Abstract
The Arabidopsis (Arabidopsis thaliana) compact inflorescence (cif) genotype causes altered adult vegetative development and a reduction in elongation of inflorescence internodes resulting in formation of floral clusters. The cif trait requires both a recessive mutation, cif1, and the activity of a naturally occurring dominant allele of an unlinked gene, CIF2D. We show here that the pseudoverticillata mutation is allelic with cif1 and that the product of the CIF1 gene is ACA10, a member of the large family of P-type Ca2+-ATPases found in higher plants. T-DNA insertion mutations in ACA10, but not in the two other Arabidopsis plasma membrane Ca2+-ATPase-encoding genes, ACA8 and ACA9, cause a cif phenotype when combined with the dominant CIF2D modifier allele. Therefore, ACA10 has a unique function in regulating adult phase growth and inflorescence development. The wild-type ACA8 and ACA10 mRNAs are present at similar levels, and the two promoter-β-glucuronidase fusion transgenes show very similar expression patterns. Moreover, transformation of the cif mutant with an extra copy of the ACA8 gene, which causes overexpression of the ACA8 transcript, can complement the cif phenotype. This suggests that these two Ca2+ pump genes have distinct but related activities and that their differential functions can be altered by relatively small changes in their patterns or levels of expression. The correspondence between cif1 and mutations in ACA10 establishes a genetic link between calcium transport, vegetative phase change, and inflorescence architecture.
Regulatory pathways that control cell growth and division in response to developmental transitions are critical to producing the often conspicuous structural changes that occur as plants progress through their life cycles. Development of the plant shoot results from activity of the shoot apical meristem (SAM), a highly organized group of dividing and differentiating cells at the shoot tip, and occurs by reiterative formation of phytomers, which consist of a node bearing a lateral organ, such as a leaf, a stem segment, or internode, and one or more axillary meristems. The activities of meristems, the determinacy or indeterminacy of those groups of cells, and the patterning and expansion growth of individual organs determine plant architectures (Sussex and Kerk, 2001; McSteen and Leyser, 2005). Internode elongation is often an important developmentally controlled component of plant architecture. For example, vegetative growth in Arabidopsis (Arabidopsis thaliana), which is characterized by production of large leaves and repressed axillary meristem development, is accompanied by little or no internode elongation and results in formation of a compact rosette. At the initiation of flowering, conversion of the SAM to an inflorescence meristem derepresses axillary meristem outgrowth and activates extensive internode elongation, resulting in an aerial branched inflorescence architecture. Variation in the number and pattern of plant inflorescence branches and in the elongation of inflorescence internodes produces much of the diversity of inflorescence morphologies observed in flowering plants (Weberling, 1989).
Development of the postembryonic plant shoot occurs in three phases: juvenile vegetative, adult vegetative, and reproductive (Poethig, 2003). Juvenile and adult vegetative identities are characterized by the absence and presence, respectively, of competence to initiate reproductive phase and, in Arabidopsis, by distinctive rosette leaf morphologies (Telfer et al., 1997). Reproductive phase is characterized by production of flowers and formation of a branched elongated inflorescence. Several Arabidopsis genes have been implicated in control of internode growth, based largely upon their effects on elongation of the inflorescence. Loss-of-function mutations in the ERECTA (ER) gene result in reduced inflorescence internode elongation and shortened aerial organs, including flowers, pedicels, and siliques (Torii et al., 1996). ER encodes an Leu-rich repeat-receptor-like kinase that is highly expressed in the SAM and in developing lateral organs (Yokoyama et al., 1998). BREVIPEDICELLUS and PENNYWISE encode, respectively, KNOTTED1-like (KNOX) and BEL1-like (BELL) homeobox proteins that physically interact with each other, show a synergistic effect on inflorescence internode patterning and elongation, and are expressed in a discrete domain of the inflorescence meristem (Douglas et al., 2002; Venglat et al., 2002; Smith and Hake, 2003; Bhatt et al., 2004; Kanrar et al., 2006). ACAULIS5, one of two Arabidopsis spermine synthase genes, is highly expressed in stem internodes, and loss-of-function mutations in this gene result in dramatically shortened inflorescence internodes and premature arrest of the inflorescence meristem (Hanzawa et al., 1997, 2000). The extent to which these different genes and gene products interact with each other in regulating stem elongation growth has not been extensively assessed, although a wild-type ER gene partially suppresses the bp phenotype (Douglas et al., 2002).
The Arabidopsis compact inflorescence (cif) mutant shows a severe lack of elongation of inflorescence internodes, resulting in the formation of tightly bunched clusters of flowers either at the ends of very short inflorescence shoots or within the center of the rosette (Goosey and Sharrock, 2001). The mutant phenotype includes slight reductions in elongation of pedicels and expansion of cauline leaves, but floral development is normal. Under most growth conditions, vegetative development of cif is indistinguishable from the wild type. However, when grown under long days of fluorescent light, the adult vegetative leaves, but not the juvenile leaves, of the cif rosette show markedly reduced expansion. This suggests that regulatory pathways involving the CIF genes are activated at the juvenile to adult vegetative phase change and function to control cell division and expansion from that phase transition up to the point of floral meristem development. The cif phenotype is inherited as a two-gene trait requiring homozygosity for a recessive allele, cif1, and either heterozygosity or homozygosity for a naturally occurring dominant modifier gene, CIF2D (Goosey and Sharrock, 2001). We describe here the identification of the CIF1 gene as ACA10, a gene encoding a P-type IIB Ca2+-ATPase. This finding suggests that regulation of internode elongation in response to phase change is mediated at least in part via calcium signaling and that this requires the activity of a specific member of the Ca2+-ATPase family.
Calcium is a ubiquitous second messenger in eukaryotic signal transduction cascades. In plants, Ca2+-binding proteins in the cytoplasm and nucleus, such as calmodulin, calcineurin B-like proteins, and Ca2+-dependent protein kinases (CDPKs), act as sensors and transduce specific calcium signatures into downstream effects. Intracellular Ca2+ levels have been shown to be modulated in response to hormones, light, mechanical disturbances, abiotic stress, and pathogen elicitors (White and Broadley, 2003). Growing evidence indicates that Ca2+ signals can be propagated as waves of Ca2+ release and uptake and that specific information is encoded in the amplitude and frequency of these oscillations within the cytosol (Evans et al., 2001; Sanders et al., 2002). Cytosolic Ca2+ homeostasis and the characteristics of perturbations, such as Ca2+ spikes or oscillations, are controlled by both influx pathways, principally involving the activities of Ca2+ channels, and efflux pathways in which Ca2+-ATPase pumps and Ca2+ exchangers remove cytosolic Ca2+ to storage sites within and outside the cell (Sanders et al., 2002; Berridge et al., 2003; Hetherington and Brownlee, 2004). The relative importance of the high-affinity Ca2+ pumps and the lower affinity H+/Ca2+ exchangers in merely maintaining intracellular Ca2+ homeostasis versus actively influencing the spatial and temporal aspects of Ca2+ perturbations is not known.
Fourteen genes encode P-type Ca2+-ATPases in Arabidopsis (Baxter et al., 2003). Four of these encode type IIA or ER-type Ca2+-ATPase (ECA) pumps and 10 encode type IIB or autoinhibited Ca2+-ATPase (ACA) pumps. The ACA ATPases contain N-terminal calmodulin-binding domains and can be activated through binding of Ca2+/calmodulin or inhibited, at least in some cases, through phosphorylation by CDPKs (Harper et al., 1998; Hwang et al., 2000b). The ACA proteins cluster into four related groups based upon sequence similarity (Baxter et al., 2003), and members of these clusters localize to different cellular membranes (Huang et al., 1993; Harper et al., 1998; Bonza et al., 2000; Geisler et al., 2000). ACA8 and ACA9 have been shown to localize predominantly to the plasma membrane (Bonza et al., 2000; Schiott et al., 2004). The function of only one of the P-type Ca2+-ATPases is known; mutations in the ACA9 gene cause defects in pollen tube growth and partial male sterility (Schiott et al., 2004). Here, we present evidence that the recessive cif1 mutations are lesions in the ACA10 gene. In addition, we show that mutations in the highly related ACA8 and ACA9 genes do not cause cif phenotypes when combined with the dominant CIF2D modifier allele. This demonstrates a unique function for the ACA10 Ca2+ pump in regulating plant elongation growth during the adult vegetative growth phase and in determining key aspects of inflorescence structure.
