissues were dried under vacuum and ground to a fine powder using a mortar and pestle. Soluble proteins were extracted in 5 mm Tris-HCl, 1 mm CaCl2 (pH 7.5) with the protease inhibitor cocktail Complete (Roche; Finnie and Svensson, 2003). Protein concentration was determined using the Popov assay with bovine serum albumin as standard (Popov et al., 1975).
Proteins (1.5 μg total protein from the aleurone layer or 6 μg total protein from the embryo) were separated on 4% to 12% Bis-Tris NuPAGE gels (Invitrogen) and stained with colloidal Coomassie Brilliant Blue G-250 (Candiano et al., 2004). Proteins were electroblotted onto a nitrocellulose membrane (Hybond-N; GE Healthcare) and probed using rabbit anti-wheat Trx h or rabbit anti-wheat NTR antibodies. Secondary antibodies conjugated to horseradish peroxidase or alkaline phosphatase (Dako Cytomation) were detected by enhanced chemiluminescence (Thorpe and Kricka, 1986) or the nitroblue tetrazolium/bromochloro indolyl phosphate colorimetric method, respectively.
2-D Gel Electrophoresis and Western Blotting
Protein extracts from 100-mg aleurone layers were desalted on NAP-5 columns (GE Healthcare). Aliquots containing 50 μg of protein were precipitated by ammonium acetate (0.1 m) in methanol (Vensel et al., 2005) and redissolved in reswelling buffer (7 m urea, 2 m thiourea, 2% [v/v] CHAPS, 0.5% [v/v] immobilized pH gradient [IPG] ampholytes 3–10 [Amersham Biosciences], 1.2% [v/v] Destreak reagent [hydroxyethyl disulfide], and a trace of bromphenol blue). First-dimension isoelectric focusing was done using 18-cm linear IPG strips, pI 3 to 10, and an IPG phor (GE Healthcare). IPG strips were subsequently equilibrated as described (Finnie et al., 2002) for second-dimension electrophoresis on Excel SDS XL (18 × 24 cm) 12% to 14% gradient gel (GE Healthcare) using Multiphor II (GE Healthcare) according to the manufacturer's recommendations and stained with either silver nitrate (Heukeshoven and Dernick, 1985) or colloidal Coomassie Brilliant Blue G-250 (Candiano et al., 2004). For electroblotting onto nitrocellulose, gels were first equilibrated for 30 min in NuPAGE transfer buffer (Invitrogen) containing 15% methanol.
In-Gel Digestion and Protein Identification
Spots cut out from Coomassie-stained gels were in-gel digested with trypsin (Promega) as described (Finnie et al., 2002). Peptides were micropurified (Gobom et al., 1999) and eluted directly onto the MALDI target in 5 μg/μL α-cyano-hydroxy-cinnamic acid in 70% acetonitrile and 0.1% trifluoroacetic acid. An Ultraflex II MALDI-TOF-TOF mass spectrometer (Bruker-Daltonics) was used in positive ion reflector mode and spectra were analyzed using FlexAnalysis software (Bruker-Daltonics). Spectra were calibrated externally using a tryptic digest of β-lactoglobulin. Internal calibration was carried out with trypsin autolysis products (m/z 842.51 and m/z 2211.10). Peptide mass data were searched against the NCBI nonredundant database using Biotools (Bruker Daltonics) software and Mascot (MatrixScience) with the following parameters: monoisotopic mass tolerance, 80 ppm; allowed missed cleavages, 1; allowed modifications, carbamidomethylation of Cys (global) and oxidation of Met (partial).
Isolation and Cloning of HvNTR1 and HvNTR2
Total RNA was extracted from embryos and contaminating genomic DNA was removed as described above. Conserved regions from plant genes encoding NTR were used to design primers (ntr1F and ntr1R). RT-PCR was performed using the one-step RT-PCR kit (Qiagen) according to the manufacturer's instructions. The amplified fragment (700 bp) was cloned in the pDrive cloning vector (Qiagen). Sequencing of cloned fragments (MWG-Biotech AG) distinguished two types of internal NTR sequences, cDNA1 and cDNA2. Because cDNA1 was 98% identical to TC142091 and cDNA2 was 100% identical to TC141301, these TC sequences were used to design gene-specific primers for RT-PCR amplification from embryo RNA and sequencing of the 5′ end (primers ntr4F and ntr4R) and 3′ end (primers ntr5F and ntr5R) of the cDNA2 coding sequence and the 3′ end of the cDNA1 coding sequence. Because TC 142091 did not include a complete coding sequence and lacked the 5′ untranslated region, the untranslated region from wheat NTR (accession no. AJ421947, 97% identical to cDNA1) was used to design primers (ntr2F and ntr2R) for isolation of the 5′ end of the cDNA1 coding sequence. Amplicons were cloned in pDrive cloning vectors and sequenced. Finally, cDNAs containing the entire coding regions of HvNTR1 and HvNTR2 were amplified by RT-PCR from embryo RNA using primers ntr2F and ntr6R and ntr4F and ntr5R, respectively, and cloned to give pDrive-HvNTR1 and pDrive-HvNTR2. Primer sequences are listed in Supplemental Table S1.
