SUMMARY
Sialic acid Ig-like binding lectins are important mediators of recognition and signaling events among myeloid cells. To investigate the molecular mechanism underlying Siglec functions, we have determined the crystal structure of the two N-terminal extracellular domains of a human myeloid cell inhibitory receptor Siglec-5 (CD170) and its complexes with two sialylated carbohydrates. The native structure revealed an unusual conformation of the CC′ ligand specificity loop and a unique inter-domain disulfide bond. The α(2,3)-sialyllactose and α(2,6)-sialyllactose complexed structures showed a conserved sialic acid recognition motif that involves both Arg 124 and a portion of the G-strand in the V-set domain forming β-sheet-like hydrogen bonds with the glycerol side chain of the sialic acid. Only few direct protein contacts to the sub-terminal sugars are observed and mediated by the highly variable GG′ linker and CC′ loop. These structural observations in conjunction with surface plasmon resonance binding assays provide mechanistic insights into the linkage-dependent Siglec carbohydrate recognition and suggest that Siglec-5 and other CD33-related Siglec receptors are more promiscuous in sialo-glycan recognition than previously understood.
Keywords: Siglec 5 inhibitory receptor; two domain structure; α(2,3) sialyllactose; α(2,6) sialyllactose; carbohydrate specificity
INTRODUCTION
The significance of lectins in human biology and diseases has resulted in considerable research interests1,2,3,4. The biological roles of lectins are diverse and include cell-cell recognition and adhesion, leukocyte trafficking, lymphocyte homing, cell proliferation and apoptosis, as well as responses to inflammation5,6. A recently discovered sialic acid Ig-like lectin (Siglec) constitutes a subfamily of lectins that are mainly expressed on myeloid lineage leukocytes7,8,9. They specifically recognize sialic acids (Sia) attached to complex carbohydrate structures found on the cell surface or on secreted glycoproteins. Based on the intracellular domain organization and sequence homology, human Siglecs can be further divided into two groups. The larger group, consisting of Siglec-3 (CD33) and Siglecs -5 to -11, and -14, is collectively called CD33-related Siglecs10. Except for Siglecs -14 and -1511, which have immunoreceptor tyrosine-based activation motifs (ITAM), all CD33-related Siglecs are characterized by the presence of two immunoreceptor tyrosine-based inhibitory motifs (ITIM) in their cytoplasmic tails12. The extracellular portion of Siglec receptors is comprised of an N-terminal V-set carbohydrate recognition domain followed by 1 to 16 constant C2-set domains. Through recognition of the sialylated carbohydrates on the target cells, the Siglecs participate in the negative regulation of immune response, including apoptosis, inhibition of cell proliferation and NK-mediated cell lysis13,14,15,16,17. CD33-related Siglecs have also been shown to serve as useful markers in both normal myelopoiesis and acute myeloid leukaemias18,19,20. While the cellular function of each Siglec has been extensively investigated, the structural basis of their carbohydrate recognition remains poorly understood.
To date, out of thirteen human Siglecs and nine mouse Siglecs identified, the structural information is only available for the V-set ligand-binding domains of a mouse sialoadhesin and human Siglec-721,22. Early structural data on mouse sialoadhesin in complex with α(2,3)-sialyllactose showed that the ligand recognition occurs primarily through interactions with the terminal sialic acid moiety9. In particular, a conserved arginine residue located on the F-strand (also called “primary” or “essential” arginine) forms a salt bridge with a carboxyl group of sialic acid, while the backbone of the G-strand provides additional ligand-stabilizing interactions. These studies, however, did not reveal the basis for linkage-dependent recognition. Recently, mutation experiments using chimeric constructs between Siglec-7 and -9, showed that a stretch of six amino acids in the CC′ loop of the variable domain is responsible for conveying the ligand-binding specificity from Siglec-7 to Siglec-923. The CC′ loop, however, was disordered in most of the ligand-bound Siglec-7 structures, making it difficult to envision the structural basis of such specificity22,24,25.
The linkage binding specificities of Siglecs have also been tested using various in vitro techniques on a wide spectrum of sialylated glycans. Due to the differences in and limitations of various assays, inconsistent results were often obtained8. By far, the most ambiguous and even contradictory result on linkage preference is reported for Siglec-5. For example, Brinkman-Van der Linden et al. observed similar affinities for both α(2,3)- and α(2,6)-linked Sia, and also demonstrated its specificity for the α(2,8)- linkage26,27. Later, Blixt et al. showed Siglec-5 to be highly specific for the α(2,3)-, but not to α(2,6)- or α(2,8)- linkages28. In contrast, Angata et al. observed high specificity of Siglec-5 toward both α(2,8)- disialic acid and sialyl-Tn structures, but non-detectable binding to either α(2,3)-, or α(2,6)-sialyllactose12. The assignment of the linkage preferences in other Siglecs is also quite tentative. Siglec-3 was reported to prefer either α(2,6)- or α(2,3)-linked sialyl-carbohydrates or both in different reports7,8,15,26,28,29. Siglec-7 appears to prefer α(2,8)-linked Sia8. An alternative splicing form of Siglec-7, though possessing the same carbohydrate binding domain, recognized preferentially the α(2,6)- linkage30. Siglec-9 was reported to specifically bind α(2,3)- over α(2,6)-linked carbohydrates, but it was also shown to recognize them with similar affinities7,8,14,15,28,31. For some Siglecs, a specific linkage recognition pattern was observed only at limited immobilization levels, whereas under saturating conditions they appeared to be linkage-promiscuous28. In contrast, the non-CD33-related Siglecs show distinct preference for glycosidic linkages8.