RESULTS
The cif1 Mutation Is Associated with a Region of Reduced Recombination
The cif mutant, although not T-DNA tagged, was identified among the progeny of a T-DNA transformation. The cif phenotype results from a recessive mutation, cif1, and a dominant modifier gene, CIF2D, the required allele of which is present in the Nossen (No-0; CIF2D:No-0) but not the Columbia (Col; CIF2Col) genetic background. In preliminary mapping experiments, the CIF1 and CIF2 genes were localized to the two ends of chromosome 1 (Goosey and Sharrock, 2001). However, further mapping showed that the recessive locus, cif1, is linked to a region on chromosome 1 that shows strongly suppressed recombination (L. George and R.A. Sharrock, unpublished data), suggesting that the cif1 mutation is associated with a chromosomal rearrangement. Consistent with this, cif1/CIF1 heterozygous plants show approximately 50% reduced pollen viability (Supplemental Fig. S1).
pseudoverticillata Is an Allele of cif1 and Requires the Activity of a CIF2D Modifier Allele
The Arabidopsis pseudoverticillata (psv) mutant, which was isolated in a screen for methyl nitrosourea-induced morphological mutants in the ecotype S96, was described as having a clustered inflorescence (Relichova, 1976). Figure 1A shows that the cif and psv mutants produce very similar floral cluster phenotypes. Progeny of a No-0 cif × S96 psv cross show no complementation of the mutant phenotype in either the F1 or the F2 generation. In addition, F2 progeny of a S96 psv × Col wild type cross segregate the psv trait in a 3:16 ratio (93:520; P = 0.3), and recombinant frequency mapping among these F2 progeny shows that a dominant gene (CIF2D:S96), linked to the same chromosome 1 markers as the dominant CIF2D:No-0 gene, is required for expression of the psv trait. Therefore, the cif and psv mutants represent independent recessive mutations in the same CIF1 gene that require the activity of naturally occurring dominant alleles of the same unlinked CIF2 modifier gene. The mutant plant lines and the recessive alleles corresponding to the cif and psv mutant phenotypes have been designated cif1-1 and cif1-2, respectively, and the dominant alleles CIF2D:No-0 and CIF2D:S96. It is possible that CIF2D:No-0 and CIF2D:S96 are the same naturally occurring CIF2 gene allele.
Figure 1.
Inflorescence and rosette phenotypes of the cif1-1 and cif1-2 (psv) mutants and transgenically complemented cif1-1 plants. Plants were grown under a 16-h photoperiod of fluorescent light. A, The cif-1 and psv/cif1-2 mutants exhibit similar floral clustering phenotypes. B and C, Transformation of the cif1-1 mutant with an ACA10 transgene complements the inflorescence internode elongation phenotype and the adult vegetative leaf expansion phenotype.
The cif Mutants Contain Mutations in the ACA10 Ca2+-ATPase Gene
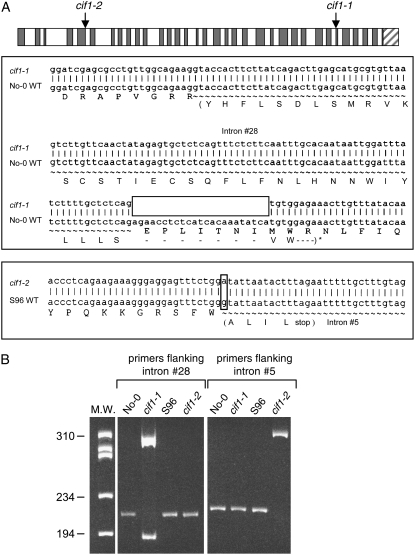
F2 progeny of the S96 cif1-2 × Col wild type cross were analyzed for segregation of the recessive cif1 and dominant CIF2D alleles as described previously for the No-0 cif1-1 × Col wild type cross (Goosey and Sharrock, 2001). Unlike the recessive cif1-1 mutation, which showed linkage to markers at the bottom of chromosome 1, the cif1-2 mutation showed linkage to markers on the top arm of chromosome 4. Subsequent reanalysis of the original No-0 cif1-1 × Col mapping population using these chromosome 4 markers showed tight linkage of these chromosome 4 sequences to both the cif phenotype and also to chromosome 1 molecular markers. These anomalous linkage relationships were not observed in crosses of No-0 wild type to Col or ecotype Landsberg erecta (Ler) wild type. These data are consistent with the interpretation that the original recessive cif1-1 mutation is associated with a translocation between chromosomes 1 and 4 and that the wild-type CIF1 gene is in fact located on the top arm of chromosome 4. A parental mapping line homozygous for the dominant CIF2D:S96 allele and heterozygous for the recessive cif1-2 allele was constructed, and analysis of 1,000 recombinant F2 chromosomes delimited the cif1-2 allele to an 80-kb region of chromosome 4. Candidate genes from the 80-kb interval were sequenced and mutations in the ACA10 Ca2+-ATPase gene were identified in both the No-0 cif1-1 and S96 cif1-2 mutants. The ACA10 gene contains 33 introns, and, as shown in Figure 2A, the cif1-1 mutant was found to have a 23-bp deletion in the exon sequence immediately following intron number 28. No evidence of a translocation breakpoint in this region was observed, indicating that the 23-bp deletion and the translocation are distinct but genetically linked mutations. A single nucleotide G-to-A transition in the first base of intron number 5 of ACA10 was identified in cif1-2 (Fig. 2A).
Figure 2.
Molecular characterization of the cif1-1 and cif1-2 mutations in the ACA10 gene. A, The locations of the cif1-1 and cif1-2 mutations are illustrated on the ACA10 gene, which contains 34 coding sequence exons and 33 introns. The cif1-1 mutation is a 23-bp deletion immediately adjacent to the intron number 28 3′ boundary. The cif1-2 mutation is an A-to-G transition of the first nucleotide of intron number 5. B, Total RNA from cif1-1 and cif1-2 mutant seedlings was subjected to RT-PCR analysis of the ACA10 transcript using primers that flank the cif1-1 mutation and amplify from exon 28 to exon 30, or primers that flank the cif1-2 (psv) mutation and amplify from exon 5 to exon 6 (see “Materials and Methods”).
The ACA10 mutations in the cif1-1 and cif1-2 mutants are both located at exon/intron boundaries. To test their effects on processing of the ACA10 pre-mRNA, cDNA samples synthesized from No-0 and S96 wild-type RNA and from cif1-1 and cif1-2 RNA were used as templates in reverse transcription (RT)-PCR. Figure 2B shows that primers flanking the region of the ACA10 gene from exon number 28 to exon number 30 amplify the expected 214-bp product from wild-type or cif1-2 cDNA, corresponding to correctly processed ACA10 mRNA, but they amplify two bands of altered size from cif1-1 cDNA. Sequencing of the upper cif1-1 RT-PCR band shows that it contains the 23-bp deletion and retains the 110-bp intron number 28, producing an insertion/deletion derivative of the ACA10 protein that regains the correct reading frame at Trp-945 (Fig. 2A). Sequencing of the lower cif1-1 band shows that intron number 28 is correctly processed from this fraction of the cif1-1 mRNA, but this mRNA contains the 23-bp deletion, which alters the reading frame so that a stop codon is encountered after the inclusion of 32 incorrect residues. Hence, the cif1-1 allele results in production of some level of both a C-terminally truncated ACA10 protein and an insertion/deletion variant of the protein. Figure 2B shows that primers flanking ACA10 intron number 5, which contains the point mutation found in cif1-2 (psv), amplify the expected 217-bp product from wild-type or cif1-1 cDNA, which corresponds to processed ACA10 mRNA lacking the intron, but they amplify a 324-bp product from cif1-2 cDNA. This band was sequenced and found to retain intron number 5 with the G-to-A transition as the first base, generating an in-frame stop codon five codons into the intron (Fig. 2A). Therefore, the cif1-2 mutation is likely an aca10 null allele.
