Abstract
UV-B signaling is an important but poorly understood aspect of light responsiveness in plants. Arabidopsis (Arabidopsis thaliana) UV RESISTANCE LOCUS8 (UVR8) is a recently identified UV-B-specific signaling component that regulates UV-protective responses. Using the uvr8 mutant, we defined genetically distinct UVR8-dependent and UVR8-independent pathways that stimulate different sets of genes in mature Arabidopsis leaf tissue. Both pathways operate at 1 μmol m−2 s−1 UV-B and above, but the UVR8-dependent pathway is able to stimulate UV-protective genes even in response to 0.1 μmol m−2 s−1 UV-B. Both pathways function in mutants lacking phytochromes, cryptochromes, or phototropins. Genes encoding the ELONGATED HYPOCOTYL5 (HY5) and HY5 HOMOLOG (HYH) transcription factors are induced at low UV-B fluence rates (0.1 μmol m−2 s−1). Experiments with hy5 and hyh mutants reveal that both these factors mediate responses of the UVR8-dependent pathway, acting with partial or complete redundancy to stimulate expression of particular genes. Furthermore, evidence is presented that all UVR8 pathway genes are likely to be regulated by HY5/HYH and that these transcription factors do not mediate UV-B responses independent of UVR8. Finally, we highlight the functions of HY5 and HYH in UV protection and show that HY5 plays the more critical role. This research provides evidence that, in UV-B signaling, UVR8, HY5, and HYH act together in a photoregulatory pathway and demonstrates a new role for HYH in UV-B responses.
Plants use different wavelengths of light as specific triggers for a variety of developmental responses. Phytochromes mediate responses to red and far-red light, cryptochromes and phototropins mediate responses to UV-A/blue light, and as yet undefined photoreception systems mediate responses to UV-B. Our lack of knowledge of the molecular mechanisms involved in UV-B photoperception contrasts strongly with advances in the understanding of other phototransduction pathways. Perhaps the main reason for this is the complexity of UV-B signaling processes. At one extreme, UV-B can cause damage and even necrosis, whereas on the other it can function as an informational signal stimulating photomorphogenesis. An added complication is that, whereas many UV-B-induced responses are concerned with reducing the levels of damage that UV-B inflicts on a plant, some are triggered directly by the light signal and others may arise as a reaction to the molecular damage caused within cells. Therefore, it may not be intuitively obvious which responses are regulated by which UV-B pathway. Thus, it is becoming clear that plants have different types of responses to UV-B and that these are regulated by distinct signaling pathways.
The type of response to UV-B is determined substantially by the fluence rate of exposure. High fluence rates of UV-B produce reactive oxygen species and may cause damage to DNA, proteins, membranes, and lipids (A-H-Mackerness et al., 2001; Brosché and Strid, 2003; Frohnmeyer and Staiger, 2003; Ulm and Nagy, 2005; Jenkins and Brown, 2007). These higher fluence rates can cause leaf curling and growth inhibition and initiate the expression of genes characteristic of stress responses via signaling pathways that are not specific to UV-B. Several lines of evidence indicate that these pathways overlap with wound and defense-signaling pathways (A-H-Mackerness et al., 2001; Stratmann, 2003; Jenkins and Brown, 2007). At low fluence rates, UV-B is capable of promoting metabolic and developmental changes, such as biosynthesis of phenolic secondary metabolites and inhibition of hypocotyl elongation (Kim et al., 1998; Jenkins et al., 2001; Suesslin and Frohnmeyer, 2003; Ulm and Nagy, 2005; Jenkins and Brown, 2007). It has been demonstrated that low fluence rates of UV-B stimulate expression of a range of genes that help protect plants against UV damage or to ameliorate its damaging effects (Jenkins et al., 2001; Frohnmeyer and Staiger, 2003; Ulm et al., 2004; Jenkins and Brown, 2007).
The wavelength of UV-B exposure also determines the nature of the response. For example, in cucumber (Cucumis sativus) hypocotyls, different UV-B wavebands elicit different growth inhibition and phototropic responses, with the short-wavelength responses most likely mediated by DNA damage signaling (Shinkle et al., 2004, 2005). Similarly, Ulm et al. (2004) found that different UV-B wavebands induce distinct gene expression responses in Arabidopsis (Arabidopsis thaliana) and that shorter wavelength UV-B antagonizes the response to longer wavelengths.
Although the signaling pathways for longer wavelength, low-fluence UV-B responses are not well characterized, there is compelling evidence, principally from studies of the expression of CHALCONE SYNTHASE (CHS) and other genes, that they are distinct from the wound/defense/stress signaling pathways (A-H-Mackerness et al., 2001; Jenkins et al., 2001; Ulm and Nagy, 2005; Jenkins and Brown, 2007). In addition, there is strong evidence from studies of gene expression and extension growth responses that the longer wavelength, low-fluence UV-B responses are not mediated by DNA damage signaling (Kim et al., 1998; Frohnmeyer et al., 1999; Boccalandro et al., 2001; Ulm et al., 2004). Nevertheless, although low-fluence, longer wavelength UV-B responses appear to be photoregulatory rather than resulting from damage or stress, no UV-B photoreceptor has been identified and little is known about the signaling components involved.
Genetic screens have played a key role in elucidating red/far-red and UV-A/blue light-signaling pathways in plants. The recent isolation of uvr8 mutant alleles (Kliebenstein et al., 2002; Brown et al., 2005) now presents a similar opportunity to advance our understanding of how plants process the UV-B light signal. UVR8 is a UV-B-specific signaling component (Brown et al., 2005). Microarray and reverse transcription (RT)-PCR analyses of uvr8 and wild-type plants have shown that UVR8 mediates the UV-B-induced expression of a range of genes, many of which have important roles in UV protection (Brown et al., 2005). For instance, these genes encode many of the enzymes involved in flavonoid biosynthesis, including CHS, the CPD photolyase PHR1, and enzymes that help to protect cells from oxidative damage. Not surprisingly, the uvr8 mutant is much more sensitive than wild type to UV-induced damage.
The UVR8 polypeptide shares approximately 50% sequence similarity with the REGULATOR OF CHROMATIN CONDENSATION1 (RCC1) family of proteins found in a variety of eukaryotes (Kliebenstein et al., 2002). RCC1 proteins are nuclear guanine nucleotide exchange factors (GEFs) for the small G-protein Ran, which is involved in regulating cell cycle, mitosis, and transport across the nuclear membrane. However, UVR8 and RCC1 differ in activity and function. The UVR8 protein has very little Ran GEF activity (Brown et al., 2005) and uvr8 mutants are indistinguishable from wild-type plants when grown in light lacking UV-B, indicating that they are not defective in nucleocytoplasmic transport, mitosis, or cell cycle regulation. UVR8 is present in the nucleus and cytosol, associates with chromatin, and mediates the regulation of transcription of target genes by UV-B (Brown et al., 2005).
UVR8 regulates expression of the HY5 transcription factor specifically in response to UV-B (Brown et al., 2005). Chromatin immunoprecipitation demonstrated a direct interaction between UVR8 and a chromatin fragment containing the HY5 gene promoter (Brown et al., 2005). HY5 is a bZIP transcription factor with a key role in photomorphogenesis (Osterlund et al., 2000). Its activity in the nucleus is regulated by phosphorylation and in dark-grown seedlings it is targeted for proteasomal degradation by COP1 (Hardtke et al., 2000; Osterlund et al., 2000). Intriguingly, it has recently been shown that, whereas COP1 negatively regulates the level of HY5 protein in dark-grown Arabidopsis seedlings, it is required for UV-B-stimulated HY5 transcript accumulation in light-grown seedlings (Oravecz et al., 2006). HY5 clearly has an important role in plant development and is believed to function downstream of multiple light-signaling pathways and other developmental signaling pathways (Hudson, 2000; Cluis et al., 2004). Extension of the microarray analysis of Arabidopsis uvr8 to a mutant deficient in HY5 revealed that the HY5 transcription factor functions downstream of UVR8 and regulates the expression of approximately one-half of the genes induced via UVR8 (Brown et al., 2005). The importance of HY5 for plant UV protection was further highlighted by the fact that hy5, like uvr8, showed much more damage than wild-type plants following exposure to supplementary UV-B (Brown et al., 2005; Oravecz et al., 2006).
The HYH protein is 49% identical to HY5, contains the critical HY5 functional domains and motifs, and mediates light-dependent transcription (Holm et al., 2002). There is evidence, particularly under blue light, that HY5 and HYH have overlapping roles and predominantly act in concert on the same set of genes. However, the phenotype of the hyh mutant is more subtle than that of hy5. For example, hyh shows an elongated hypocotyl only under blue light, whereas hy5 has a long hypocotyl also under white, red, and far-red light (Holm et al., 2002). No studies of the potential role of HYH in UV-B responses have previously been reported.