Expression, Purification, and Chemical Properties of Recombinant HvNTR1 and HvNTR2
The restriction sites NdeI and BamHI were introduced at the ends of the HvNTR1 coding sequence by primers ntr10F and ntr10R, using HotStar HiFidelity PCR (Qiagen) and pDrive-HvNTR1 as template. pDrive-HvNTR2 was used as template with primers ntr8F and ntr8R to introduce NdeI and XhoI sites. After cleavage by the appropriate restriction enzymes, the inserts were ligated into pET15b (Novagen) to give pET15b-HvNTR1 and pET15b-HvNTR2. Sequences were verified and constructs were used to transform E. coli strain Rosetta (DE3). Cells were grown at 37°C in Luria-Bertani medium supplemented with 100 μg/mL ampicillin and 5 μg/mL chloramphenicol to an OD600 of 0.6. Cultures were induced by 100 μm IPTG for 3.5 h. Cells were harvested by centrifugation and frozen at −20°C until use.
Frozen pellets were resuspended in Bugbuster protein extraction reagent including Benzonase (Novagen) and shaken for 20 min at room temperature. After centrifugation (14,000g for 20 min, 4°C), recombinant protein in the supernatant was purified by a His-Trap HP column (Amersham Biosciences) as described (Maeda et al., 2006) and eluted fractions were analyzed by SDS-PAGE. Purified proteins were desalted on PD-10 columns (GE Healthcare) in 10 mm Tris-HCl (pH 8.0).
Protein concentration was determined by amino acid analysis. The absorption spectrum was recorded for 8.7 μm NTR in 10 mm Tris-HCl (pH 8.0) at room temperature. For cleavage of the N-terminal His-tag, purified His6-tag HvNTR2 (0.1 μg/μL) was treated with immobilized thrombin (Calbiochem) at 1:100 (w/w) thrombin:fusion in 0.6 mm NaCl, 50 mm Tris-HCl (pH 6.0) for 24 h at 24°C. Mass spectrometric analysis of intact proteins was performed using 20 pmol His-HvNTR2 or cleaved HvNTR2 after micropurification of samples as described above, but using Poros 20 R1 (Applied Biosystem) as column material and eluting in sinapinic acid (10 mg/mL) in 70% acetonitrile and 0.1% trifluoroacetic acid. External calibration of spectra was performed using protein standard II (Bruker-Daltonics).
Enzyme Activity Assay
Activity of NTR was measured at 25°C using DTNB as the final disulfide substrate as described (Miranda-Vizuete et al., 1997) with slight modifications. The reaction mixture contained 100 mm potassium phosphate (pH 5.0–7.4), 10 mm EDTA, 200 μm DTNB (Sigma-Aldrich), 200 μm NADPH (Sigma-Aldrich), and Trx (1–10 μm) from barley or E. coli (Promega). The reaction was started by the addition of NTR (40 nm) and the rate of reduction of DTNB was calculated from the A412. As 1 mol Trx-(SH)2 reduces 1 mol DTNB to yield 2 mol TNB (2-nitro-5-thiobenzoic acid) with a molar extinction coefficient of 13,600 m−1 cm−1 (Ellman, 1959), a molar extinction coefficient of 27,200 m−1 cm−1 was applied for quantification. The extinction coefficient of TNB varied by 1% to 4% from this value in the pH range 5.0 to 7.4 employed in this work (data not shown).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EU314717, EU250021, ABY27300, and ABX09990.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Multiple alignment of primary sequences of NTRs from different sources using ClustalW.
Supplemental Table S1. List of forward (F) and reverse (R) primers.
Supplementary Material
Acknowledgments
We thank B. Buchanan (University of California, Berkeley) for rabbit anti-wheat Trx h, F.J. Cejudo (University of Seville, Spain) for rabbit anti-wheat NTR, and J.P. Jacquot (INRA, Nancy, France) for recombinant Arabidopsis NTR. We also wish to thank P. Hägglund and K. Maeda for helpful discussions, B.C. Bønsager for assistance with embryo samples, and K. Rasmussen, A. Blicher, and B. Andersen for technical assistance.