To confirm the identity of ACA10 as the CIF1 gene, a 9.3-kb DNA fragment containing the wild-type allele of ACA10 was transformed into cif1-1 plants. Figure 1, B and C, shows that cif1-1 plants transformed with the ACA10 transgene have a wild-type phenotype, with normal development of adult vegetative organs, including adult leaves of the rosette and inflorescence internodes.
T-DNA Mutations in ACA10, But Not ACA8 or ACA9, Cause a cif Phenotype When Combined with the CIF2D Modifier Allele
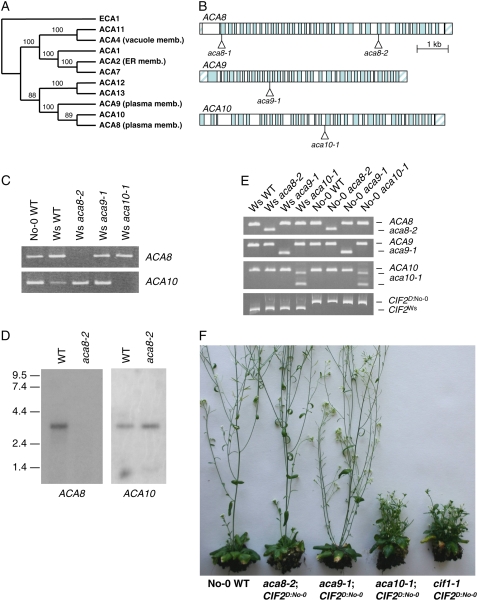
The ACA8, ACA9, and ACA10 Ca2+-ATPases are approximately 70% identical in amino acid sequence and comprise a discrete subfamily within the family of 10 Arabidopsis ACA proteins (Fig. 3A). ACA8 and ACA9 have previously been shown to localize to the plasma membrane (Bonza et al., 2000; Schiott et al., 2004). T-DNA insertion mutations in the ACA8, ACA9, and ACA10 genes were identified in Arabidopsis ecotype Wassilewskija (Ws; Fig. 3B). The location and molecular properties of the aca9-1 allele have been described previously (Schiott et al., 2004). The aca10-1 T-DNA is inserted in intron number 17 of the ACA10 gene, with one border at 4,041 bp and the other at 4,067 bp downstream of the start codon. The aca9-1 and aca10-1 T-DNA alleles result in lack of expression of their respective transcripts downstream of the insertion sites (Schiott et al., 2004; Fig. 3C). Two aca8 T-DNA alleles were analyzed: aca8-1, which is inserted 35 bp downstream of the ATG in the first coding sequence exon, and aca8-2, which is inserted in the 25th coding sequence exon with borders at 5,101 bp and 5,112 bp. In RT-PCR analysis of RNA isolated from the aca8-1 mutant line, low levels of ACA8 transcript sequences from downstream of the insertion site were detected (L. George and R.A. Sharrock, unpublished data). This indicates that transcription of the 3′ end of the ACA8 coding sequence, beginning downstream of nucleotide +35, may occur in this line, perhaps initiated from an internal T-DNA promoter. The aca8-2 mutation blocks transcription of mRNA sequences downstream of the T-DNA insertion site (Fig. 3C), and RNA-blot analysis confirms the absence of full-length ACA8 transcript in this line (Fig. 3D). In the experiments described below, both the aca8-1 and aca8-2 mutations were analyzed and gave the same results, but only data for the presumptive null aca8-2 are presented. Table I and Supplemental Figure S2 show that, in the Ws genetic background, the aca8, aca9, and aca10 T-DNA mutants all lack an adult phase leaf expansion phenotype, flower at the same time as the wild type, and have raceme inflorescences very similar to Ws wild-type plants.
Figure 3.
Phenotypic effects of aca8, aca9, and aca10 T-DNA insertion mutations when combined with the CIF2D:No-0 modifier gene. A, Phylogenetic analysis of the 10 Arabidopsis ACA Ca2+-ATPase protein sequences, with the ECA1 Ca2+-ATPase protein as outgroup, produces a single most parsimonious tree. Bootstrap values from 2,000 replications are shown. B, The locations of T-DNA insertions in the transcribed regions of the ACA8, ACA9, and ACA10 genes are indicated. Exons are shown as shaded boxes and introns as unshaded boxes. C, The aca8-2 and aca10-1 mutations block transcription downstream of the insertion sites. Total RNA was prepared from seedlings of the original Ws aca8-2, aca9-1, and aca10-1 T-DNA lines, and RT-PCR was performed with primers that amplify sequences in the 3′ ends of the ACA8 and ACA10 transcripts. D, Blot analysis of poly(A)-selected RNA from wild type and the aca8-2 mutant probed with gene-specific fragments from the 5′ coding regions of ACA8 and ACA10. E, Genotypes at the T-DNA insertion loci and at the CIF2 locus were determined for the original lines in the Ws genetic (CIF2Ws) background and for representative BCII F3 lines following introgression of the T-DNA insertions into the No-0 genetic background (CIF2D:No-0). DNA of the indicated lines was analyzed by PCR to test for homozygosity for the respective insertion mutation and for the allele state at the CIF2 modifier gene. F, Representative BCII F3 plants of the aca8-2, aca9-1, and aca10-1 lines containing the CIF2D:No-0 allele grown under a 16-h photoperiod of fluorescent light.
Table I.
Inflorescence phenotypes of aca T-DNA mutants and transgenic lines
Values given are averages of measurements from 6 to 10 individual plants. Values in parentheses are ses of the means. Plants were grown under a 16-h photoperiod of fluorescent light. Internode lengths, total inflorescence lengths, and number of secondary inflorescences were measured on day 45 for the Ws lines and on day 50 for the No-0 lines. The No-0 aca8-2, aca9-1, and aca10-1 lines were BCII F3 plants derived from representative homozygous BCII F2 parents. The cif1-1 lines containing chimeric transgenes were T3 plants derived from representative homozygous T2 parents. *, P < 0.05; **, P < 0.0001 (Student's t test).