Here we investigate UV-B signaling pathways that stimulate gene expression in mature Arabidopsis leaf tissue. We show that the UVR8 pathway acts at low fluence rates of UV-B to stimulate expression of a number of genes. In contrast, we show that a genetically distinct, UVR8-independent signaling pathway, promotes expression of genes in response to higher fluence rates of UV-B. We report a function for the HYH transcription factor in UV-B responses and show that HY5 and HYH have overlapping functions in effecting gene expression from the UVR8 pathway. The UVR8-independent pathway does not require HY5 or HYH. Finally, we report that, of the two bZIP transcription factors, HY5 plays the more critical role in orchestrating UV protection.
RESULTS
Genetically Distinct UV-B Signaling Pathways Stimulate Gene Expression in Mature Arabidopsis Leaf Tissue, But Only the UVR8-Dependent Pathway Operates at Low UV-B Fluence Rates
Our previous transcriptome analysis with wild-type and uvr8 mutant plants (Brown et al., 2005), carried out using a fluence rate of UV-B within the ambient range (3 μmol m−2 s−1), identified a set of 72 UV-B-stimulated genes (at a false discovery rate [FDR] of 5%) that were regulated by UVR8. However, the analysis additionally showed that many UV-B-stimulated genes do not require UVR8. In fact, 639 genes (listed in Supplemental Table S1) showed UV-B stimulation at 5% FDR in wild-type plants exposed to 3 μmol m−2 s−1 UV-B and most of these were expressed normally in uvr8. We wanted to extend these experiments using RT-PCR analysis of individual genes to characterize the UVR8-dependent and UVR8-independent UV-B pathways with respect to the fluence rates over which they operate. RT-PCR experiments were undertaken with a selection of genes chosen because they were likely to be UVR8 dependent or UVR8 independent, based on their rank products (RP) score and FDR values (Breitling et al., 2004) in the microarray analysis (Table I). For each gene, RT-PCR conditions were tested and carefully selected to give product amplification over a linear range under the relevant light conditions (data not shown) so that transcript levels could be quantified.
Table I.
Microarray analysis data for genes selected for this study
The microarray data shown for the genes listed suggests they are all UV-B stimulated (wild-type UV-B versus wild-type LW column; LW represents low fluence rate [25 μmol m−2 s−1] white light) and likely to be regulated by both UVR8 and HY5 (gene group A), neither UVR8 nor HY5 (gene group B), UVR8 only (gene group C), or HY5 only (gene group D). The HY5 and HYH transcription factors are shown in gene group E. Figures provided are FDR for the given comparisons (the higher expressing condition [i.e. wild-type UV-B in each case] is listed first) and indicate the expected percentage of false positives. For simplicity, RP scores are not shown. Complete microarray data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE3533). Full sets of data for columns 1 and 3 are presented in Supplemental Tables S1 and S2, respectively, and for column 2 in Brown et al. (2005).
| Gene | FDR
|
||
|---|---|---|---|
| Wild-Type UV-B versus Wild-Type LW | Wild-Type UV-B versus uvr8 UV-B | Wild-Type UV-B versus hy5 UV-B | |
| Group A: Representative UVR8- and HY5-regulated genes | |||
| At5g13930 (CHS) | 0.67 | 0.00 | 0.00 |
| At3g22840 (ELIP1) | 0.00 | 0.00 | 0.00 |
| At5g24850 (CRYD) | 0.60 | 0.00 | 0.00 |
| At1g12370 (PHR1) | 2.98 | 0.04 | 0.75 |
| Group B: Representative UVR8- and HY5-independent genes | |||
| At5g24110 (WRKY) | 0.05 | >50 | >50 |
| At1g26380 (FAD oxred) | 0.00 | >50 | >50 |
| At1g05680 (UDPgtfp) | 0.00 | >50 | >50 |
| Group C: Candidate UVR8-regulated and HY5-independent genes | |||
| At4g31870 (GPX7) | 0.05 | 0.00 | 23.32 |
| At1g16260 (WAKL8) | 2.14 | 0.06 | >50 |
| At5g24120 (SIG5) | 0.59 | 0.00 | >50 |
| Group D: Candidate HY5-regulated and UVR8-independent genes | |||
| At1g71330 | 0.60 | >50 | 0.27 |
| At3g13910 | 0.62 | >50 | 1.11 |
| At1g17180 | 0.74 | >50 | 2.72 |
| At5g26030 | 0.93 | >50 | 1.83 |
| At3g16330 | 1.31 | 43.90 | 0.96 |
| At1g22180 | 1.43 | >50 | 3.37 |
| At2g36220 | 1.65 | >50 | 6.47 |
| At3g21690 | 1.74 | >50 | 4.03 |
| At1g28190 | 1.94 | >50 | 2.51 |
| At2g17500 | 2.00 | >50 | 4.86 |
| At4g34710 | 2.18 | >50 | 0.40 |
| At1g12320 | 2.22 | >50 | 9.79 |
| Group E: bZIP transcription factors | |||
| At5g11260 (HY5) | 1.05 | 0.05 | Not applicable |
| At3g17610 (HYH) | 0.15 | 0.04 | 35.72 |
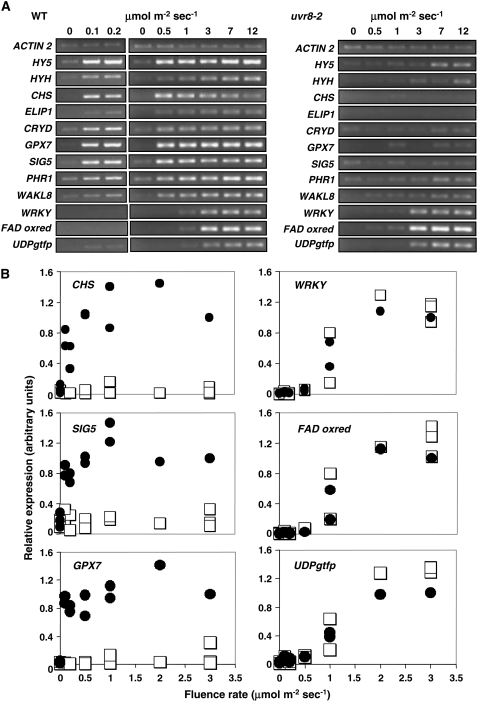
Wild-type and mutant plants were grown for 3 weeks in a low fluence rate of white light that does not stimulate accumulation of UV-B-induced transcripts in leaf tissue, as shown in Figure 1A (first lane of each image). When transferred to UV-B light of various fluence rates, expression of the selected genes is stimulated in wild-type plants. Only three of the genes shown (WRKY, FAD oxidoreductase, and UDPgtfp) are stimulated normally in the uvr8-2 mutant and these were predicted to be UVR8 independent from the microarray analysis. However, these genes are only induced, in both wild type and mutant, at UV-B fluence rates of 1 μmol m−2 s−1 or greater. By contrast, the genes that fail to be stimulated by UV-B in the uvr8-2 mutant (including CHS, ELIP1, CRYD, SIG5, GPX7, PHR1, and WAKL8) are induced in wild-type plants at fluence rates below 1 μmol m−2 s−1 and, in most cases, as low as 0.1 μmol m−2 s−1. It is important to note that the genes encoding the HY5 and HYH transcription factors, which regulate several of the other genes shown, are induced at the lowest fluence rate. The UV-B induction of both HY5 and HYH is UVR8 dependent; the low level of expression of HY5 in uvr8-2 at the highest, above-ambient fluence rates was not observed consistently.
Figure 1.
Transcript accumulation in wild-type and uvr8 mutant under different UV-B fluence rates. A, Wild-type Ler and uvr8-2 mutant plants grown for 3 weeks in a low fluence rate (20 μmol m−2 s−1) of white light were transferred to various fluence rates of UV-B light for 4 h. Leaf tissue was harvested, RNA isolated, and cDNA synthesized. Transcript levels for the genes indicated were measured by RT-PCR. ACTIN2 transcript levels are shown as a loading control. The data shown are representative of up to four independent experiments. B, Levels of selected transcripts were quantified from RT-PCR products obtained as in A using imaging software. The images show combined data from three independent experiments. Each point shows the transcript level relative to ACTIN2 in the same experiment. In each image, the data from the three experiments are normalized relative to the transcript level in wild type at 3 μmol m−2 s−1 UV-B, set at 1.0. Black circles, Wild type; white squares, uvr8-2.