To further understand the ligand-binding preferences and their biological implications of the Siglec family of lectins, we have expressed and crystallized a two-domain N-terminal construct of human Siglec-5 that includes both the V-type and C2-type Ig domains. Through the structural studies of Siglec-5 and its complexes with two carbohydrate ligands, we examined the glycosidic linkage preference of Siglec-5 and the contribution of the C2-type domain to the binding of carbohydrate ligands. We also investigated the role of the inter-domain disulfide bond, an unusual and highly conserved feature among Siglec family members. This structural data provided further insight on the mechanism of linkage-dependent carbohydrate recognition by Siglecs.
RESULTS AND DISCUSSION
Overall Structure of Siglec-5
Although previous studies indicate that the sialic acid binding site of Siglecs resides within the V-set domain, the contribution from the adjoining C2-set domain can not be excluded32,33,34. We determined the crystal structure of a human Siglec-5 at 2.85 Å resolution (Table 1, Figure 1(a)); this represents the first two-domain structure of the Siglec family. The N-terminal V-set domain displays an Ig-like fold characteristic to other Siglecs21,22. Briefly, the two opposing anti-parallel β-sheets are formed by strands ABED and C′CFG, respectively (Figure 1(a)). The β-sandwich in Siglec-5 is wider than in a typical Ig-V fold. This is due to the formation of a disulfide bond between strands B and E, as opposed to B and F found in the standard Ig-V fold. Additionally, the G-strand is split into two shorter strands, G and G′, which are connected by a GG′ linker. The superposition of 103 Cα atoms of the V-domain of Siglec-5 with that of mouse sialoadhesin and Siglec-7 results in a root-mean-square deviation (r.m.s.d.) of 1.2 Å and 0.9 Å, respectively. The main differences lie in their BC, CC′ and C′D loops. Most noticeable, is the position of the specificity-determining CC′ loop (Figure 1(b)). In the structure of the sialoadhesin and native Siglec-7, the CC′ loop points “down and away” from the ligand-binding groove, assuming an “open” conformation. In contrast, the CC′ loop in Siglec-5 is oriented “up and toward” binding site in a “closed” conformation. An intermediate conformation of the CC′ loop is found in the structure of Siglec-7 in complex with GT1b35. Notably, in both the native and ligand-bound structures of Siglec-5, the CC′ loop is well-ordered. This is not the case for the majority of reported Siglec-7 structures. The observed CC′ loop conformation in Siglec-5 is partly attributed to four intramolecular hydrogen bonds between C′D and CC′ loops (Figure 1(c)). They are between the carbonyl oxygen of Pro 72 and NH2 of Arg 85 (2.8 Å), the carbonyl oxygen of Ala 75 and NE of Arg 85 (2.9 Å), the amide nitrogen of Val 77 and the carbonyl oxygen of Arg 86 (3.1 Å), and the carbonyl oxygen of Val 77 and the amide nitrogen of Lys 88 (3.0 Å). A single intermolecular hydrophilic contact occurs between the carbonyl oxygen of Tyr 73 and the Tyr 62 hydroxyl of the symmetry-related molecule.
TABLE 1.
Data collection, phasing, and refinement statistics.
| Native | Complex | Pt derivative | |||
|---|---|---|---|---|---|
| α(2,3)-sialyllactose | α(2,6)-sialyllactose | Peak | Remote | ||
| Data collection | |||||
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0000 | 1.0716 | 1.0624 |
| Resolution (Å) | 2.75 | 3.00 | 2.70 | 3.2 | 3.2 |
| Space Group | R32 | R32 | R32 | R32 | R32 |
| Unit Cell | a=93.15 | a=93.14 | a=94.04 | a=91.82 | a=91.91 |
| Parameters (Å) | c=205.82 | c=213.57 | c=209.89 | c=206.31 | c=206.72 |
| Unique reflections | 9235 | 7405 | 10119 | 12015 | 11940 |
| Completeness (%) | 91.5 (82.6) | 99.8 (98.8) | 99.8 (100) | 99.0 (92.5) | 98.7 (91.2) |
| I/σ(I) | 23.0 (2.7) | 34.3 (4.6) | 38.4 (4.7) | 20.4 (3.9) | 23.0 (4.1) |
| Rsym1 (%) | 6.7 (47) | 4.6 (36) | 5.4 (55) | 8.6 (44.6) | 7.3 (30.6) |
| Phasing statistics | |||||
| Resolution range (Å) | 15 – 3.8 | ||||
| FOM (centric/acentric) | 0.15/0.22 | ||||
| Refinement | |||||
| Resolution range (Å) | 30 – 2.85 | 25 – 3.0 | 25 – 2.8 | ||
| Rcryst (%) | 24.9 (36.1) | 23.5 (36.3) | 24.4 (34.6) | ||
| Rfree2 (%) | 28.4 (32.3) | 27.0 (43.2) | 27.0 (36.7) | ||
| r.m.s.d.bond lengths (Å) | 0.008 | 0.008 | 0.007 | ||
| r.m.s.d bond angles (°) | 1.49 | 1.37 | 1.36 | ||
| Total Number of Atoms | 1728 | 1757 | 1722 | ||
| Protein Atoms | 1697 | 1680 | 1661 | ||
| Carbohydrate atoms | - | 43 | 20 | ||
| Water Molecules | 31 | 34 | 41 | ||
| Average B factor (protein/carbohydrate/water) | 88/69/69 | 109/100/65 | 107/147/81 | ||
| Ramachadran statistics | |||||
| Favored/Allowed (%) | 76.8/22.7 | 77.4/22.6 | 80.3/19.7 | ||
| Disallowed (%) | 0.6 | – | – | ||
Rsym= Σ∣Ihkl−〈Ihkl〉∣/Σ〈Ihkl〉, where Ihkl is the observed intensity and 〈Ihkl〉 is the average intensity from multiple measurements. Values in parentheses refer to the highest resolution shell.