| Line | Days to Flower | Average Length of First Six Internodes | Total Length of Inflorescence | No. of 2° Rosette Inflorescences |
|---|---|---|---|---|
| mm | mm | |||
| Ws wild type | 17.6 (0.4) | 22.9 (3.4) | 364 (9) | 4.5 (0.3) |
| Ws aca8-2 | 17.6 (0.9) | 20.9 (2.6) | 339 (15) | 4.2 (0.4) |
| Ws aca9-1 | 17.3 (0.7) | 24.0 (2.8) | 449 (8)* | 4.2 (0.6) |
| Ws aca10-1 | 16.8 (0.8) | 25.4 (3.5) | 361 (7) | 4.0 (0.5) |
| Ws aca8-2 aca10-1 | 19.0 (0.4)* | 21.3 (1.7) | 275 (8)** | 4.4 (0.4) |
| No-0 wild type | 26.6 (1.0) | 23.3 (2.4) | 383 (12) | 4 (0.3) |
| No-0 cif1-1 | 27.1 (0.8) | 0.94 (0.32)** | 7.5 (1.6)** | 11.5 (1.1)** |
| No-0 aca8-2 | 26.6 (1.2) | 22.6 (2.2) | 382 (9) | 5.6 (0.4)* |
| No-0 aca9-1 | 24.5 (0.8) | 21.8 (2.2) | 412 (13) | 5.2 (0.5) |
| No-0 aca10-1 | 25.3 (0.6) | 4.0 (1.0)** | 29 (7)** | 12.1 (1.1)** |
| No-0 wild type | 23.7 (0.4) | 22.8 (2.4) | 315 (16) | 5 (0.3) |
| No-0 cif1-1 | 24.4 (0.4) | 1.6 (0.6)** | 16 (1.3)** | 14.7 (1.5)** |
| cif1-1 (P10-10) | 24.9 (0.4) | 22.0 (1.8) | 340 (17) | 3.5 (0.4)* |
| cif1-1 (P10-8) | 24.2 (0.5) | 24.2 (2.7) | 398 (41) | 4.8 (0.3) |
| cif1-1 (P8-10) | 25.7 (0.4)* | 21.1 (2.1) | 363 (30) | 3.7 (0.7) |
| cif1-1 (P8-8) | 24.2 (0.5) | 23.1 (2.4) | 342 (13) | 4.7 (0.3) |
The cif1-1 and cif1-2 mutations are reduction-of-function or loss-of-function aca10 alleles, so the observation that aca10 T-DNA mutations do not cause a cif phenotype in the Ws background indicates that, like CIF2Col and CIF2Ler, CIF2Ws does not function as a dominant modifier gene similar to CIF2D:No-0 and CIF2D:S96. It is predicted, therefore, that combining an aca10 T-DNA mutation with CIF2D:No-0 should result in a cif phenotype. To confirm this and to test whether loss of function of either ACA8 or ACA9 causes cif-like characteristics in combination with the CIF2D:No-0 modifier gene, Ws lines homozygous for the aca8-1, aca8-2, aca9-1, and aca10-1 T-DNA alleles were crossed to No-0 wild type and then backcrossed twice to No-0 to introgress the T-DNA insertion mutations into the No-0 genetic background. BCII F1 progeny, heterozygous for the T-DNA allele and homozygous for CIF2D:No-0, were selfed, and the BCII F2 progeny were scored for their T-DNA insertion genotype and their inflorescence phenotypes. All aca10-1/aca10-1 CIF2D:No-0/CIF2D:No-0 F2 homozygotes derived from these crosses exhibited a cif phenotype, including reduced elongation of adult leaves and inflorescence internodes and reduced apical dominance. In contrast, aca8-1/aca8-1 CIF2D:No-0/CIF2D:No-0, aca8-2/aca8-2 CIF2D:No-0/CIF2D:No-0, and aca9-1/aca9-1 CIF2D:No-0/CIF2D:No-0 F2 segregants showed wild-type adult vegetative and inflorescence development phenotypes. Figure 3, E and F, and Table I present the analysis of representative F3 progeny from these crosses. These results demonstrate that, among the mutations in these three highly related Ca2+ pump genes, only aca10 loss-of-function alleles interact with the dominant CIF2D modifier allele to produce a cif phenotype. As expected from previous studies (Schiott et al., 2004), plants homozygous for the aca9-1 mutation in the No-0 genetic background show reduced fertility.
The Expression Patterns of the ACA10 and ACA8 Genes Are Similar and Are Not Regulated by Growth Phase or the Allele State at CIF2
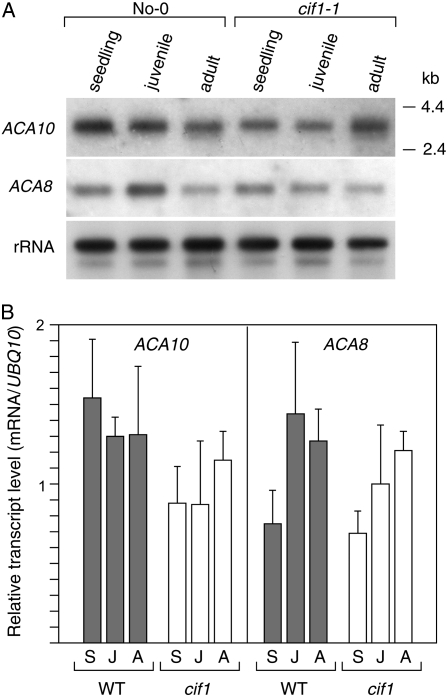
Onset of the cif mutant phenotype correlates closely with the juvenile to adult vegetative phase transition (Goosey and Sharrock, 2001). To determine whether this phase change and the onset of the cif phenotype also correlate with alteration in the expression of the ACA10 gene, northern-blot and quantitative RT-PCR (qRT-PCR) analyses were performed on RNA extracted from seedlings and from isolated juvenile and adult leaves of No-0 wild-type and cif1-1 plants. Figure 4, A and B, shows that the ACA10 mRNA is present at levels varying less than 2-fold at these stages of vegetative development in both wild-type and mutant plants. Schiott et al. (2004) showed that the ACA9 transcript is present almost exclusively in pollen and, consistent with this, no signal for the ACA9 mRNA was detected on RNA blots of vegetative tissues (L. George and R.A. Sharrock, unpublished data). However, northern-blot and qRT-PCR analyses show that, like the ACA10 mRNA, the ACA8 mRNA is expressed evenly over the course of seedling, juvenile, and adult vegetative development and at a level very similar to that of the ACA10 transcript (Fig. 4, A and B).
Figure 4.
RNA-blot and qRT-PCR analysis of the developmental regulation of the ACA10 and ACA8 mRNAs. A, Poly(A)-selected RNA was prepared from seedlings, juvenile rosette leaves, and adult rosette leaves of No-0 wild-type and cif1-1 plants. Blots were probed with gene-specific probes from the 5′ nonconserved coding regions of the ACA8 and ACA10 mRNAs. B, Quantitative RT-PCR analysis was performed on total RNA prepared from the same tissue samples as in part A, above: seedlings (S), juvenile (J), and adult (A) leaves. Error bars represent sds from the mean values for replicate assays.
To further assess the expression patterns of the ACA10 and ACA8 genes and the possible effect of the CIF2 gene or the cif1 genotype on those patterns, the promoter regions from these genes were transcriptionally fused to the GUS coding sequence and introduced into No-0 wild-type (CIF2D:No-0), Col wild-type (CIF2Col), and cif1-1 mutant plants. Figure 5, A and B, shows that, in No-0 (ACA10:GUS) and No-0 (ACA8:GUS) lines, both promoters are active throughout roots, leaves, and stems of seedlings and rosette-stage plants, including cotyledons and juvenile and adult rosette leaves. The two promoters also exhibit very similar expression patterns in inflorescences, with strong expression in developing inflorescence internodes, cauline leaves, and flowers (Fig. 5, C–F). Surprisingly, as the inflorescence internodes expand, they exhibit reduced GUS staining, as shown for the ACA10:GUS plants in Figure 5, C to E. This is also true for ACA8:GUS lines (Fig. 5F). This may reflect dilution of the GUS stain in rapidly expanding cells, as initial RT-PCR analyses of the ACA10 mRNA in dissected inflorescence stems did not show a strong down-regulation of transcript level in those regions (L. George and R.A. Sharrock, unpublished data). The ACA10:GUS and ACA8:GUS genes were also introduced into Col wild-type and cif1-1 host plants, and staining patterns very similar to those of the No-0 lines were observed throughout seedling, vegetative, and reproductive growth (L. George and R.A. Sharrock, unpublished data). Therefore, neither the allele state at the CIF2 gene nor the presence or absence of an aca10 mutation has a strong effect on these ACA promoter expression patterns. In the cif1-1(ACA10:GUS) and cif1-1(ACA8:GUS) lines, the one or two elongated inflorescence internodes subtending the floral clusters show low GUS staining, like the elongated internodes of the wild-type lines. However, in the floral clusters, although individual internodes are not distinguishable, the point from which pedicels originate in those clusters stains darkly for GUS activity (Fig. 5G), consistent with relatively strong expression of these genes in unexpanded inflorescence internodes. The expression patterns determined here for the ACA10 and ACA8 genes are in agreement with those obtained from microarray data sets presented in Genevestigator (http://www.genevestigator.ethz.ch).