The results of three independent RT-PCR experiments with wild-type and uvr8-2 plants were combined to produce quantitative data for transcript levels of selected genes over a range of fluence rates relative to control ACTIN2 transcripts (Fig. 1B). The difference in the fluence-rate threshold for induction of the UVR8-dependent and UVR8-independent genes is clear.
We therefore conclude that (1) at least two genetically distinct UV-B signaling pathways stimulate gene expression in mature Arabidopsis leaf tissue, only one of which requires UVR8; and (2) the pathways operate over different fluence ranges and only the UVR8-dependent pathway is stimulated at low UV-B fluence rates.
Mutants Lacking Phytochromes, Cryptochromes, or Phototropins Are Not Altered in Either UVR8-Dependent or UVR8-Independent UV-B-Induced Gene Expression
Over the last two decades, a number of blue/UV photoreceptors have been uncovered in Arabidopsis (Ahmad and Cashmore, 1993; Huala et al., 1997; Guo et al., 1998; Jarillo et al., 2001; Kagawa et al., 2001). However, a UV-B photoreceptor has not yet been identified and there is no evidence that the UVR8 protein functions in UV-B photoreception (C. Cloix, E. Kaiserli, and G.I. Jenkins, unpublished data). Thus, it is possible that one or more previously characterized photoreceptors may mediate plant responses to UV-B because the studies to date indicate that each is likely to have some capacity for UV-B absorption (Hartmann, 1966; Lin et al., 1995; Malhotra et al., 1995; Salomon et al., 2000). We wanted to test whether the phytochromes, cryptochromes, or phototropins could be primary photoreceptors in either the UVR8-dependent or UVR8-independent UV-B signaling pathways in mature leaf tissue.
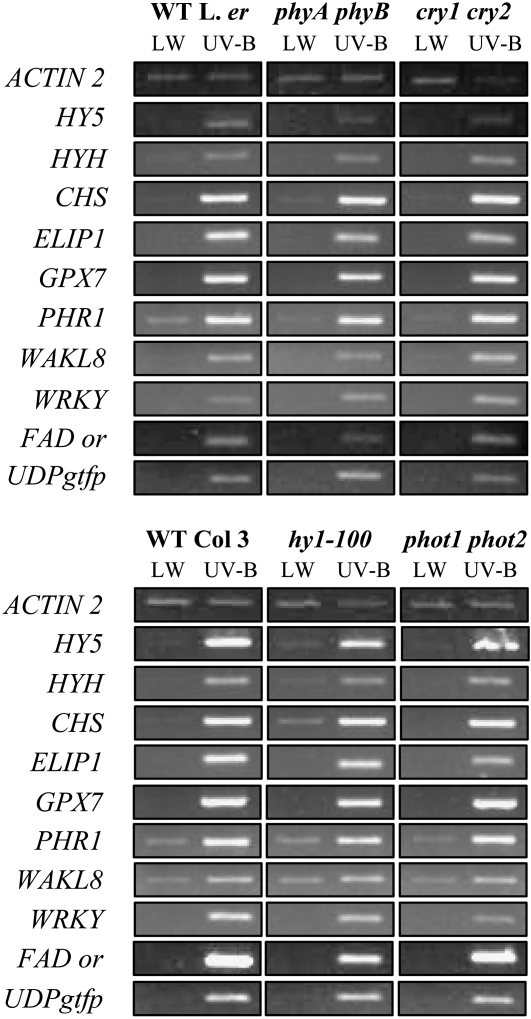
Wild-type and photoreceptor mutant plants were grown for 3 weeks in noninductive white light as described above and given sufficient UV-B to stimulate both the UVR8-dependent and UVR8-independent pathways. Stimulation of gene expression by both UV-B signaling pathways is retained in each of the photoreceptor mutants shown in Figure 2. The mutants chosen cover deficiencies in phytochromes A and B (phyA phyB) and in the phytochrome chromophore itself (hy1-100), as well as in the cryptochromes and phototropins. Note that only results from representative genes chosen from each UV-B signaling pathway are shown because the other genes were found to behave similarly. We conclude that mutation of the phytochromes, cryptochromes, or phototropins does not impede expression from the UVR8-dependent and UVR8-independent UV-B signaling pathways.
Figure 2.
Transcript accumulation in response to UV-B in photoreceptor mutants. Wild-type, phyA phyB, cry1 cry2, hy1-100, and phot1 phot2 mutant plants grown for 3 weeks in a low fluence rate (25 μmol m−2 s−1) of white light were transferred to 3 μmol m−2 s−1 UV-B light for 4 h. Leaf tissue was harvested and transcript levels measured as in Figure 1.
The UVR8-Dependent UV-B Signaling Pathway Is Effected by bZIP Transcription Factors with Overlapping Roles
Previously, it was shown that UVR8 regulates the expression of the HY5 gene specifically in response to UV-B. Transcriptome analysis had indicated that some UV-B-stimulated genes were regulated by HY5 and others were not (Brown et al., 2005). Genes that are stimulated by UV-B in wild type but not in hy5 are listed in Supplemental Table S2. Moreover, approximately one-half of the UVR8-regulated genes were additionally regulated by HY5. In addition, we noted that the gene encoding another bZIP transcription factor, HYH, which is very similar in sequence to HY5 (Holm et al., 2002), was found to be UV-B induced in the microarray experiment (Brown et al., 2005), and we decided to examine whether or not HYH had a role to play in the UV-B-UVR8-HY5 pathway. UV-B induction of HYH transcripts is confirmed in Figures 1 and 2.
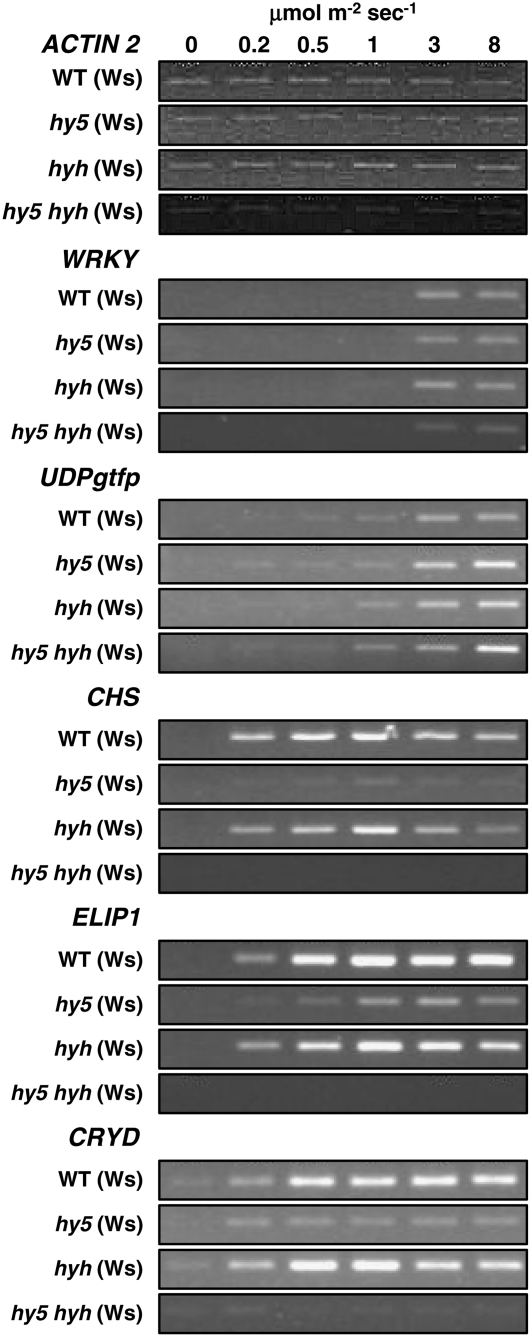
Wild-type, hy5, hyh, and hy5 hyh mutant plants were grown for 3 weeks in noninductive white light and then exposed to a range of fluence rates of UV-B as described above. Stimulation of the expression of genes from the UVR8-independent pathway was unaffected by the loss of either or both of the HY5 and HYH transcription factors, as exemplified in Figure 3 by expression of the WRKY and UDPgtfp genes; the FAD oxidoreductase gene behaved similarly (data not shown). By contrast, expression of the CHS, ELIP1, and CRYD genes from the UVR8-dependent pathway was substantially reduced in hy5 and completely abolished in the hy5 hyh double mutant. However, some expression was observed in hy5 and there was little or no reduction in transcript levels in the hyh mutant for CHS, ELIP1, or CRYD (Fig. 3). We conclude that (1) neither HY5 nor HYH is required for the UVR8-independent UV-B signaling pathway; (2) the bZIP transcription factors HY5 and HYH have overlapping roles in the UVR8-dependent UV-B signaling pathway; (3) HY5 has a more important role than HYH in the UVR8-dependent pathway that stimulates expression of the CHS, ELIP1, and CRYD genes; and (4) in the absence of HY5, HYH allows some UV-B stimulation of CHS, ELIP1, and CRYD gene expression.