Rfree was calculated using 5% of the total reflections chosen randomly and omitted from refinement.
Figure 1.
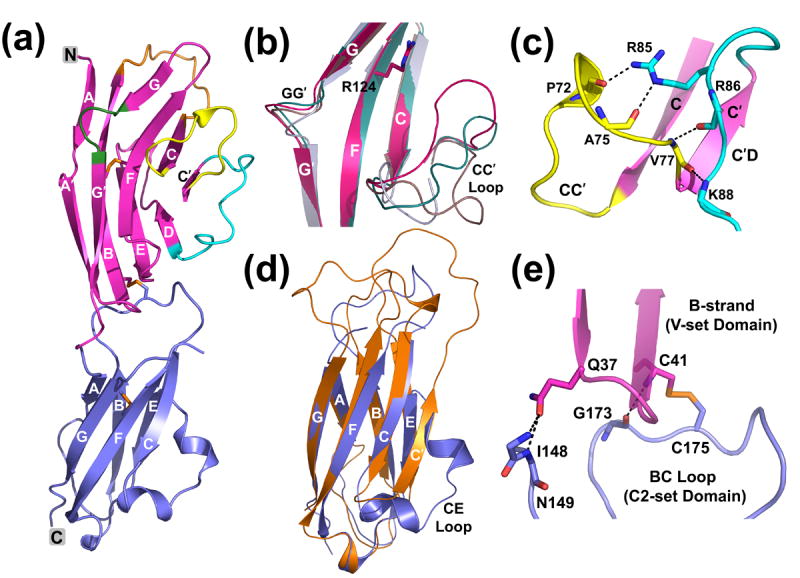
(a) Ribbon diagram showing the native Siglec-5 structure with V-domain in magenta and C2-domain in blue. The disulfides are depicted with sticks. BC and C′D loops and the specificity-determining GG′ and CC′ regions are depicted in orange, cyan, green and yellow, respectively. (b) The superposition among the V-set domains of Siglec-5 (magenta), sialoadhesin (grey; 1QFO), the native Siglec-7 (brown, 1O7V) and the Siglec-7 from its GT1b complex (teal, 2HRL) illustrates the differences in the CC′ and GG′ regions. Arg 124 marks the ligand-binding site. (c) Intramolecular hydrogen-bond interactions involving CC′ loop facilitating a “closed” loop conformation. The C′D and CC′ loops are colored in yellow and cyan, respectively. (d) The superposition of the C2-set domains of Siglec-5 (blue) and VCAM-1 (orange; 1VCA). (e) Hydrophilic interactions and the inter-domain disulfide bridge at the VC-domain interface.
The closest structural homolog (DALI search) for the C2-type domain of Siglec-5 is a vascular cell adhesion molecule-1 (VCAM-1)36. A superposition of the C2-set domains of Siglec-5 and VCAM-1 results in an r.m.s.d. of 1.4 Å for 65 Cα atoms. A distinctive feature of the Siglec-5 C2-type domain lies in the fact that the β-sandwich consists of only six strands, ABE and CFG, as opposed to a classic seven-stranded C2-set containing strands ABE and C′CFG (Figure 1(d)). The C′-strand in Siglec-5 is absent and replaced with an extended CE loop that coils into two one-turn helices. The thermal displacement parameters of all 15 residues in the CE loop are higher than average, indicating an intrinsic flexibility of this loop.
The N-terminal V-set domain (residues 25–145) joins to the C2-set domain (residues 151–238) via a short linker. Sequence-based analysis predicted a potential inter-domain disulfide bond between the N-terminal V- and C2-set domains8,9 The electron density at the domain junction in Siglec-5 clearly revealed the inter-domain disulfide bridge connecting Cys 41 from the B-strand in the V-set to Cys 175 from the BC loop in the C2-set domain (Figure 1(e)). The inter-domain interface is mostly hydrophobic with only a few hydrogen bonds present. The hinge angle formed by V-set and C2-set domains is roughly 140°, as calculated by the program HINGE37. It is substantially larger than that found in Fc (52°-70°), Fab (~86°), TCR (76°), KIR (66°-80°) receptors, but is comparable to the hinge angle found in adhesion molecules, such as VCAM-1 (141°) or ICAM-1 (170°)36,37.