Figure 5.
Histochemical analysis of ACA10:GUS and ACA8:GUS expression patterns. ACA10:GUS and ACA8:GUS transgenes were introduced into No-0 and Col wild-type plants and into the cif1-1 mutant. A, Leaf, stem, and root tissues of seedlings (inset) and 28-d-old rosette stage plants of No-0 (ACA10:GUS) stain for GUS activity. Leaves with clear juvenile identities in the stained rosettes are labeled with a star and leaves with clear adult identities are labeled with a dot. B, No-0 (ACA8:GUS) seedling and rosette expression patterns, as in part A. C to E, Progressive pattern of GUS expression in the developing and elongating primary inflorescence of No-0 (ACA10:GUS) plants. F, The primary inflorescence of a No-0 (ACA8:GUS) plant shows a similar staining pattern to that of ACA10:GUS plants. G, The unexpanded internodes of a cif1-1(ACA10:GUS) floral cluster stain strongly for GUS activity.
Overexpression of ACA8 Can Complement the cif1 Mutation
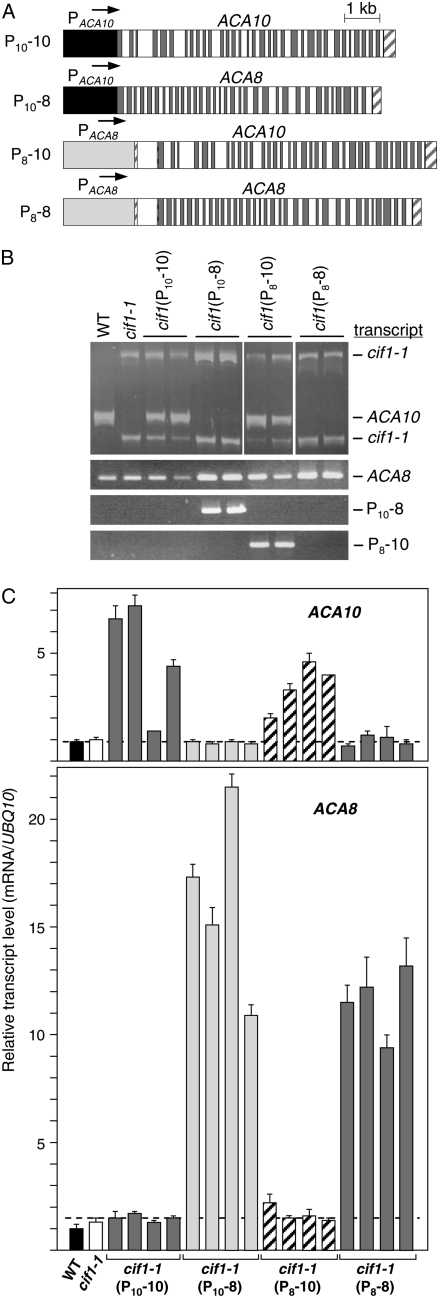
Although the ACA8 and ACA10 mRNAs encode related proteins and are expressed in very similar patterns, only aca10 mutations cause the cif phenotype when combined with the CIF2D:No-0 modifier gene (Fig. 3D). This suggests either that the two Ca2+ pumps have diverged in function at the protein level or that there are aspects of the regulation of the genes, not apparent in RNA quantification or GUS fusion analyses, that underlie their functions. To determine whether the differential functions of ACA8 and ACA10 can be specifically attributed to their transcription regulatory regions or to their protein coding regions, transgenes were constructed in which the genomic coding sequences for the ACA8 and ACA10 proteins were fused to the ACA8 and ACA10 promoters at their ATG start codons (Fig. 6A). In all cases, the same 5′ upstream promoter sequences were used to make these constructs as the ACA10:GUS and ACA8:GUS fusion genes analyzed in Figure 5 (see “Materials and Methods”). The P10-10, P10-8, P8-10, and P8-8 genes were introduced into cif1-1 host plants, and multiple independent transgenic lines that segregated as single insertion events in the T2 generation were isolated for each construct. Figure 6B shows that T3 lines that are homozygous for the various transgene inserts express the transgene-encoded mRNAs and are not strongly altered in expression of the endogenous ACA10 and ACA8 genes. Table I gives flowering time and inflorescence morphology data for representative homozygous T3 lines containing each construct. As expected, the P10-10 gene complements the cif1 phenotype in the T1 and subsequent T2 and T3 generations (Table I). The P10-8 transgene also very efficiently complements the cif trait, suggesting that, with regard to their capacities to transgenically regulate the cif trait, the ACA8 and ACA10 proteins are interchangeable. This is similar to the observed ability of a P9-8 transgene to complement the aca9-reduced fertility phenotype (Schiott et al., 2004) and confirms that all three of these genes likely encode plasma membrane-localized pumps with similar Ca2+-transporting activities.
Figure 6.
Gene expression analysis of cif1-1 mutant lines complemented with ACA8, ACA10, and chimeric transgenes. A, The ACA10 and ACA8 promoter regions were fused to the genomic sequences encoding the ACA10 and ACA8 ATPases at their ATG start codons. These transgenes were introduced into the cif1-1 mutant, and quantitative analysis of the inflorescence phenotypes of representative homozygous T3 lines are given in Table I. B, Samples of cDNA from homozygous T3 transgenic lines were assayed for expression of endogenous and transgene-encoded mRNAs by RT-PCR using primer sets that distinguish the wild-type ACA10 and mutant cif1-1 mRNAs, detect total ACA8 mRNA, or detect only the mRNAs produced from the chimeric genes. C, Quantitative PCR was performed to assess the total level of ACA10 and ACA8 transcript present in RNA from four independent T3 transgenic lines for each construct. Dotted lines show the mean of the values for all lines that do not contain a transgene encoding the mRNA being assayed.
Surprisingly, the P8-10 and P8-8 transgenes were also found to be capable of complementing the cif phenotype in homozygous T3 lines. Initial T1 plants carrying these transgenes were observed to be variable for the phenotype, with some elongated inflorescence stems and some floral clusters, but all cif1-1(P8-8) and cif1-1(P8-10) lines bred to homozygosity in the T2 and T3 generations were strongly complemented (Table I). This indicates that, when incorporated in transgenes, both the ACA10 and ACA8 promoter regions are capable of producing a Ca2+-ATPase expression pattern needed to complement the cif phenotype. To further investigate this, the level of total ACA8 and ACA10 mRNA present in several of each of the complemented T3 transgenic lines was determined by qRT-PCR. Figure 6C shows that the P10-10 and P8-10 lines contain total amounts of ACA10 transcript, including both the endogenous mutant transcript and the transgene product, which is from 1.5- to 7-fold over that of the control lines. The P10-8 and P8-8 lines overexpress the ACA8 mRNA from 6- to 15-fold relative to the endogenous ACA8 gene. The reason that an increased gene dosage of ACA8 causes such a disproportionately large increase in the ACA8 transcript is not known. Nevertheless, it appears that, while the diploid copy of the endogenous ACA8 gene present in nontransgenic cif1 lines cannot compensate for loss of the ACA10 gene, overexpression of ACA8 in the P8-8 transgenic lines is sufficient to do so.