Figure 3.
Transcript accumulation in hy5, hyh, and hy5 hyh mutants under different UV-B fluence rates. Wild-type, hy5, hyh, and hy5 hyh mutant plants grown for 3 weeks in a low fluence rate (25 μmol m−2 s−1) of white light were transferred to various fluence rates of UV-B light for 4 h. Leaf tissue was harvested and transcript levels measured as in Figure 1.
UVR8, HY5, and HYH Appear to Act in a Single UV-B Signaling Pathway
Our previous microarray analysis had suggested that there might be four classes of UV-B-regulated genes in Arabidopsis: genes regulated by both UVR8 and HY5, genes regulated by UVR8 but not HY5, genes regulated by HY5 but not UVR8, and genes regulated independently of UVR8 and HY5. As indicated above, this model can now be extended to include HYH, which overlaps in function with HY5 in the regulation of some UVR8 pathway genes. To test the model, we attempted to identify UV-B-induced genes regulated by UVR8 but not HY5, and vice versa.
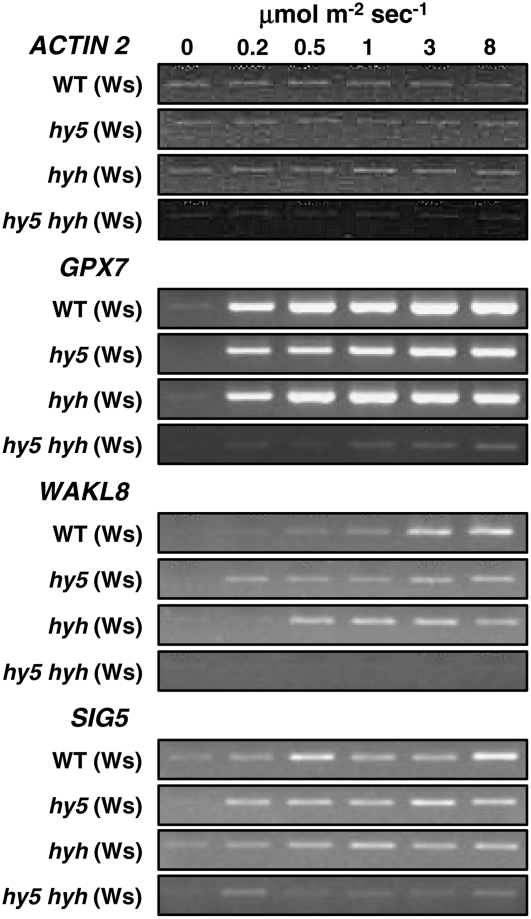
Statistical analysis of the microarray data of Brown et al. (2005) using the RP method (Breitling et al., 2004) identified candidate genes that were highly likely to be regulated by UVR8, but unlikely to be regulated by HY5. In this method, genes with the greatest differential expression between two treatments have the lowest RP scores and FDR values. Three genes were identified (GPX7, WAKL8, and SIG5) that had low RP scores and FDR values when wild-type plants exposed to UV-B were compared with uvr8 plants exposed to UV-B, but relatively high RP scores and FDR values when wild-type plants exposed to UV-B were compared with hy5 plants exposed to UV-B (Table I). Expression of these genes was examined in wild-type and mutant plants grown for 3 weeks in low-fluence-rate white light and treated with UV-B. Their expression is much reduced in uvr8 compared to wild type (Fig. 1) but, as shown in Figure 4, is essentially unaltered in the hy5 and hyh single mutants, consistent with microarray analysis of the hy5 mutant. However, expression of all three genes was greatly reduced in the hy5 hyh double mutant compared to the corresponding single mutants and wild type. Thus, HY5 and HYH act in a completely redundant manner in UV-B stimulation of these UVR8-regulated genes. Therefore, a likely explanation for the apparent lack of involvement of HY5 in the regulation of approximately 50% of the UVR8 pathway genes identified in our microarray analysis is either partial or complete functional redundancy of HY5 and HYH.
Figure 4.
Redundancy of HY5 and HYH in the regulation of gene expression by UV-B. Wild-type, hy5, hyh, and hy5 hyh mutant plants grown for 3 weeks in a low fluence rate (25 μmol m−2 s−1) of white light were transferred to various fluence rates of UV-B light for 4 h. Leaf tissue was harvested and transcript levels measured as in Figure 1.
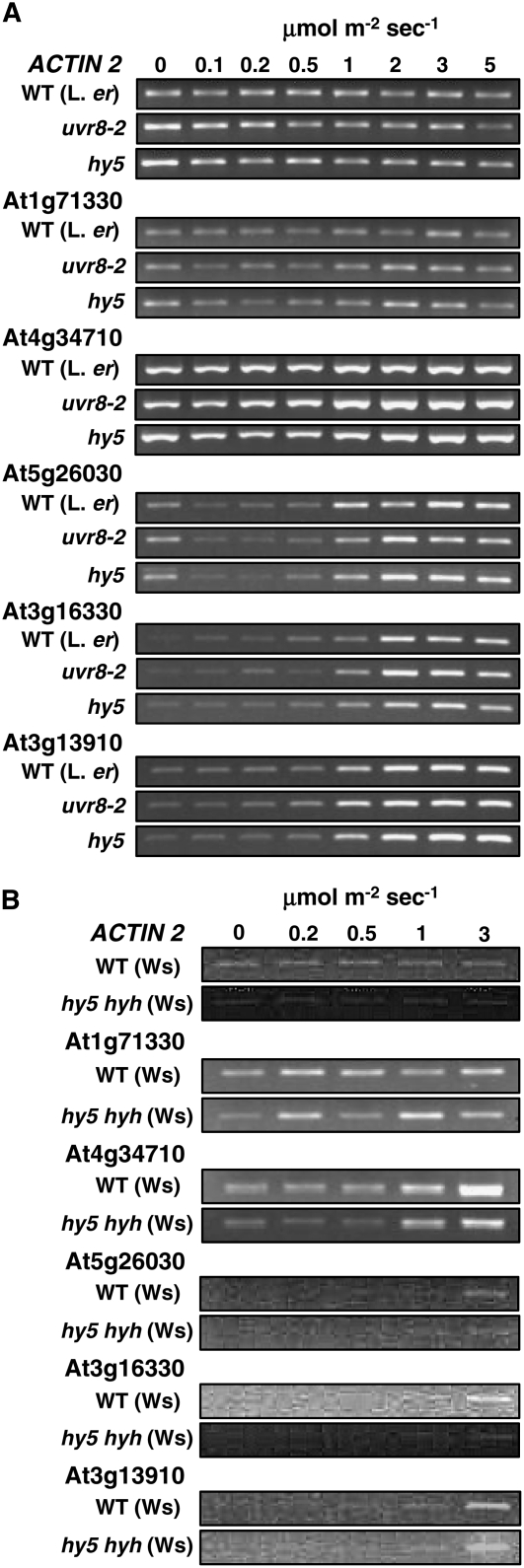
A similar statistical analysis was undertaken to search for UV-B-induced genes that required HY5, but were independent of UVR8. Candidate genes were identified that were likely to be UV-B induced in wild-type (relatively low RP scores and FDR values for wild type exposed to UV-B versus wild-type in low-fluence-rate white light) and likely to be regulated by HY5 (relatively low RP scores and FDR values for wild type in UV-B versus hy5 in UV-B), but unlikely to be regulated by UVR8 (high RP scores and FDR values for the comparison of wild type in UV-B versus uvr8 in UV-B). The most likely candidates are shown in Table I. RT-PCR primers specific for five of these genes (At1g71330, At4g34710, At5g26030, At3g16330, and At3g13910) were designed and expression examined in UV-B-treated wild-type, uvr8, hy5, and hy5 hyh leaf tissue. However, despite predictions from transcriptome analysis data (Table I), none of the genes was found to be stimulated by UV-B and regulated by HY5, but not UVR8 (Fig. 5). Moreover, there was no evidence that the unaltered expression in hy5 was due to redundancy between HY5 and HYH. Our failure to identify any genes regulated in this manner from the statistically most likely candidates leads us to conclude that the functions of UVR8 and the bZIP transcription factors HY5 and HYH may be inextricably linked in a single UV-B signaling pathway.