The Carbohydrate Binding Site
The sialic acid binding site is situated on the face of the CFG β-sheet (Figure 1(b)). Arg 124 from the F-strand is a key residue in chelating the negatively charged sialic acid (Sia, in short). The bottom of the carbohydrate binding site is lined with polar residues from the CC′ loop and GG′ linker. Both contain variable residues that could define the shape and charge of the binding groove, thus, contributing to the diversity of carbohydrate recognition (Figure 2(a)). The CC′ loop assumes a “closed” conformation, creating the deepest binding groove among known Siglecs (Figure 2(b)). Comparison between the native and carbohydrate complexed Siglec-5 structures shows minor conformational changes associated with the ligand binding, which are not quite the same as observed in sialoadhesin or Siglec-725. In particular, the Sia-coordinating Arg 124 is hydrogen-bonded to Glu 126 (2.5 Å) and Ser 134 (3.1 Å) through its primary and secondary amines in the native structure (Figure 3). Upon ligand binding, the guanidino group of Arg 124 flips such that the primary amines form a salt bridge with the carboxylic group of sialic acid and a hydrogen bond to Ser 134, leaving the secondary amine to coordinate Glu 126. Another notable difference between Siglec-5 and other Siglecs is the side chain conformation of Lys 132 upon sialic acid binding. In Siglec-7, for example, the lysine side-chain blocks the entry to the essential arginine in the native form, and moves ~4 Å upon ligand binding25. In Siglec-5, the binding site remains unobstructed in both native as well as in complexed forms. Other ligand-induced conformational changes involve residues from the G-strand, GG′ and CC′ loops. This is in contrast to a recently reported structure of Siglec-7 and its complex with disialylganglioside analog, GT1b, where the CC′ loop adopts an open conformation in the native Siglec-7 structure but becomes closed upon GT1b binding35. The same loop assumes the closed conformation in both the native and ligand bound Siglec-5 structures.
Figure 2.
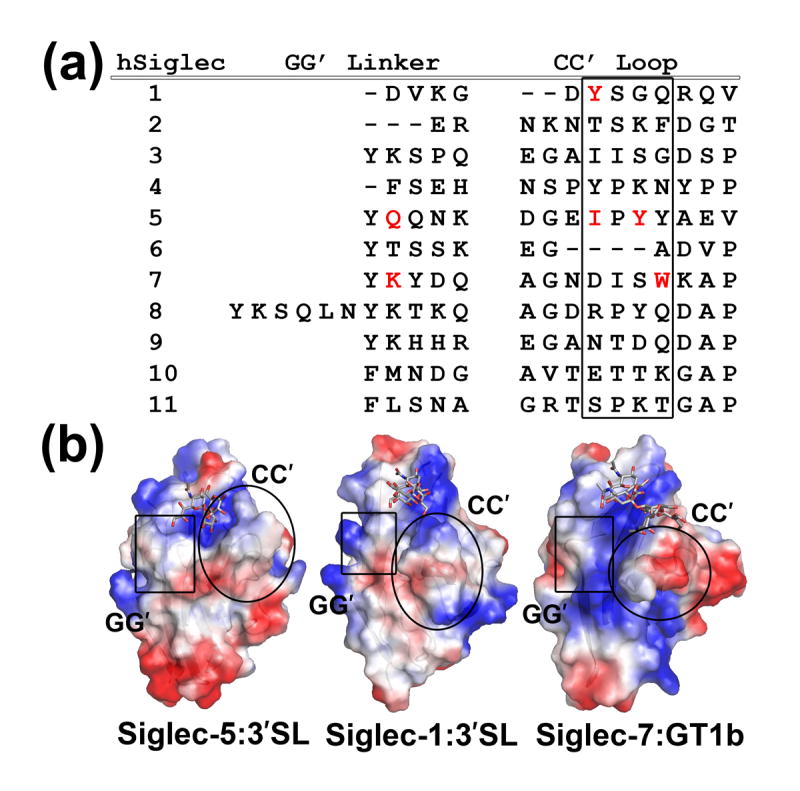
(a) The amino acid sequence alignment of the GG′ and CC′ variable regions in human Siglecs. Residues participating in the ligand-stabilizing interactions (according to X-ray data) are shown in red. The region in the CC′ loop participating in ligand binding is boxed. (b) Electrostatic surface potential representation of the sialoadhesin, Siglec-5, and -7 complexes with carbohydrate moieties shown in grey balls-and-sticks. Areas corresponding to the specificity determining GG′ and CC′ regions are outlined.
Figure 3.
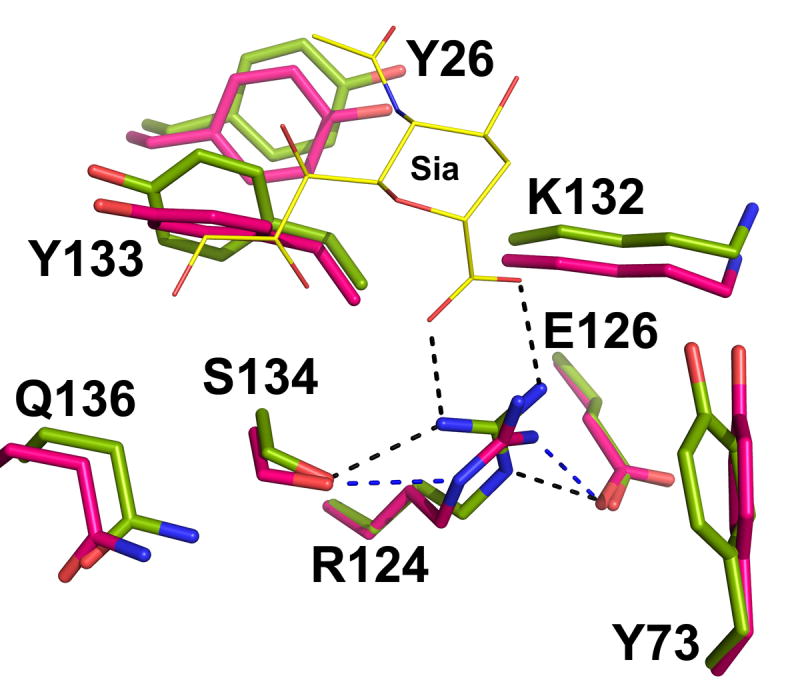
The structural comparison at the binding site between the native (magenta) and liganded (green) Siglec-5. Only sialic acid part of α(2,3)-sialyllactose is depicted (yellow lines).