It is possible that the ACA8 and ACA10 ATPases differ in their transport activities, the types of complexes they form at the plasma membrane, or their regulation by Ca2+/calmodulin or other mechanisms. Nevertheless, expression of at least one of these two pumps above a threshold level in the membranes of cells of adult leaves and inflorescence stems appears to be sufficient to mediate correct intracellular Ca2+ dynamics and CIF growth signaling. In a CIF2D genetic background, aca10 mutations presumably result in pump levels falling below this threshold, whereas aca8 mutations do not. This suggests that there may be functional redundancy between the ACA8 and ACA10 genes. One hypothesis for the activity of the CIF2 gene might be that a dominant CIF2D allele down-regulates expression of the ACA8 gene in critical cells, resulting in a situation where loss of ACA10 gene expression triggers the cif phenotype. To test this hypothesis and assess the interaction of these two Ca2+ pump mutations, aca8-2 aca10-1 double T-DNA mutants were constructed in the No-0 (CIF2D) background and in the Ws genetic background, which does not carry a dominant CIF2D allele. The aca8-2 aca10-1 CIF2D line has a cif inflorescence phenotype similar to the aca10-1 CIF2D line (R.A. Sharrock, unpublished data). In contrast, the aca8-2 aca10-1 CIF2Ws line has a raceme inflorescence with a slightly shorter overall length than wild type but clearly elongated internodes (Table I; Supplemental Fig. S2). Therefore, in a genetic background homozygous for a recessive CIF2 allele, there is no requirement for either the ACA10 or ACA8 Ca2+ pump to mediate CIF signaling and functional redundancy of the ACA8 and ACA10 genes likely does not explain their respective activities in regulating inflorescence internode elongation.
DISCUSSION
Inflorescence Structure, Calcium Signaling, and the Control of Plant Phase-Regulated Development
Specification of the structure of the inflorescence begins with changes in the developmental potential and activity of the apical meristem triggered at the onset of reproductive phase but depends critically upon subsequent control of expansion and elongation of newly formed organs and stem tissues. Extensive inflorescence internode elongation, along with the production of secondary inflorescence meristems in the axils of cauline leaves, generates the open aerial reproductive architecture of Arabidopsis. The cif mutant shows a striking reduction in inflorescence internode elongation, due to both lower cell numbers and reduced cell expansion, resulting in a suppressed bolt and closely clustered bracts and flowers (Goosey and Sharrock, 2001). This mutant architecture is functional and fertile and resembles umbel-like architectures, with pedicels originating from a common point, found in other plant species. It is therefore possible that at least some elements of the cif pathway have been targets of selection for diverse inflorescence types during plant evolution. The cif mutant phenotype is dependent upon the interaction of a recessive cif1 mutation and a naturally occurring dominant modifier allele of a second gene, CIF2D. The demonstration here that the CIF1 gene encodes the ACA10 Ca2+ pump suggests that loss of function of a specific Ca2+ efflux pump, presumably leading to changes in the activities of one or more Ca2+-signaling pathways, strongly alters reproductive architecture. How such a Ca2+-signaling mechanism may relate to internode elongation regulatory mechanisms involving the ER kinase (Shpak et al., 2004), KNOX/BELL homeobox proteins (Kanrar et al., 2006), or spermine synthase (Hanzawa et al., 2000) is not known. However, it is notable that the mutations that defined the roles of those proteins in inflorescence development were all isolated in the Ler or the Col genetic background, so no interactions of those pathways with the CIF2 gene are indicated.
Although the most striking aspect of the cif phenotypes is the effect on bolting, under certain light conditions, cif leaves with adult phase identities also show reduced elongation and expansion, while cif leaves with juvenile identities are completely normal (Goosey and Sharrock, 2001). Therefore, the effects of the cif mutant genotype first become apparent at the juvenile to adult phase transition. The molecular mechanism that regulates the timing of the juvenile to adult phase transition in Arabidopsis has been shown to involve the activities of small interfering RNAs and microRNAs (Hunter et al., 2006; Wu and Poethig, 2006), but downstream effectors and pathways that mediate morphological changes and developmental responses that are triggered in phase-specific patterns are less well understood. Identification of CIF1 as the gene encoding the ACA10 Ca2+ pump suggests that intracellular Ca2+ levels may be an important integration point for such pathways. Analysis of ACA10 transcript levels and ACA10:GUS transgene activity shows that the ACA10 gene is expressed in many parts of the plant and is not strongly up- or down-regulated at the juvenile to adult phase transition. Moreover, because different plasma membrane Ca2+-ATPase isoforms are expressed in developmentally regulated patterns in animal cells through alternative splicing (Strehler and Zacharias, 2001; Zylinska et al., 2002), we also tested for possible developmentally controlled alternative splicing of the ACA10 mRNA. RT-PCR analysis of seedling, juvenile leaf, and adult leaf RNA with primers for small sequential fragments covering the entire length of the ACA10 transcript revealed no evidence for this (L. George and R.A. Sharrock, unpublished data). Therefore, alteration in expression of the ACA10 gene itself is not currently implicated in the regulation of leaf expansion at the adult phase transition. More likely, adult and reproductive developmental programs, most strikingly inflorescence internode elongation, involve signaling pathways that trigger Ca2+ movement across the plasma membrane and, in the absence of the ACA10 pump, the cif mutant has abnormal cellular Ca2+ dynamics that specifically affect these programs.
Diversity of Regulatory Function among Plant Ca2+ Pumps
A family of 14 ACA and ECA genes encodes Ca2+ pumps in Arabidopsis, and different ATPase subfamilies are localized to different cellular membranes (Baxter et al., 2003). The biological activity of only one of these pumps has been previously determined. T-DNA mutations in the ACA9 gene were shown to cause defects in pollen cell elongation and, consistent with this, the ACA9 transcript was shown to be predominantly expressed in pollen (Schiott et al., 2004). Based upon sequence similarity and intron position in their respective genes, the ACA8, ACA9, and ACA10 proteins constitute a well-defined subfamily of Ca2+ pumps. We have shown here that, in contrast to aca9 mutants, aca8 and aca10 knockout lines and the cif1-1 and cif1-2 lines are unaffected in fertility. Indeed, in the Ws, Col, or Ler background, no strong effects on the morphology of the plants are seen with any of the aca10 mutations, and it is only in the presence of the CIF2D modifier allele that reduction of function of this pump results in a strong inflorescence development phenotype. The aca8 T-DNA mutants show neither reduced fertility nor an effect on inflorescence structure in the presence or absence of the CIF2D modifier allele. Therefore, the functions of these three highly sequence-related Ca2+ pumps are divergent. Unlike the ACA9 mRNA, the ACA8 and ACA10 mRNAs are quite strongly expressed in most plant organs over the course of plant development. Recently, the transcription of ACA8 and ACA9, but not ACA10, has been shown to be stimulated by abscisic acid (Cerana et al., 2006). The assignment of discrete functions and expression patterns to the ACA8, ACA9, and ACA10 genes suggests that a wide array of regulatory Ca2+-signaling pathways in plants may be dependent upon efflux mechanisms involving specific pumps or exchange proteins.
The ACA8 and ACA9 pumps are plasma membrane localized (Bonza et al., 2000; Schiott et al., 2004) and transport Ca2+ ions to the apoplast. Although plasma membrane targeting has not been directly shown for ACA10, the observation that the aca9 and aca10 mutant phenotypes can be complemented by transgenes containing the ACA8 coding sequence suggests that these three proteins are very similar in intracellular localization and activity. In addition, all three have protein sequences characteristic of autoregulated ATPases that may be activated through binding of Ca2+/calmodulin (Harper et al., 1998; Geisler et al., 2000; Hwang et al., 2000a). We have shown that introduction of a transgene containing any combination of the ACA8 or ACA10 promoters and coding regions can complement the cif phenotype. For unknown reasons, these P10-10, P10-8, P8-10, and P8-8 transgenes drive overexpression of the transgene-encoded mRNAs, severalfold higher than the expected gene dosage effect. An aca8 null mutation does not cause a cif phenotype when combined with the dominant CIF2D:No-0 gene, but overexpression of the ACA8 mRNA is able to complement the cif phenotype. It is likely, therefore, that the unique role for ACA10 in regulating reproductive development reflects either subtle differences in the expression patterns or levels of the normal ACA8 and ACA10 genes, or differences in the properties of the two pumps themselves relating to their transport activities, autoregulation by Ca2+, or abilities to form membrane complexes with other proteins. However, these differences can be overcome or mitigated by overexpression.