Figure 5.
Failure to identify genes regulated by HY5/HYH, but not UVR8. A, Wild-type (Ler), uvr8-2, and hy5 mutant plants grown for 3 weeks in a low fluence rate (20 μmol m−2 s−1) of white light were transferred to various fluence rates of UV-B light for 4 h. B, Wild-type (Ws) and hy5 hyh mutant plants grown for 3 weeks in a low fluence rate (25 μmol m−2 s−1) of white light were transferred to various fluence rates of UV-B light for 4 h. Leaf tissue was harvested and transcript levels measured as in Figure 1.
HY5 Is More Important Than HYH in Conferring UV Protection
Exposure of plants to high fluence rates of UV-B radiation can result in significant levels of damage (A-H-Mackerness, 2000; Frohnmeyer and Staiger, 2003). Two of the key symptoms of UV-induced damage, leaf necrosis and plant growth inhibition, can be assessed visually. We previously demonstrated that both UVR8 and HY5 are important in conferring UV-B protection (Brown et al., 2005). Having shown in this study that HYH overlaps in function with HY5 in the stimulation of gene expression by UV-B, we wanted to investigate whether HYH also has a role in UV-B protection.
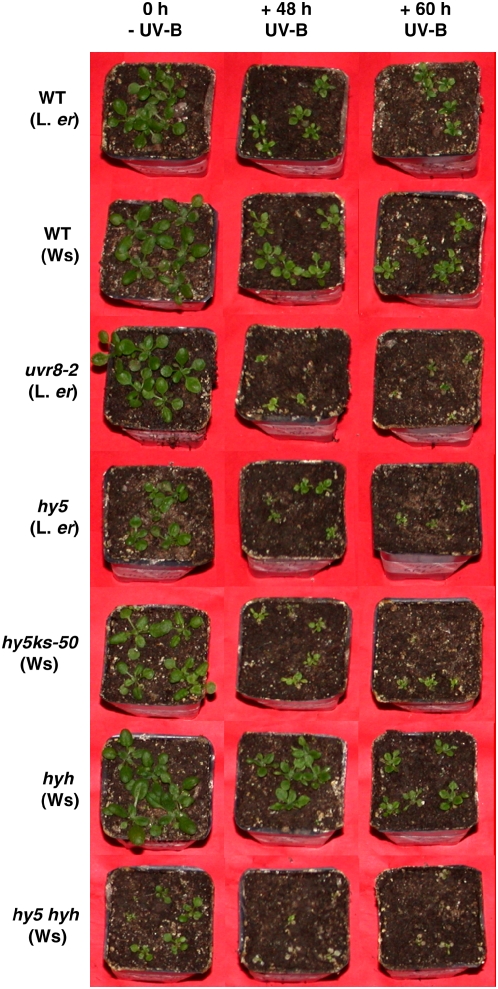
Wild-type and mutant plants were grown for 12 d in a moderate fluence rate of white light. Plants were then exposed to above-ambient levels of supplementary UV-B for the durations shown, returned to white light for several days, and then photographed (Fig. 6). The importance of HY5 and HYH in UV-B protection was assessed visually by comparing each of the single mutants with both the double mutant and wild-type plants. Although hy5 hyh double-mutant plants not treated with UV-B were generally smaller than wild-type or single-mutant plants, they also showed a slight increase in UV sensitivity when compared to hy5 single mutants. However, the hy5 hyh double mutant was much more sensitive than the hyh single mutant (Fig. 6). Furthermore, in contrast to hy5, the hyh single mutant did not appear to be significantly more UV-B sensitive than the corresponding wild-type Wassilewskija-2 (Ws-2). This finding reiterates the importance of HY5 in UV protection reported previously (Brown et al., 2005; Oravecz et al., 2006). We conclude that HY5 is the more critical bZIP transcription factor for providing UV-B protection, whereas HYH appears to play a minor role in the absence of HY5.
Figure 6.
UV-B sensitivity assay. Wild-type and mutant plants were grown in 120 μmol m−2 s−1 white light for 12 d and then exposed to above-ambient (5 μmol m−2 s−1) UV-B with supplementary 120 μmol m−2 s−1 white light for the durations shown. Untreated control plants (0 h, - UV-B) were left in 120 μmol m−2 s−1 white light throughout. Plants were photographed after return to white light for approximately 5 d.
DISCUSSION
In plants, low levels of UV-B stimulate transcription of genes involved in UV protection. UVR8 orchestrates these responses and is the only known UV-B-specific signaling component. It is therefore important to define the nature of the UVR8 pathway and to understand how it relates to other UV-B signaling pathways. In this study, we extend characterization of the UVR8 pathway and show that at least two genetically distinct UV-B signaling pathways stimulate gene expression in mature Arabidopsis leaf tissue. We demonstrate that each of these pathways has a different fluence rate response profile and regulates a distinct set of genes. We reveal that HY5 and HYH have overlapping functions that effect responses to UV-B, thus demonstrating a novel role for HYH. In addition, we provide evidence that, in UV-B signaling, the functions of UVR8, HY5, and HYH are linked in a single, low-fluence pathway. Finally, we highlight the functions of HY5 and HYH in UV protection and show that HY5 plays the more critical role.
A High-Fluence UV-B Signaling Pathway, Genetically Distinct from the UVR8-Dependent Pathway, Stimulates Gene Expression in Mature Arabidopsis Leaf Tissue
Large numbers of genes are regulated by UV-B in Arabidopsis, maize (Zea mays), and other species (Casati and Walbot, 2003; Izaguirre et al., 2003; Ulm et al., 2004; Brown et al., 2005). Despite our incomplete knowledge of the underlying mechanisms, it is clear that plants utilize multiple UV-B signaling pathways to regulate gene expression (Brosché and Strid, 2003; Frohnmeyer and Staiger, 2003; Ulm and Nagy, 2005; Jenkins and Brown, 2007). The identification of UVR8 has enabled specific UV-B signaling pathways to be defined genetically.
Previously, we used transcriptome analysis to study UV-B signaling in the uvr8 and hy5 mutants (Brown et al., 2005), and in this work we have employed RT-PCR measurements to examine the expression of selected genes. For each gene, RT-PCR conditions were chosen so that product amplification occurred within the linear range necessary for quantification of transcript levels. Results generated by our RT-PCR procedure have been compared to corresponding real-time PCR data for some of the genes and the two sets of data have proved to be consistent (L.R. Headland and G.I. Jenkins, unpublished data).
Here, we have shown that at least two genetically distinct UV-B signaling pathways stimulate gene expression in mature Arabidopsis leaf tissue, but only one of these pathways requires UVR8. Additionally, we have demonstrated that neither HY5 nor HYH is required for the UVR8-independent UV-B signaling pathway and that the pathway is functional in mutants lacking phytochromes, cryptochromes, or phototropins. Our microarray analysis, using 3 μmol m−2 s−1 UV-B, revealed that 639 genes (Supplemental Table S1) showed UV-B stimulation at 5% FDR, of which 72 were regulated by UVR8 (Brown et al., 2005). Therefore, it is clear that non-UVR8 pathways regulate a large number of UV-B-stimulated genes. The particular UVR8-independent pathway identified here operates only at higher UV-B fluence rates. There is good evidence from other studies that genes we have selected here as representative of this UVR8-independent pathway are also regulated by oxidative stress and wound-signaling pathways (Gechev and Hille, 2005; Taki et al., 2005), so the UVR8-independent pathway may overlap with pathways that are not specific to UV-B.
The UVR8-Dependent UV-B Signaling Pathway Operates at Low Fluence Rates and Regulates UV Protection
We previously showed that the UVR8 pathway is a UV-B-specific pathway with a vital role in UV protection, promoting plant survival under high fluence rates of UV-B. Here, we show that the UVR8-dependent and UVR8-independent UV-B signaling pathways have different fluence rate response profiles and regulate distinct sets of genes. Both pathways operate at relatively high ambient fluence rates of UV-B. However, the UVR8-dependent pathway is able to stimulate UV-protective gene expression at low UV-B fluence rates, even approximately 1/40 the level in full sunlight. Evidently, low fluence rates of UV-B are biologically important in that they are able to establish UV protection via the UVR8 pathway.