This work was supported by the Iranian Ministry of Science, Research and Technology, the Danish Agriculture and Veterinary Research Council, and the Danish Centre for Advanced Food Studies.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Christine Finnie (csf@biocentrum.dtu.dk).
The online version of this article contains Web-only data.
References
- Alkhalfioui F, Renard M, Montrichard F (2007. a) Unique properties of NADP-thioredoxin reductase C in legumes. J Exp Bot 58 969–978 [DOI] [PubMed] [Google Scholar]
- Alkhalfioui F, Renard M, Vensel WH, Wong J, Tanaka CK, Hurkman WJ, Buchanan BB, Montrichard F (2007. b) Thioredoxin-linked proteins are reduced during germination of Medicago truncatula seeds. Plant Physiol 144 1559–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse I, Wong JH, Kobrehel K, Buchanan BB (1996) Thiocalsin: a thioredoxin-linked, substrate-specific protease dependent on calcium. Proc Natl Acad Sci USA 93 3169–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke PC, Fath A, Spiegel YN, Hwang YS, Jones RL (2002) Abscisic acid, gibberellin and cell viability in cereal aleurone. Euphytica 126 3–11 [Google Scholar]
- Bønsager BC, Finnie C, Roepstorff P, Svensson B (2007) Germination and radicle elongation in barley tracked using proteome analysis of dissected embryo, aleurone layer and endosperm tissues. Proteomics 7 4538–4540 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56 187–220 [DOI] [PubMed] [Google Scholar]
- Cabrillac D, Cock JM, Dumas C, Gaude T (2001) The S-locus receptor kinase is inhibited by thioredoxins and activated by pollen coat proteins. Nature 410 220–223 [DOI] [PubMed] [Google Scholar]
- Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG (2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25 1327–1333 [DOI] [PubMed] [Google Scholar]
- Cazalis R, Pulido P, Aussenac T, Pérez-Ruiz JM, Cejudo FJ (2006) Cloning and characterization of three thioredoxin h isoforms from wheat showing differential expression in seeds. J Exp Bot 57 2165–2172 [DOI] [PubMed] [Google Scholar]
- Cho MJ, Wong JH, Marx C, Jiang W, Lemaux PG, Buchanan BB (1999) Overexpression of thioredoxin h leads to enhanced activity of starch debranching enzyme (pullulanase) in barley grain. Proc Natl Acad Sci USA 96 14641–14646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Saarinen M, Ramaswamy S, Meyer Y, Jacquot JP, Eklund H (1996) Crystal structure of Arabidopsis thaliana NADPH dependent thioredoxin reductase at 2.5 Å resolution. J Mol Biol 264 1044–1057 [DOI] [PubMed] [Google Scholar]
- Day DA, Tuite MF (1998) Post-transcriptional gene regulatory mechanisms in eukaryotes: an overview. J Endocrinol 157 361–371 [DOI] [PubMed] [Google Scholar]
- Dominguez F, Cejudo FJ (1999) Patterns of starchy endosperm acidification and protease gene expression in wheat grains following germination. Plant Physiol 119 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellman GL (1959) Tissue sulfhydryl groups. Arch Biochem Biophys 82 70–77 [DOI] [PubMed] [Google Scholar]
- Fath A, Bethke PC, Jones RL (2001) Enzymes that scavenge reactive oxygen species are down-regulated prior to gibberellic acid-induced programmed cell death in barley aleurone. Plant Physiol 126 156–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnie C, Melchior S, Roepstorff P, Svensson B (2002) Proteome analysis of grain filling and seed maturation in barley. Plant Physiol 129 1308–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnie C, Svensson B (2003) Feasibility study of a tissue-specific approach to barley proteome analysis: aleurone layer, endosperm, embryo and single seeds. J Cereal Sci 38 217–227 [Google Scholar]
- Gautier MF, Lullien-Pellerin V, de Lamotte-Guéry F, Guirao A, Joudrier P (1998) Characterization of wheat thioredoxin h cDNA and production of an active Triticum aestivum protein in Escherichia coli. Eur J Biochem 252 314–324 [DOI] [PubMed] [Google Scholar]