Structures of Siglec-5 and Carbohydrate Complexes
The structures of Siglec-5 carbohydrate complexes were obtained with two model carbohydrate compounds: α(2,3)-sialyllactose and α(2,6)-sialyllactose (Figures 4(a), (b)). Similar to other known Siglec structures, the majority of the interactions in the Siglec-5 binding site occurred through the Sia portion of the ligand (Table 2). In addition to Arg 124, the main chain atoms of Lys 132 and Ser 134 form hydrogen bonds with the N5 amide, the O8 and O9 hydroxyl groups of Sia (Figure 4(a)). The aromatic ring of Tyr 133 makes a van der Waals contact with Sia C9 methylene. In sialodhesin and Siglec-7 complexes, residue equivalent to the Tyr 133 is a trptophan that stacks against the aliphatic part of glycerol side chain in a similar manner (Figure 4(c)). Further comparisons between the α(2,3)-sialyllactose complexed sialoadhesin and Siglec-5 show a similar ligand orientation (Figure 4(c)). The superposition between the bound α(2,3)-sialyllactose results in a 0.92 Å r.m.s.d., with the Glc moiety, Sia glycerol side chain and N-acetyl group showing larger displacement. Compared to the sialoadhesin, the sialic acid N-acetyl group is rotated about 120° in Siglec-5 but 180° in Siglec-7. In both Siglec-5 and -7, the N-acetyl carbonyl oxygen makes a weak hydrogen bond to the hydroxyl of Tyr 26, whereas a van der Waals contact was observed in sialoadhesin between the N-acetyl methyl of Sia and Trp 2.
Figure 4.
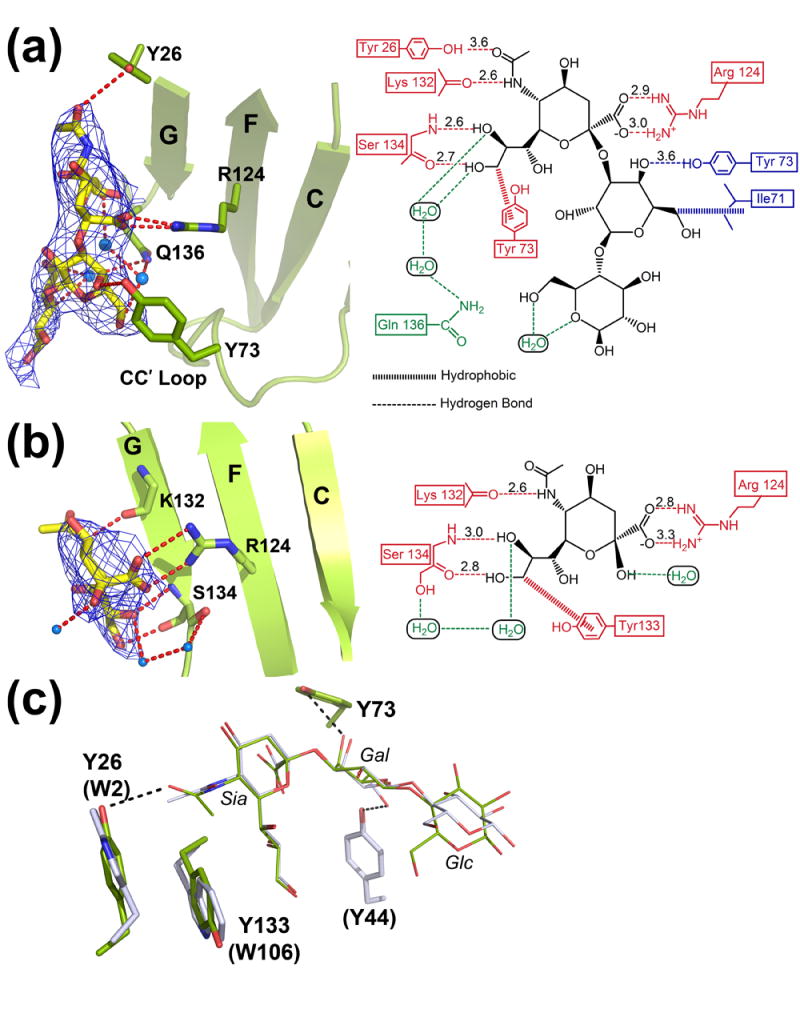
Siglec-5 in complex with (a) α(2,3)-sialyllactose, and (b) α(2,6)-sialyllactose. Left panels: Sections of the unbiased (before the inclusion of the ligand) ∣Fo∣-∣Fc∣ electron density maps contoured at 2σ. Siglec-5 receptor is shown in ribbon diagram (green) with the Arg 124 depicted in the balls-and-sticks. The carbohydrate moieties are colored in yellow. Right panels: Interactions between the respective receptor and carbohydrate at the binding site. Direct protein-ligand interactions with the sialic acid and the galactose are shown in red and blue, respectively. Water-mediated interactions are colored in green. (c) Superposition of the ligand binding sites of the sialoadhesin (grey) and Siglec-5 (green) in their complex structures with α(2,3)-sialyllactose. Residues corresponding to sialoadhesin are given in parenthesis.