Previous studies on the UV-B induction of CHS, a gene regulated by UVR8 (Fig. 1), provide information on the nature of the UVR8 pathway. The UV-B induction of CHS is not mediated by DNA damage signaling because it occurs in response to very brief (even millisecond) UV-B exposures that do not cause detectable DNA damage (Frohnmeyer et al., 1999); it is unaltered in DNA repair mutants (Ulm et al., 2004; B.A. Brown and G.I. Jenkins, unpublished data) and is not diminished by UV-A/blue light, which would repair DNA damage (Wade et al., 2001). In addition, CHS expression is not stimulated by oxidative stress and UV-B induction of CHS is not impaired by treatment with antioxidants (Jenkins et al., 2001). Moreover, CHS induction by UV-B does not appear to be mediated by wound/defense pathways involving jasmonic acid or ethylene because it is not impaired in mutants altered in signaling via these molecules (C.M. Pidgeon and G.I. Jenkins, unpublished data). Furthermore, Brown et al. (2005) showed that the uvr8 mutant is not altered in the induction of CHS by several non-UV-B stimuli. Together, these observations indicate that the UVR8 pathway that mediates the UV-B induction of CHS and other genes is UV-B specific and that it does not overlap with nonspecific UV-B signaling pathways. In this sense, the UVR8-dependent UV-B signaling pathway may be regarded as photoregulatory, meaning that it mediates a specific regulatory response to UV-B light rather than a nonspecific UV stress response. Thus, the UVR8 pathway is analogous to previously defined photoreceptor-mediated pathways, although clearly a UV-B photoreceptor has not been identified.
Partly because of similarities between low-fluence UV-B responses in plants and existing photomorphogenic pathways, it has often been postulated that one or more of the previously characterized photoreceptors may mediate responses to UV-B. Studies to date indicate that each of these photoreceptors is likely to have some capacity for UV-B absorption (Hartmann, 1966; Lin et al., 1995; Malhotra et al., 1995; Salomon et al., 2000). Furthermore, as it is becoming clear that plants utilize several distinct UV-B signaling pathways, it is plausible that at least some of these may be mediated by existing photoreceptors. Previous work in Arabidopsis has provided evidence that UV-B stimulation of the CHS gene in mature leaf and of six genes in light-grown seedlings is not mediated by phyA, phyB, cry1, or cry2 (Wade et al., 2001; Ulm et al., 2004). To test the possibility of phytochromes, cryptochromes, or phototropins mediating the UV-B responses studied in this work, we examined the stimulation of gene expression in a series of photoreceptor mutants. The mutants chosen for study cover deficiencies in phytochromes A and B (phyA phyB) and in the phytochrome chromophore itself (hy1-100), as well as in the cryptochromes and phototropins. As with the UVR8-independent pathway, expression mediated by the UVR8-dependent UV-B signaling pathway was not altered in mutants lacking phytochromes, cryptochromes, or phototropins. The data suggest that these photoreceptors are not required for the pathway to operate, although we cannot exclude the possibility of multiple redundancy between classes of photoreceptors. Moreover, this finding does not mean that existing photoreceptors do not play an important role in modulating these UV-B signaling pathways; indeed, this has already been established for the UV-B regulation of CHS gene expression (Wade et al., 2001), which is mediated by UVR8.
HY5 and HYH Play Overlapping Roles in UV-B Signaling and Are Tightly Linked to the UVR8-Dependent Pathway
UVR8 regulates expression of the HY5 gene specifically in response to UV-B. In addition, we noted that the gene encoding the HY5 homolog HYH appeared to be UV-B induced in our microarray experiments (Brown et al., 2005). The UV-B stimulation of HYH gene expression was confirmed in this study by RT-PCR. HYH is 49% identical to HY5 at the amino acid level and shares many of the HY5 functional domains and motifs. Previous work had suggested that HYH may be specific to blue-light signaling and that HY5 and HYH had overlapping functions in blue- and white-light responses in Arabidopsis (Holm et al., 2002). Our finding that both HY5 and HYH are stimulated by as little as 0.1 μmol m−2 s−1 UV-B is consistent with these transcription factors having a key role in regulating low-fluence UV-B responses.
Our transcriptome analysis had indicated that approximately one-half of the UV-B stimulated, UVR8-regulated genes were also regulated by HY5 (Brown et al., 2005). In this work, we have shown that genes controlled by the UVR8-dependent UV-B signaling pathway are regulated by the bZIP transcription factors HY5 and HYH acting together with partial (in the case of CHS, ELIP1, and CRYD) or complete (in the case of GPX7, WAKL8, and SIG5) redundancy. This finding extends our understanding of the function of HY5 and demonstrates a novel role for HYH in UV-B signaling. It is likely that the overlapping functions of HY5 and HYH in UV-B signaling explains why only one-half of the UV-B-stimulated, UVR8-regulated genes appear to be regulated by HY5 in the previous transcriptome analysis. For example, Table I suggests that, whereas ELIP1 expression is regulated by HY5, GPX7 expression is not. However, the RT-PCR data in Figures 3 and 4, respectively, show that both genes are regulated by HY5. The differences in observed expression levels seen in the hy5 single mutant (Figs. 3 and 4) result from the greater capacity of HYH to compensate for the absence of HY5 in the stimulation of GPX7 expression. We hypothesize that all UVR8-regulated genes are controlled in this way by HY5/HYH, although we cannot exclude the possibility that there are UV-B-stimulated genes that are regulated by UVR8, but not HY5/HYH, because only three candidates (GPX7, WAKL8, and SIG5), albeit very good candidates (see Table I), were examined here by RT-PCR.
Furthermore, we were unable to find any UVR8-independent, UV-B-stimulated genes that were regulated by HY5/HYH despite an RT-PCR examination of the strongest candidates from our previous transcriptome analysis. Because none of the five genes (selected from the most likely candidates shown in Table I, group D) was stimulated by UV-B and regulated by HY5 but not UVR8 and since there was no evidence that unaltered expression in hy5 was due to redundancy between HY5 and HYH, we conclude that UVR8, HY5, and HYH appear to function together in a single UV-B signaling pathway. Once more, it remains theoretically possible that there are UV-B-stimulated genes that are regulated by HY5/HYH, but not UVR8, because some genes that are regulated redundantly by HY5/HYH may not be highlighted by transcriptome analysis of the hy5 mutant.
The Genetically Distinct UV-B Signaling Pathways May Have Separate Roles in Planta
Previously, we showed that both UVR8 and HY5 are important in conferring UV protection (Brown et al., 2005). In this work, we demonstrate that HY5 is the more critical bZIP transcription factor for providing UV-B protection, whereas HYH plays a minor role as seen in UV sensitivity experiments with the corresponding mutants (Fig. 6).
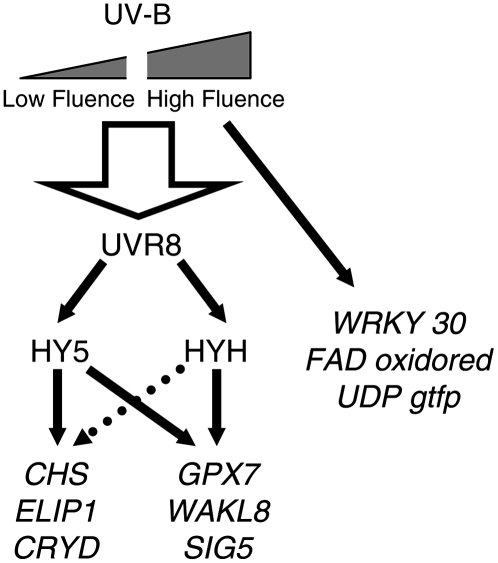
We highlighted previously that many UVR8-regulated genes are directly involved in UV protection (e.g. function in flavonoid biosynthesis, DNA repair, and protection from oxidative stress), accounting for the increased UV sensitivity of the uvr8 and hy5 mutants (Brown et al., 2005). It will be interesting to discover whether genes regulated by the high-fluence, UVR8-independent UV-B signaling pathway also have protective functions. One intriguing possibility is that the UVR8-dependent UV-B signaling pathway functions primarily in UV acclimation, whereas the UVR8-independent pathway acts to counteract the effects of unusually high doses of UV-B. The two UV-B signaling pathways and the genes they regulate are represented diagrammatically in Figure 7.
Figure 7.
Model showing the UVR8-dependent and UVR8-independent UV-B signaling pathways and representative genes they regulate in mature Arabidopsis leaf tissue. The broken arrow leading from HYH to the CHS, ELIP1, and CRYD genes indicates that HYH cannot completely compensate for the absence of HY5 in this pathway. The large arrow leading from UV-B to UVR8 overlaps the low fluence and high fluence range, indicating that both high and low UV-B fluence rates can activate UVR8.