- Gelhaye E, Navrot N, Macdonald IK, Rouhier N, Raven EL, Jacquot JP (2006) Ascorbate peroxidase-thioredoxin interaction. Photosynth Res 89 193–200 [DOI] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Jacquot JP (2003) Evidence for a subgroup of thioredoxin h that requires GSH/Grx for its reduction. FEBS Lett 555 443–448 [DOI] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Jacquot JP (2004) The thioredoxin h system of higher plants. Plant Physiol Biochem 42 265–271 [DOI] [PubMed] [Google Scholar]
- Gobom J, Nordhoff E, Mirgorodskaya E, Ekman R, Roepstorff P (1999) Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom 34 105–116 [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R (1999) Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heukeshoven J, Dernick R (1985) Improved silver staining procedure for fast staining in PhastSystem Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis 9 28–32 [DOI] [PubMed] [Google Scholar]
- Hirasawa M, Schürmann P, Jacquot JP, Manieri W, Jacquot P, Keryer E, Hartman FC, Knaff DB (1999) Oxidation-reduction properties of chloroplast thioredoxins, ferredoxin:thioredoxin reductase, and thioredoxin f-regulated enzymes. Biochemistry 38 5200–5205 [DOI] [PubMed] [Google Scholar]
- Hynek R, Svensson B, Jensen ON, Barkholt V, Finnie C (2006) Enrichment and identification of integral membrane proteins from barley aleurone layers by reversed-phase chromatography, SDS-PAGE, and LC-MS/MS. J Proteome Res 5 3105–3113 [DOI] [PubMed] [Google Scholar]
- Ishiwatari Y, Honda C, Kawashima I, Nakamura S, Hirano H, Mori S, Fujiwara T, Hayashi H, Chino M (1995) Thioredoxin h is one of the major proteins in rice phloem sap. Planta 195 456–463 [DOI] [PubMed] [Google Scholar]
- Jacquot JP, Lancelin JM, Meyer Y (1997) Thioredoxins: structure and function in plant cells. New Phytol 136 543–570 [DOI] [PubMed] [Google Scholar]
- Jacquot JP, Rivera-Madrid R, Marinho P, Kollarova M, Le Maréchal P, Miginiac-Maslow M, Meyer Y (1994) Arabidopsis thaliana NAPHP thioredoxin reductase. cDNA characterization and expression of the recombinant protein in Escherichia coli. J Mol Biol 235 1357–1363 [DOI] [PubMed] [Google Scholar]
- Kallis GB, Holmgren A (1980) Differential reactivity of the functional sulfhydryl groups of cysteine-32 and cysteine-35 present in the reduced form of thioredoxin from Escherichia coli. J Biol Chem 255 10261–10265 [PubMed] [Google Scholar]
- Kobrehel K, Wong JH, Balogh A, Kiss F, Yee BC, Buchanan BB (1992) Specific reduction of wheat storage proteins by thioredoxin h. Plant Physiol 99 919–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobrehel K, Yee BC, Buchanan BB (1991) Role of the NADP/thioredoxin system in the reduction of alpha-amylase and trypsin inhibitor proteins. J Biol Chem 266 16135–16140 [PubMed] [Google Scholar]
- Krnajski Z, Gilberger TW, Walter RD, Müller S (2001) The malaria parasite Plasmodium falciparum possesses a functional thioredoxin system. Mol Biochem Parasitol 112 219–228 [DOI] [PubMed] [Google Scholar]
- Lacey BM, Hondal RJ (2006) Characterization of mitochondrial thioredoxin reductase from C. elegans. Biochem Biophys Res Commun 346 629–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C, Rayapuram N, Chartier Y, Grienenberger JM, Bonnard G, Meyer Y (2001) Identification and characterization of a mitochondrial thioredoxin system in plants. Proc Natl Acad Sci USA 98 14144–14149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano RM, Wong JH, Yee BC, Peters A, Kobrehel K, Buchanan BB (1996) New evidence for a role for thioredoxin h in germination and seedling development. Planta 200 100–106 [Google Scholar]
- Maeda K, Finnie C, Østergaard O, Svensson B (2003) Identification, cloning and characterization of two thioredoxin h isoforms, HvTrxh1 and HvTrxh2, from the barley seed proteome. Eur J Biochem 270 2633–2643 [DOI] [PubMed] [Google Scholar]
- Maeda K, Hägglund P, Finnie C, Svensson B, Henriksen A (2006) Structural basis for target protein recognition by the protein disulphide reductase thioredoxin. Structure 14 1701–1710 [DOI] [PubMed] [Google Scholar]