TABLE 2.
Hydrophilic interactions at the carbohydrate-binding site.
| Interaction
type |
Ligand
atom |
Protein
atom |
Distance (Å) | |||
|---|---|---|---|---|---|---|
| α(2,3)-Sialyllactose | α(2,6)-Sialyllactose | |||||
| Salt Bridge | O1A | Sia | Arg 124 | NH2 | 3.0 | 3.3 |
| Salt Bridge | O1B | Sia | Arg 124 | NH1 | 2.9 | 2.8 |
| H-Bond | N5 | Sia | Lys 132 | O | 2.6 | 2.6 |
| H-Bond | O8 | Sia | Ser 134 | N | 2.6 | 3.0 |
| H-Bond | O9 | Sia | Ser 134 | O | 2.7 | 2.8 |
| H-Bond | O5 | Sia | Tyr 26 | OH | 3.6 | |
| H-Bond | O5 | Gal | Tyr 73 | OH | 3.6 | |
Siglec-5 receptor showed limited interactions with the sub-terminal sugars in the α(2,3)-sialyllactose complex. A weak hydrogen bond is observed between the Gal O4 hydroxyl and Tyr 73 of the CC′ loop (Figure 4(a), Table 2). A tyrosine located in the CC′ loop of sialoadhesin was involved in contacting the Gal moiety (Figure 3). However, because of the diametrically different orientation of the CC′ loop in these two receptors, the interactions occur from the opposite sides of the Gal hexose ring. In the case of Siglec-5, Tyr 73 interacts with Gal O4 hydroxyl, whereas Tyr 44 in sialoadhesin interacts with the Gal O6 hydroxyl (Figure 4(c)). In addition to the direct protein-ligand interaction, three water molecules were found at the binding site of α(2,3)-sialyllactose complex to mediate H-bond interactions between the Gal, Glc and Sia of the carbohydrate, and the amide of Gln 136 in the GG′ linker (Figure 4(a)). The glucose moiety in α(2,3)-sialyllactose complex is poorly ordered. This is most likely due to both the rotational flexibility around the Gal-Glc β(1-4) linkage and a lack of interactions between the receptor and the sugar.
Since α(2,6)- and α(2,3)-sialyllactose differ only in their glycosidic linkage, the coordination of the sialic acid was found conserved in the two complexes (Figure 4(b), Table 2). However, no density is observed in the ∣Fo∣-∣Fc∣ map beyond the sialic acid moiety in the α(2,6)-sialyllactose complex (Figure 4(b), left panel). No direct protein interactions was observed to the Gal and Glc sugars in the α(2,6)-sialyllactose complex. Thus, the lactose part of the α(2,6)-sialyllactose appear to rotate freely around the Sia-Gal α(2-6) linkage.
Sialic Acid Mimics a β -Strand to Pair with the G-Strand
The structural scaffold of Siglec molecules has evolved to specifically recognize sialylated carbohydrates. Unlike that of a classic Ig-V domain, the G-strand in Siglec-5 is split into two strands, G and G′, separated by a short GG′ linker (Figure 5(a)). The presence of the GG′ linker is a critical structural feature of the Siglec-specific Ig fold, and is directly implicated in the ligand recognition. While the GG′ linker is responsible for linkage specificity, the G-strand mediates the linkage-independent Sia recognition. Interestingly, the pattern of the hydrogen bonds between Sia and the G-strand main chain atoms mimics that between β-strands in a classical pleated antiparallel β sheet (Figure 5(b)). In other words, sialic acid appears to Siglecs as a “short β strand” with matching hydrogen bonding pattern to the G-strand. This backbone-mediated sialic acid hydrogen bonding recognition underscores the need of an immunoglobulin-like fold for sialic acid specific lectins (Figure 5(a)). Modifications that disrupt the hydrogen bonding pattern, such as truncation of the glycerol side chain (O8, O9 hydroxyls) abrogated the binding in most Siglecs8. On the contrary, substitution to O9 hydroxyl that maintains hydrogen-bond pattern preserved sialic acid binding25.
Figure 5.
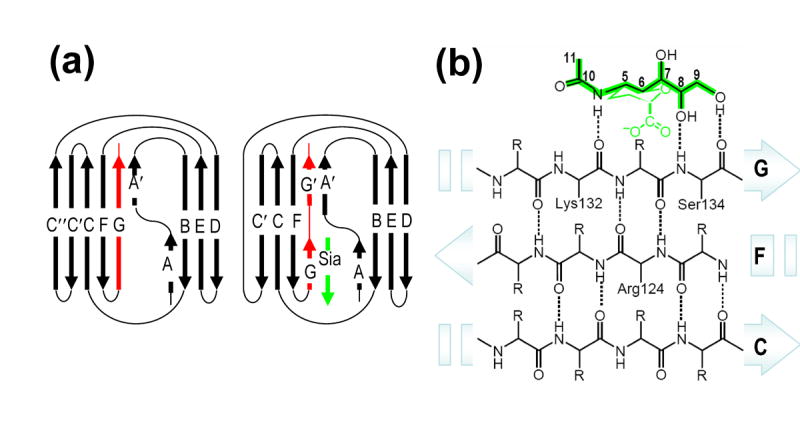
(a) A schematic diagram featuring differences in the G-strand (red) between classic V-set immunoglobulin fold (left panel) and Siglec-specific Ig fold (right panel). Sialic acid is shown as a β strand (green). (b) Schematic representation of conserved main-chain interactions between Siglec CFG β sheet and sialic acid (green). The mode in which sialic acid interacts with main-chain atoms on the G strand mimics hydrogen bond interactions (dashed lines) in classic antiparallel pleated β sheet.