An intriguing aspect of the genetics of the cif trait is that the requirement for ACA10 Ca2+ pump activity to mediate adult vegetative growth and inflorescence internode elongation is completely dependent upon the allele state of the unlinked CIF2 gene. Preliminary mapping of CIF2 shows that it does not correspond to any of the ACA Ca2+-ATPase genes (L. George and R.A. Sharrock, unpublished data). At least two alleles of the CIF2 gene are present in different wild accessions of Arabidopsis. The CIF2D allele present in the No-0 and S96 ecotypes acts in a dominant way in conjunction with aca10 mutations to alter inflorescence development, whereas the CIF2 allele found in Col, Ler, and Ws acts in a recessive manner to suppress the effects of aca10 mutations. Apparently, dominant CIF2D:No-0, S96 alleles predispose the plant to requiring a minimum level of efflux of Ca2+ ions by ACA10 in particular cells, whereas recessive CIF2Col, Ler, Ws alleles obviate the need for that efflux. In a genetic background homozygous for the recessive CIF2Ws allele, even inactivation of both the ACA8 and ACA10 genes does not result in a cif phenotype. Therefore, the recessive CIF2 allele effectively disconnects regulation of adult leaf expansion and inflorescence internode elongation from the activities of ACA8 or ACA10. The molecular mechanism for this genetic phenomenon is currently not known, but we have shown that the two different naturally occurring CIF2 alleles do not strongly influence the pattern or level of ACA10 or ACA8 transcription. If the ACA10 and ACA8 pumps are regulated through binding of Ca2+/calmodulin, or through phosphorylation by CDPKs as has been shown for ACA2 (Hwang et al., 2000b), it is possible that CIF2 encodes a component of a transport regulatory mechanism. The striking inflorescence elongation phenotype elicited by mutations in the CIF1(ACA10) gene establishes a link between Ca2+ transport, vegetative phase change, and plant reproductive architecture. Further analysis of the interaction of the CIF1 and CIF2 gene, including identification of CIF2 and characterization of its polymorphic alleles, will advance our understanding of this novel role for Ca2+ ion transport in plant development.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Histochemical Assays
The Arabidopsis (Arabidopsis thaliana) cif mutant has been described (Goosey and Sharrock, 2001). The psv mutant was obtained from the Arabidopsis Biological Resource Center (The Ohio State University). Plants were grown under a 16-h photoperiod of fluorescent light at 21°C unless otherwise indicated. T-DNA insertion mutants in Arabidopsis Ws were identified in the ACA8, ACA9, and ACA10 genes by PCR screening pools of insertion lines from the University of Wisconsin Arabidopsis knockout facility (Krysan et al., 1996). Two aca8 lines (aca8-1 = JHss106, with a kanamycin resistance marker; and aca8-2 = JHss107, with a Basta-resistance marker), one aca10 line (aca10-1 = JHss110, with a kanamycin resistance marker), and the previously described aca9-1 line (Schiott et al., 2004) were used. Plant lines homozygous for the T-DNA insertions were identified by the inability to amplify a wild-type band using primers from the respective genes flanking the inserts. T-DNA border fragments were amplified and sequenced for each insertion to verify the site of insertion. Histochemical GUS assays were performed as described (Goosey and Sharrock, 2001). Pollen staining was performed with malachite green-fuschin-orange G (Alexander, 1969).
Genetic Markers and Mapping
For mapping the cif1-1 and cif1-2 mutations, Ler/Col simple sequence length polymorphism or insertion/deletion DNA markers from the Monsanto Arabidopsis Polymorphism and Ler Sequence Collections (Jander et al., 2002) were screened to identify those that are also polymorphic between Col and the No-0 and/or S96 ecotypes. Supplemental Table S1 contains the sequences of PCR primers used to detect mutations and T-DNA insertions in various mutant lines. The cif1-1 mutation was detected in crosses and transgenic lines by PCR amplification with primers LG593F and LG593R. The aca10-1 T-DNA mutation was followed by PCR using a mixture of primers 346, 347, and 348. The aca9-1 T-DNA mutation was followed by PCR using a mixture of primers 346, 349, and 350. The aca8-2 T-DNA mutation was followed by PCR using a mixture of primers 346, 500, and 501. The CIF2D:No-0 and CIF2D:S96 alleles were distinguished from the CIF2Col and CIF2Ws alleles by assaying for a linked simple sequence length polymorphism marker, located on bacterial artificial chromosome F3C3, which was detected with primers LG231 and LG232.
Construction of ACA8 and ACA10 Transgenes and Plant Transformation
All Agrobacterium transformation plasmids were constructed as derivatives of pBI101.1 and confer kanamycin resistance (Jefferson et al., 1987). A 9.3-kb DNA fragment containing the wild-type allele of ACA10, extending from 1,460 bp upstream of the ATG to 275 bp downstream of the TAA stop codon, was PCR amplified from Col wild-type genomic DNA and cloned to create pBI-ACA10. To construct the P10-10, P10-8, P8-10, and P8-8 transgenes, the ACA8 and ACA10 coding regions, beginning at the start codons and including the introns, and the ACA8 and ACA10 promoter regions (2,700 bp and 1,460 bp) were PCR amplified from Col wild-type genomic DNA with restriction sites inserted at their ATG start codons. The various transgene fusion constructs were assembled and transformed into cif1-1 host plants by the floral dip method (Clough and Bent, 1998). ACA8:GUS and ACA10:GUS transgenes were constructed by inserting the promoter fragments described above in front of the GUS coding sequence in the pBI101.1 vector. These GUS fusion genes were transformed into No-0 wild-type, Col wild-type, and cif1-1 host plants by the floral dip method.
RNA Isolation, RNA Blots, and RT-PCR Analyses
Total RNA for northern-blot analysis was isolated from seven day-old light-grown seedlings, juvenile rosette leaves, and adult rosette leaves of No-0 wild-type and cif1-1 plants as described (Clack et al., 1994). Poly(A)-enriched RNA was selected using an Oligotex Maxi kit (Qiagen). Hybridization probes were derived from nonconserved regions of the 5′ ends of the ACA8 and ACA10 cDNA sequences delimited by primer pairs 384/385 and LG592F/LG592R (Supplemental Table S1). Samples containing 1 μg of poly(A)-selected RNA were separated on agarose-formaldehyde gels, blotted, and hybridized as described (Clack et al., 1994). Total RNA for RT-PCR was isolated using an RNeasy kit (Qiagen). Samples containing 1.1 μg of total RNA were digested with RQ1 DNase I (Promega), and oligo(dT)-primed cDNA was synthesized using Superscript RNase H− reverse transcriptase (Invitrogen). RT-PCR analysis of the cif1-1 and cif1-2 transcripts was performed with primer pairs LG589F/LG589R, which amplify from ACA10 exon 28 to exon 30, and LG588F/LG588R, which amplify from ACA10 exon 5 to exon 6. RT-PCR analysis of ACA10 and ACA8 transcript levels in T-DNA lines was performed using primer pairs LG589F/LG589R and 461/462, respectively. RT-PCR analysis of lines containing chimeric transgenes utilized primer sets that distinguish the wild-type ACA10 and mutant cif1-1 mRNAs (LG589F/LG589R), detect total ACA8 mRNA (395/395), or detect only the mRNAs produced from the chimeric P10-8 (412/413) or P8-10 (579/580) transgene. Real-time qRT-PCR was performed in 25-μL reactions using 2× SYBR Green PCR master mix (Applied Biosystems) and a Rotorgene 3200 thermocycler (Corbett Life Science). Samples of cDNA were synthesized from total RNA as described above. Primer pairs q468/q469, q464/q465, and q551/q552 were used to quantify the ACA8, ACA10, and UBQ10 cDNAs, respectively. The thermal profile was: 95°C for 10 min followed by 40 cycles of 95°C for 20 s, 55°C for 20 s, and 72°C for 20 s. Data were analyzed using Rotor-gene Version 5.0.61 software (Corbett Life Science). A relative calibrator standard curve was generated for each primer pair in each experiment from a control wild-type seedling cDNA sample. The expression level of the ACA8 or ACA10 transcript in each cDNA sample assayed was calculated based on the standard curve for that primer pair and was subsequently normalized to that of the control UBQ10 transcript for that sample. Each experiment was repeated twice with independent biological samples and each sample was represented in triplicate in each experiment.