- Marx C, Wong JH, Buchanan BB (2003) Thioredoxin and germinating barley: targets and protein redox changes. Planta 216 454–460 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Reichheld JP, Vignols F (2005) Thioredoxins in Arabidopsis and other plants. Photosynth Res 86 419–433 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Vignols F, Reichheld JP (2002) Classification of plant thioredoxins by sequence similarity and intron position. Methods Enzymol 347 394–402 [DOI] [PubMed] [Google Scholar]
- Miranda-Vizuete A, Damdimopoulos AE, Gustafsson J, Spyrou G (1997) Cloning, expression, and characterization of a novel Escherichia coli thioredoxin. J Biol Chem 272 30841–30847 [DOI] [PubMed] [Google Scholar]
- Montrichard F, Renard M, Alkhalfioui F, Duval FD, Macherel D (2003) Identification and differential expression of two thioredoxin h isoforms in germinating seeds from pea. Plant Physiol 132 1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustacich D, Powis G (2000) Thioredoxin reductase. Biochem J 346 1–8 [PMC free article] [PubMed] [Google Scholar]
- Popov N, Schmitt M, Schulzeck S, Matthies H (1975) Eine störungsfreie Mikromethode zur Bestimmung des Proteinghaltes in Gewbehomogenaten. Acta Biol Med Ger 34 1441–1446 [PubMed] [Google Scholar]
- Reichheld JP, Meyer E, Khafif M, Bonnard G, Meyer Y (2005) AtNTRB is the major mitochondrial thioredoxin reductase in Arabidopsis thaliana. FEBS Lett 579 337–342 [DOI] [PubMed] [Google Scholar]
- Serrato AJ, Cejudo FJ (2003) Type-h thioredoxins accumulate in the nucleus of developing wheat seed tissues suffering oxidative stress. Planta 217 392–399 [DOI] [PubMed] [Google Scholar]
- Serrato AJ, Crespo JL, Florencio FJ, Cejudo FJ (2001) Characterization of two thioredoxins h with predominant localization in the nucleus of aleurone and scutellum cells of germinating wheat seeds. Plant Mol Biol 46 361–371 [DOI] [PubMed] [Google Scholar]
- Serrato AJ, Pérez-Ruiz JM, Cejudo FJ (2002) Cloning of thioredoxin h reductase and characterization of the thioredoxin reductase-thioredoxin h system from wheat. Biochem J 367 491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrato AJ, Pérez-Ruiz JM, Spínola MC, Cejudo FJ (2004) A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J Biol Chem 279 43821–43827 [DOI] [PubMed] [Google Scholar]
- Setterdahl AT, Chivers PT, Hirasawa M, Lemaire SD, Keryer E, Miginiac-Maslow M, Kim SK, Mason J, Jacquot JP, Longbine CC, et al (2003) Effect of pH on the oxidation-reduction properties of thioredoxins. Biochemistry 42 14877–14884 [DOI] [PubMed] [Google Scholar]
- Tamura T, Stadtman TC (1996) A new selenoprotein from human lung adenocarcinoma cells: purification, properties, and thioredoxin reductase activity. Proc Natl Acad Sci USA 93 1006–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe GH, Kricka LJ (1986) Enhanced chemiluminescent reactions catalyzed by horseradish peroxidase. Methods Enzymol 133 331–353 [DOI] [PubMed] [Google Scholar]
- Vensel WH, Tanaka CK, Cai N, Wong JH, Buchanan BB, Hurkman WJ (2005) Developmental changes in the metabolic protein profiles of wheat endosperm. Proteomics 5 1594–1611 [DOI] [PubMed] [Google Scholar]
- Wong JH, Cai N, Tanaka CK, Vensel WH, Hurkman WJ, Buchanan BB (2004) Thioredoxin reduction alters the solubility of proteins of wheat starchy endosperm: an early event in cereal germination. Plant Cell Physiol 45 407–415 [DOI] [PubMed] [Google Scholar]
- Wong JH, Kim YB, Ren PH, Cai N, Cho MJ, Hedden P, Lemaux PG, Buchanan BB (2002) Transgenic barley grain overexpressing thioredoxin shows evidence that the starchy endosperm communicates with the embryo and the aleurone. Proc Natl Acad Sci USA 99 16325–16330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D, Motohashi K, Kasama T, Hara Y, Hisabori T (2004) Target proteins of the cytosolic thioredoxins in Arabidopsis thaliana. Plant Cell Physiol 45 18–27 [DOI] [PubMed] [Google Scholar]
- You L, Yin J (2000) Patterns of regulation from mRNA and protein time series. Metab Eng 2 210–217 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.