Carbohydrate Linkage Specificities
While all CD33-related Siglecs recognize terminal sialic acid in a conserved manner, they seem to display differential specificity for the α-glycosidic linkage between the sialic acid and the connecting sugar. Previously reported binding experiments8 showed that the CD33-related Siglecs tend to recognize multiple glycosidic linkages. On the contrary, Siglec-1, and other non-CD33-related Siglecs have a more distinct preference for a particular linkage. The linkage promiscuity of the CD33-related Siglecs may be a result of the structural variability of the CC′ and GG′ regions, as these two critical regions define the shape and charge of the binding pocket for the sub-terminal carbohydrate moieties (Figure 2(b)). Whereas the importance of the CC′ loop and the GG′ linker in ligand binding has been previously recognized through mutagenesis and structural data, their exact involvement in governing carbohydrate specificity has not been sufficiently investigated35. The GG′ linker is an uncommon structural feature in the immunoglobulin superfamily and is unique to Siglecs. The distribution of charged residues in the GG′ linker, and thus the electrostatic potential of the carbohydrate binding site, varies substantially among Siglecs. The length of the GG′ linker within human and mouse Siglec family also varies between 2 and 11 residues (Figure 2(a)). The reduced length of the GG′ and CC′ regions may result in a narrower binding groove, thus playing a role in restricting the carbohydrate recognition in Siglecs. This is, perhaps, why Siglecs with shorter GG′ linker (such as Siglec-1, -2, and -4) or CC′ loop (such as Siglec-1 and -6) have been shown to be mostly linkage-specific. Additionally, it appears that the receptors abundant with polar residues in their critical variable regions are likely to promote favorable hydrophilic interactions with sub-terminal sugar hydroxyls. For example, abundance of polar and charged residues in the CC′ and GG′ regions of Siglec-9 (Figure 2(a)) may contribute to a network of stabilizing interactions to the sub-terminal sugars, resulting in overall a tighter binding of this receptor.
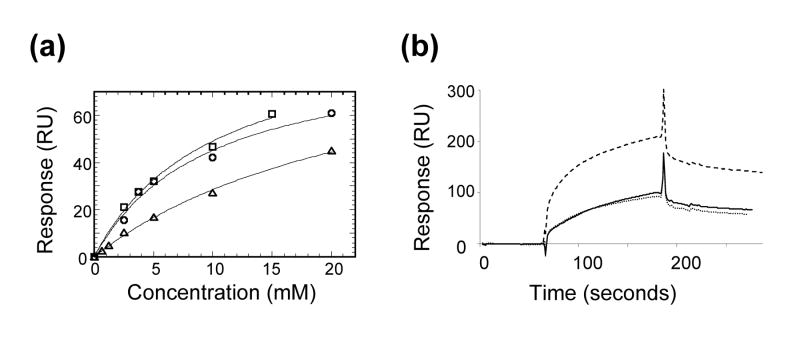
Siglec-5 linkage specificity has been a matter of controversy and the mechanism of its linkage-specific recognition has not been well established. From the structural point of view, the plasticity of Siglec-5 binding site allows for recognition of all three commonly occurring α(2,3)-, α(2,6)- and α(2,8)-linkage types. Whereas we were successful in obtaining crystal structures of complexes with α(2,3)- and α(2,6)-linked Sia, the stability of α(2,8)- complex was predicted through molecular modeling (results are not shown). To further define the linkage-dependent carbohydrate Siglec-5 recognition, we carried out a series of SPR solution binding experiments between Siglec-5 receptor and α(2,3)-, α(2,6)-sialyllactose and α(2,8)-disialic acid. This particular choice of probes enabled adequate assessment of glycosidic linkage specificities while eliminating the effects of underlying sugar structures. Additionally, it enabled a direct comparison of the binding affinities of different carbohydrates to the same receptor on an immobilized sensor chip. The results showed that Siglec-5 recognizes both α(2,3)- and α(2,6)-linked Sia with similar affinity with dissociation constants of Kd = 8.7 mM and 8.0 mM, respectively, while exhibits weaker binding to α(2,8)-linked disialic acid (Kd = 25 mM) (Figure 6 (a)). Similar binding affinity toward α(2,3)- and α(2,6)-linked sialyllactose is consistent with our structural data and results from the lack of interactions to the sub-terminal sugars (Figure 4(a), (b), Table 2). When using PAA-conjugated sialoglycans, equal binding of both α(2,3) and α(2,6) isomers (2-4 μM) was observed, whereas α(2,8)-linked disialic acid bound tighter (0.4 μM) (Figure 6 (b)). Similar effect of multivalency and carbohydrate distribution on ligand affinity has been observed earlier for other Siglecs28. The differences in binding response of Siglec-5 to multimeric sialoglycan probes versus monomeric sialoglycans indicates that the specificity of this receptor derives to a greater extent from the distribution of carbohydrates on the substrate surface and not as much from individual ligand binding affinities. In vivo, the specificity-determining factor may also include interactions with the subterminal sugars remote from the sialic acid binding site. At the same time, such interactions are unlikely to govern linkage specificity to a larger extent. Crystal structure of Siglec-7 with a GT1b analog may serve as an example35. The GT1b septa-saccharide forms ten H-bonds to the Siglec-7 receptor, seven of which involve terminal sialic acid and only three the underlying hexa-saccharide (average of ½ hydrogen bond per subterminal sugar residue). A steric effect could also play a role, as the α-glycosydic linkage sets up a directional vector to which the subterminal sugars project out; yet it is difficult to factor in, given extreme conformational flexibility of glycans35. Therefore, the mechanism linkage-dependent carbohydrate recognition by Siglec-5 (and perhaps other CD33-related Siglecs), most likely lies beyond the glycosidic linkage of the monomeric sialoglycan, but rather in their distribution, presentation and the structure of the oligosaccharide underlying the terminal Sia.