Phylogenetic Analysis
Protein sequences were aligned using Clustal (Chenna et al., 2003), and a branch and bound parsimony search was performed using PAUP* (Swofford, 2002). Bootstrap analysis implementing the branch and bound search option and a random addition of terminal taxa for each of 2,000 bootstrap replicates was performed.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_125093 (ACA8), NM_113013 (ACA9), and NM_119136 (ACA10).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Viability staining of pollen in the anthers of a cif1/CIF1 heterozygous plant.
Supplemental Figure S2. Inflorescence phenotypes of the aca T-DNA mutants in the Ws (CIF2Ws recessive) background.
Supplemental Table S1. Oligonucleotide primers used in experiments.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center (The Ohio State University) for providing genetic stocks and Mike Faul for technical assistance.
This work was supported by the National Research Initiative of the U.S. Department of Agriculture (grant no. 2004–35304–14933 to R.A.S.), by the Department of Energy (grant no. DE–FG03–94ER20152 to S.M.R. and J.F.H.), and by the National Institutes for Health (grant no. GM070813–01).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Robert A. Sharrock (sharrock@montana.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alexander MP (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44 117–122 [DOI] [PubMed] [Google Scholar]
- Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren MG, Gribskov M, Harper JF, Axelsen KB (2003) Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol 132 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ, Bootman MD, Roderick HL (2003) Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol 4 517–529 [DOI] [PubMed] [Google Scholar]
- Bhatt AM, Etchells JP, Canales C, Lagodienko A, Dickinson H (2004) VAAMANA: a BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 328 103–111 [DOI] [PubMed] [Google Scholar]
- Bonza MC, Morandini P, Luoni L, Geisler M, Palmgren MG, De Michelis MI (2000) At-ACA8 encodes a plasma membrane-localized calcium-ATPase of Arabidopsis with a calmodulin-binding domain at the N terminus. Plant Physiol 123 1495–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerana M, Bonza MC, Harris R, Sanders D, De Michelis MI (2006) Abscisic acid stimulates the expression of two isoforms of plasma membrane Ca2+-ATPase in Arabidopsis thaliana seedlings. Plant Biol (Stuttg) 8 572–578 [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25 413–427 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Douglas SJ, Chuck G, Dengler RE, Pelecanda L, Riggs CD (2002) KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans NH, McAinsh MR, Hetherington AM (2001) Calcium oscillations in higher plants. Curr Opin Plant Biol 4 415–420 [DOI] [PubMed] [Google Scholar]
- Geisler M, Frangne N, Gomes E, Martinoia E, Palmgren MG (2000) The ACA4 gene of Arabidopsis encodes a vacuolar membrane calcium pump that improves salt tolerance in yeast. Plant Physiol 124 1814–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosey L, Sharrock R (2001) The Arabidopsis compact inflorescence genes: phase-specific growth regulation and the determination of inflorescence architecture. Plant J 26 549–559 [DOI] [PubMed] [Google Scholar]
- Hanzawa Y, Takahashi T, Komeda Y (1997) ACL5: an Arabidopsis gene required for internodal elongation after flowering. Plant J 12 863–874 [DOI] [PubMed] [Google Scholar]
- Hanzawa Y, Takahashi T, Michael AJ, Burtin D, Long D, Pineiro M, Coupland G, Komeda Y (2000) ACAULIS5, an Arabidopsis gene required for stem elongation, encodes a spermine synthase. EMBO J 19 4248–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JF, Hong B, Hwang I, Guo HQ, Stoddard R, Huang JF, Palmgren MG, Sze H (1998) A novel calmodulin-regulated Ca2+-ATPase (ACA2) from Arabidopsis with an N-terminal autoinhibitory domain. J Biol Chem 273 1099–1106 [DOI] [PubMed] [Google Scholar]
- Hetherington AM, Brownlee C (2004) The generation of Ca2+ signals in plants. Annu Rev Plant Biol 55 401–427 [DOI] [PubMed] [Google Scholar]
- Huang L, Berkelman T, Franklin AE, Hoffman NE (1993) Characterization of a gene encoding a Ca2+-ATPase-like protein in the plastid envelope. Proc Natl Acad Sci USA 90 10066–10070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutierrez-Nava M, Poethig SR (2006) Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133 2973–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Harper JF, Liang F, Sze H (2000. a) Calmodulin activation of an endoplasmic reticulum-located calcium pump involves an interaction with the N-terminal autoinhibitory domain. Plant Physiol 122 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sze H, Harper JF (2000. b) A calcium-dependent protein kinase can inhibit a calmodulin-stimulated Ca2+ pump (ACA2) located in the endoplasmic reticulum of Arabidopsis. Proc Natl Acad Sci USA 97 6224–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanaugh TA, Bevan MW (1987) GUS fusions: B-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanrar S, Onguka O, Smith HM (2006) Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta 224 1163–1173 [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Tax F, Sussman MR (1996) Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc Natl Acad Sci USA 93 8145–8150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P, Leyser O (2005) Shoot branching. Annu Rev Plant Biol 56 353–374 [DOI] [PubMed] [Google Scholar]
- Poethig RS (2003) Phase change and the regulation of developmental timing in plants. Science 301 334–336 [DOI] [PubMed] [Google Scholar]
- Relichova J (1976) Some new mutants. Arabidopsis Inf Serv 13 25–28 [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signaling. Plant Cell (Suppl) 14 S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiott M, Romanowsky SM, Baekgaard L, Jakobsen MK, Palmgren MG, Harper JF (2004) A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proc Natl Acad Sci USA 101 9502–9507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, Berthiaume CT, Hill EJ, Torii KU (2004) Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131 1491–1501 [DOI] [PubMed] [Google Scholar]
- Smith HM, Hake S (2003) The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehler EE, Zacharias DA (2001) Role of alternative splicing in generating isoform diversity among plasma membrane calcium pumps. Physiol Rev 81 21–50 [DOI] [PubMed] [Google Scholar]
- Sussex IM, Kerk NM (2001) The evolution of plant architecture. Curr Opin Plant Biol 4 33–37 [DOI] [PubMed] [Google Scholar]
- Swofford D (2002) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0 beta 10. Sinauer Associates, Sunderland, MA
- Telfer A, Bollman KM, Poethig RS (1997) Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124 645–654 [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venglat SP, Dumonceaux T, Rozwadowski K, Parnell L, Babic V, Keller W, Martienssen R, Selvaraj G, Datla R (2002) The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc Natl Acad Sci USA 99 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weberling F (1989) Morphology of Flowers and Inflorescences. Cambridge University Press, Cambridge, UK
- White PJ, Broadley MR (2003) Calcium in plants. Ann Bot (Lond) 92 487–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Takahashi T, Kato A, Torii KU, Komeda Y (1998) The Arabidopsis ERECTA gene is expressed in the shoot apical meristem and organ primordia. Plant J 15 301–310 [DOI] [PubMed] [Google Scholar]
- Zylinska L, Kawecka I, Lachowicz L, Szemraj J (2002) The isoform- and location-dependence of the functioning of the plasma membrane calcium pump. Cell Mol Biol Lett 7 1037–1045 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.