It is important to recognize that the overlapping functions of the bZIP transcription factor homologs HY5 and HYH demonstrated here for UV-B and by Holm et al. (2002) for blue-light signaling suggest that HYH may play a role in other important responses involving HY5. HY5 has a variety of functions in plants, not only in photomorphogenesis, but also in processes such as auxin signaling (Oyama et al., 1997; Cluis et al., 2004). HY5 is thought to act downstream of convergence points between signals from multiple photoreceptors and other developmental signals (Hudson, 2000). It is particularly significant that HYH was able to completely compensate for the loss of HY5 in the UV-B regulation of several genes examined in this work. Our findings suggest that a complete picture of the significance of HY5 over the range of processes it regulates will only emerge after studies on the hy5 hyh double mutant have also been completed.
MATERIALS AND METHODS
Plant Material
Seeds of wild-type Arabidopsis (Arabidopsis thaliana) ecotype Landsberg erecta (Ler) and those of Columbia-3 (Col-3) and Ws-2 were obtained from the European Arabidopsis Stock Centre. Also provided by the European Arabidopsis Stock Centre in the Ler ecotype were the hy5-1 mutant and the hy4-2.23N and fha1 mutants used to make the cry1 cry2 double mutant (as described in Wade et al., 2001). The phyA-1 phyB-1 mutant (Ler) was donated by Prof. Garry Whitelam (University of Leicester) and the uvr8-2 mutant (also Ler) was generated in a screen carried out by Brown et al. (2005). The hy1-100 mutant in the Col ecotype was provided by Dr. Enrique Lopez-Juez (Royal Holloway, University of London) and the phot1-5 phot2-1 double mutant (also Col) was from Dr. John Christie (University of Glasgow) and Prof. Winslow Briggs (Carnegie Institution of Washington). The hy5-ks50, hyh, and hy5-ks50 hyh mutants, all in the Ws ecotype, were kindly provided by Prof. Xing Wang Deng and Lia Yao (Yale University).
Seeds were sown on compost and stratified at 4°C for several days before transfer to a low fluence rate of white light (20–25 μmol m−2 s−1) at 20°C where they were grown for 21 d before UV-B treatments were applied to stimulate gene expression.
UV-B Treatments
UV-B illuminations were carried out in controlled-environment rooms at 20°C. Plants were exposed to either UV-B alone or, in the case of the UV-B sensitivity assay, to UV-B supplemented with white light. White light was provided by Osram warm-white fluorescent tubes. UV-B was obtained from UVB-313 UV fluorescent tubes (Q-Panel) covered with cellulose acetate (West Design Products), which was changed every 24 h. This source has maximal emission at 313 nm and no emission below 290 nm; it emits very low levels of UV-A and blue light, which have been found to be insufficient to induce CHS expression (Christie and Jenkins, 1996). Fluence rates of white light (photosynthetically active radiation, 400–700 nm) were measured using a Skye RS232 meter equipped with a Quantum sensor (Skye Instruments). Fluence rates of UV-B (280–315 nm) were measured either by a Skye RS232 meter equipped with a SKU 430 sensor or using a Macam spectroradiometer (model SR9910; Macam Photometrics).
RNA Isolation and Transcript Assays
Samples of leaf tissue were harvested into liquid nitrogen, ground with a mortar and pestle, and RNA extracted using the Purescript kit (Flowgen) with an additional chloroform extraction. Following RNA extraction, a DNase treatment (DNA-free; Ambion) was used to eliminate contamination with genomic DNA. Complementary DNA was then synthesized using an oligo(dT) primer and avian myeloblastosis virus reverse transcriptase (Promega) at 48°C for 45 min. Equivalent amounts of cDNA, estimated using reactions with ACTIN2 primers, were used as template in the following PCR reaction: 25-μL volume containing 1.5 mm MgCl2, 0.2 mm each dNTP, 0.5 μm each gene-specific primer, and 0.625 units Taq DNA polymerase together with the manufacturer's buffer (Promega) using the following protocol: 2 min 30 s at 94°C, 1 min at 55°C, 2 min at 72°C; 45 s at 94°C, 1 min at 55°C, 1 min at 72°C to the appropriate number of cycles for each primer pair (shown in Table II); and 5 min at 72°C. PCR products were visualized by electrophoresis on agarose gels containing ethidium bromide.
Table II.
Primers used for RT-PCR in this study
| Gene/AGI No. | Sequence 1 | Sequence 2 | No. of Cycles | PCR Product |
|---|---|---|---|---|
| bp | ||||
| ACTIN2 (At3g18780) | 5′-CTTACAATTTCCCGCTCTGC-3′ | 5′-GTTGGGATGAACCAGAAGGA-3′ | 24 | 500 |
| HY5 (At5g11260) | 5′-GCTGCAAGCTCTTTACCATC-3′ | 5′-AGCATCTGGTTCTCGTTCTG-3′ | 28 | 404 |
| HYH (At3g17610) | 5′-TGACCAGACTCAAAATGGAG-3′ | 5′-CATCAGTTTTAGGCCTTGTG-3′ | 24 | 272 |
| CHS (At5g13930) | 5′-ATCTTTGAGATGGTGTCTGC-3′ | 5′-CGTCTAGTATGAAGAGAACG-3′ | 26 | 337 |
| ELIP1 (At3g22840) | 5′-GTAGCTTCCCTAACCTCAAG-3′ | 5′-GAATCCAACCATCGCTAAAC-3′ | 24 | 239 |
| CRYD (At5g24850) | 5′-CTTTTTCATCTAGGTGGCCC-3′ | 5′-TCCCGTGGATCATTTCCAAC-3′ | 28 | 305 |
| GPX7 (At4g31870) | 5′-TGCAGCAGAGAAGTCTGTTC-3′ | 5′-ATCACCAAGGAAACCACCAG-3′ | 28 | 370 |
| SIG5 (At5g24120) | 5′-TCCTTCGTGTTCGTTAGGAG-3′ | 5′-CAGTCCAAGCTCACTATATC-3′ | 26 | 369 |
| WAKL8 (At1g16260) | 5′-TTCTGAGACGGCAAGAAGTC-3′ | 5′-TGTGTCTTGTGAGGCATTAG-3′ | 28 | 382 |
| WRKY (At5g24110) | 5′-TGCACACCAGTTTGGATCAG-3′ | 5′-CAGCGTTCTATCAACACCAG-3′ | 26 | 256 |
| FAD oxidoreductase (At1g26380) | 5′-CGAAAAACACGAGGTTCTCG-3′ | 5′-CCTCATCGATCTTCACGTAG-3′ | 28 | 291 |
| UDPgtfp (At1g05680) | 5′-TGGAAAGAGTAGAAACCAGC-3′ | 5′-CTAATGTCGAGTGACCGTAC-3′ | 26 | 249 |
| PHR1 (At1g12370) | 5′-TGACCCGAGTGGATATGTTG-3′ | 5′-CAACACAGGGCAAAGTAGTC-3′ | 30 | 296 |
| At1g71330 | 5′-CCAAGATGCGGACATCTATC-3′ | 5′-CAGTATACATCCAAAGCGAC-3′ | 34 | 474 |
| At4g34710 | 5′-GAAGCAGAGGTGTGTTGAAG-3′ | 5′-GCCACATTGTTGAACTCATC-3′ | 28 | 636 |
| At5g26030 | 5′-GAGAGTCTATGACGATCACG-3′ | 5′-TTTCGGAGCACGAACAACAG-3′ | 26 | 302 |
| At3g16330 | 5′-CTAGCCGACTTTAATGCCAC-3′ | 5′-GTGTTACTTCCCTGATCATG-3′ | 26 | 296 |
| At3g13910 | 5′-CAGTGAAAGGTTACAACAGC-3′ | 5′-GAAGCTCTTTCGAATGGATC-3′ | 24 | 230 |
For each gene, several combinations of primers were designed and tested. The preferred combination was then assayed over a range of cycle numbers to select optimal conditions for visualization of the PCR product and quantification. Transcript levels in different RNA samples were compared using cycle numbers within the linear range of amplification. Each experiment was repeated up to four times and the data shown are representative of the results obtained.
RT-PCR products were quantified using Quantity One software of the Bio-Rad gel doc system. A numerical value was obtained for the brightness of each band and adjusted by subtracting the value obtained for an adjacent background region of the gel image. In each experiment, the adjusted numerical value obtained for each UV-B-regulated transcript band was divided by that of the ACTIN2 transcript band at the same fluence rate. In each of three independent experiments, the resultant values for the UV-B-regulated transcripts were normalized relative to wild-type Ler at 3 μmol m−2 s−1 UV-B, set at 1.0. Combined data are shown in Figure 1B.
Data from the transcriptome analysis presented in Table I were generated as described by Brown et al. (2005).
UV-B Sensitivity Assay
For the UV-B sensitivity assay, seeds were sown on compost and stratified at 4°C for several days before transfer to 120 μmol m−2 s−1 white light at 20°C. Seedlings were grown for 12 d and then exposed to 120 μmol m−2 s−1 white light plus 5 μmol m−2 s−1 UV-B for the durations shown. Plants were then returned to 120 μmol m−2 s−1 white light for 5 d to determine survival. The sensitivity assay was repeated at least three times and the data shown are representative of the results obtained.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers UVR8 (AF130441), ACTIN2 (AK230311), HY5 (NM_121164), HYH (AF453477), CHS (BT000596), ELIP1 (NM_113183), CRYD (NM_122394), GPX7 (NM_119337), SIG5 (NM_122317), WAKL8 (NM_101492), WRKY (NM_122316), FAD oxidoreductase (NM_102402), UDPgtfp (NM_100448), PHR1 (NM_101109), At1g71330 (NM_105802), At4g34710 (NM_119637), At5g26030 (NM_122504), At3g16330 (NM_112504), and At3g13910 (NM_112245).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Genes induced by UV-B in wild-type Ler.
Supplemental Table S2. Genes requiring HY5 for UV-B induction.
Supplementary Material
Acknowledgments
We would like to thank the European Arabidopsis Stock Centre, Prof. Garry Whitelam, Dr. Enrique Lopez-Juez, Dr. John Christie, and Prof. Winslow Briggs for providing mutant seeds. We are especially grateful to Prof. Xing-Wang Deng and Lia Yao for providing the hy5-ks50, hyh, and hy5-ks50 hyh mutants. We would also like to thank members of the Jenkins and Christie laboratories for helpful discussions.
This work was supported by a research grant from the Biotechnology and Biological Sciences Research Council.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Gareth I. Jenkins (g.jenkins@bio.gla.ac.uk).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- A-H-Mackerness S (2000) Plant responses to ultraviolet-B (UV-B: 280-320 nm) stress: What are the key regulators? Invited review. Plant Growth Regul 32 27–39 [Google Scholar]
- A-H-Mackerness S, John CF, Jordan B, Thomas B (2001) Early signaling components in ultraviolet-B responses: distinct roles for different reactive oxygen species and nitric oxide. FEBS Lett 489 237–242 [DOI] [PubMed] [Google Scholar]
- Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366 162–166 [DOI] [PubMed] [Google Scholar]
- Boccalandro HE, Mazza CA, Mazzella MA, Casal JJ, Ballare CL (2001) Ultraviolet-B radiation enhances a phytochrome-B-mediated photomorphogenic response in Arabidopsis. Plant Physiol 126 780–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573 83–92 [DOI] [PubMed] [Google Scholar]
- Brosché N, Strid A (2003) Molecular events following perception of ultraviolet-B radiation by plants. Physiol Plant 117 1–10 [Google Scholar]
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI (2005) A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA 102 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Walbot V (2003) Gene expression profiling in response to ultraviolet radiation in maize genotypes with varying flavonoid content. Plant Physiol 132 1739–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Jenkins GI (1996) Distinct UV-B and UV-A blue light signal transduction pathways induce chalcone synthase gene expression in Arabidopsis cells. Plant Cell 8 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluis CP, Mouchel CF, Hardtke CS (2004) The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J 38 332–347 [DOI] [PubMed] [Google Scholar]
- Frohnmeyer H, Loyall L, Blatt MR, Grabov A (1999) Millisecond UV-B irradiation evokes prolonged elevation of cytosolic-free Ca2+ and stimulates gene expression in transgenic parsley cell cultures. Plant J 20 109–117 [DOI] [PubMed] [Google Scholar]
- Frohnmeyer H, Staiger D (2003) Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol 133 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev T, Hille J (2005) Hydrogen peroxide as a signal controlling plant programmed cell death. J Cell Biol 168 17–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HW, Yang WY, Mockler TC, Lin CT (1998) Regulations of flowering time by Arabidopsis photoreceptors. Science 279 1360–1363 [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW (2000) HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J 19 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann KM (1966) A general hypothesis to interpret “high energy phenomena” of photomorphogenesis on the basis of phytochrome. Photochem Photobiol 5 349–366 [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR (1997) Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278 2120–2123 [DOI] [PubMed] [Google Scholar]
- Hudson ME (2000) The genetics of phytochrome signaling in Arabidopsis. Semin Cell Dev Biol 11 475–483 [DOI] [PubMed] [Google Scholar]
- Izaguirre MM, Scopel AL, Baldwin IT, Ballare CL (2003) Convergent responses to stress. Solar ultraviolet-B radiation and Manduca sexta herbivory elicit overlapping transcriptional responses in field-grown plants of Nicotiana longiflora. Plant Physiol 132 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo JA, Gabrys H, Capel J, Alonso JM, Ecker JR, Cashmore AR (2001) Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature 410 952–954 [DOI] [PubMed] [Google Scholar]
- Jenkins GI, Brown BA (2007) UV-B perception and signal transduction. In GC Whitelam, KJ Halliday, eds, Light and Plant Development, Vol 30. Blackwell Publishing, Oxford, pp 155–182
- Jenkins GI, Long JC, Wade HK, Shenton MR, Bibikova TN (2001) UV and blue light signaling: pathways regulating chalcone synthase gene expression in Arabidopsis. New Phytol 151 121–131 [DOI] [PubMed] [Google Scholar]
- Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M (2001) Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science 291 2138–2141 [DOI] [PubMed] [Google Scholar]
- Kim BC, Tennessen DJ, Last RL (1998) UV-B-induced photomorphogenesis in Arabidopsis thaliana. Plant J 15 667–674 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lim JE, Landry LG, Last RL (2002) Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human REGULATOR OF CHROMATIN CONDENSATION1. Plant Physiol 130 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, Dutton PL, Cashmore AR (1995) Association of flavin adenine-dinucleotide with the Arabidopsis blue-light receptor CRY1. Science 269 968–970 [DOI] [PubMed] [Google Scholar]
- Malhotra K, Sang-Tae K, Batschauer A, Dawut L, Sancar A (1995) Putative blue-light photoreceptors from Arabidopsis thaliana and Sinapis alba with a high degree of sequence homology to DNA photolyase contain the 2 photolyase cofactors but lack DNA repair activity. Biochemistry 34 6892–6899 [DOI] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Mate Z, Brzezinska A, Molinier J, Oakeley EJ, Adam E, Schafer E, Nagy F, Ulm R (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 462–466 [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR (2000) Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry 39 9401–9410 [DOI] [PubMed] [Google Scholar]
- Shinkle JR, Atkins AK, Humphrey EE, Rodgers CW, Wheeler SL, Barnes PW (2004) Growth and morphological responses to different UV wavebands in cucumber (Cucumis sativum) and other dicotyledonous seedlings. Physiol Plant 120 240–248 [DOI] [PubMed] [Google Scholar]
- Shinkle JR, Derickson DL, Barnes PW (2005) Comparative photobiology of growth responses to two UV-B wavebands and UV-C in dim-red-light- and white-light-grown cucumber (Cucumis sativus) seedlings: physiological evidence for photoreactivation. Photochem Photobiol 81 1069–1074 [DOI] [PubMed] [Google Scholar]
- Stratmann J (2003) Ultraviolet-B radiation co-opts defense signaling pathways. Trends Plant Sci 8 526–533 [DOI] [PubMed] [Google Scholar]
- Suesslin C, Frohnmeyer H (2003) An Arabidopsis mutant defective in UV-B light-mediated responses. Plant J 33 591–601 [DOI] [PubMed] [Google Scholar]
- Taki N, Sasaki-Sekimoto Y, Obayashi T, Kikuta A, Kobayashi K, Ainai T, Yagi K, Sakurai N, Suzuki H, Masuda T, et al (2005) 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol 139 1268–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Mate Z, Adam E, Oakeley EJ, Schafer E, Nagy F (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Nagy F (2005) Signaling and gene regulation in response to ultraviolet light. Curr Opin Plant Biol 8 477–482 [DOI] [PubMed] [Google Scholar]
- Wade HK, Bibikova TN, Valentine WJ, Jenkins GI (2001) Interactions within a network of phytochrome, cryptochrome and UV-B phototransduction pathways regulate chalcone synthase gene expression in Arabidopsis leaf tissue. Plant J 25 675–685 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.