Figure 6.
Results of Surface Plasmon Resonance binding between Siglec-5 and model sialoglycans. (a) SPR binding response (response units, RU) is plotted against concentration of analyte (mM) for α(2,3) sialyllactose (open circles); α(2,6) sialyllactose (open squares); and α(2,8) disialic acid (open triangles). Solid line represents an ideal curve corresponding to the straight line in the double reciprocal plot. The dissociation constants (Kd) are 8.7 mM, 8.0 mM and 25 mM, for α(2,3)-, α(2,6)- and α(2,8)-linked sialoglycans, respectively. (b) The SPR sensorgrams of Siglec-5 Fc binding to the PAA-conjugated probes: α(2,3)- sialyllactose (solid line); α(2,6)-sialyllactose (dotted line), and α(2,8)-disalic acid (dashed line).
CONCLUSION
This work provides a structural examination of the two extracellular domains of Siglec-5, a member of the CD33-related inhibitory Siglecs. The V-set and C2-set Ig domains form a nearly linear tandem arrangement, linked by an inter-domain disulfide bond. To our knowledge, this is the first account of inter-domain disulfide association in the Ig superfamily and a unique feature of Siglecs. Whereas the inter-domain disulfide does not directly participate in ligand binding, it likely serves to reduce conformational flexibility of the carbohydrate-binding domain. Structural comparisons among the α(2,3)-sialyllactose and α(2,6)-sialyllactose complexed Siglec-5 structures showed that despite a very different arrangement of the sub-terminal sugars, the receptor recognizes both carbohydrates primarily through the terminal sialic acid. The conserved structural elements for sialic acid recognition include Arg 124, which forms a salt bridge with the Sia carboxylic group, and the G-strand, which forms β-sheet like hydrogen bonds with the Sia glycerol side chain. Both Arg 124 and the G-strand are important for Sia recognition by Siglecs. For instance, a natural killer cell receptor, NKp44, possessing the equivalent Arg residue, but having a different G strand, failed to bind sialylated carbohydrates38. Conversely, the lack of the primary Arg in the presence of the Siglec-specific Ig fold abrogated or markedly reduced Sia binding8. This may also explain why human Siglec-XII (Siglec-like molecule-1), which lacks the essential Arg in its V-set domains, is inefficient in Sia binding9,15.
The linkage-specific recognition, on the other hand, arises from the interactions with the residues located on two highly variable GG′ and CC′ regions of the V-set domain. Previous biochemical binding results showed that the recognition by the CD33-related Siglecs is less restricted to a particular carbohydrate linkage. This carbohydrate promiscuity is reflected in the structure of both Siglec-5 complexes and results from the lack of strong protein carbohydrate interactions beyond the sialic acid moiety. These structural observations together with the solution binding studies provide mechanistic insights into the Siglec-glycan recognition, and suggest that members of CD33-related Siglecs are more promiscuous for the glycosidic linkages than previously recognized and that the carbohydrate preference seems to correlate with the polarity and length of their GG′ and CC′ variable regions. Additionally, the distribution of carbohydrates on the substrate surface seems to play a significant role in determining linkage preference and may override the effect of individual ligand binding specificity.
MATERIALS AND METHODS
Expression and Purification of the Recombinant Soluble Siglec-5
The cDNA encoding the extracellular V-set and C2-set domains (residues 24-238) of Siglec-5 was subcloned into a pET30a vector (Novagen, Inc.). The recombinant Siglec-5 was expressed in the BL21 (DE3) strain of E.coli and purified using a method similar to previously described39. In brief, Siglec-5 transformed cells were cultured at 37°C in LB broth containing 50 μg/ml kanamycin, and induced at OD596 ~0.9 with 0.5 mM IPTG for 4 hours. The inclusion bodies were isolated by repeated washes in a 2 M urea-containing wash buffer. The refolding was carried out by a quick dilution method into a buffer containing 100 mM Tris-HCl (pH 8.0), 0.5 M L-arginine, 2.5 mM cystamine, 5 mM cysteamine and 10 μg/ml 4-2[-aminoethyl]-benzenesulfonyl fluoride hydrochloride. The refolded Siglec-5 was captured using a Ni-NTA affinity column (Qiagen) and separated from a soluble aggregate using a Superdex-200 gel-filtration column (GE Healthcare) with 10 mM Tris-HCl (pH 9.0), 10 mM imidazole and 50 mM NaCl as the running buffer. Inclusion of small amounts of imidazole in the buffer appeared to be critical for the protein solubility. The identity and purity of the protein was confirmed by SDS-PAGE, N-terminal sequencing and TOF mass-spectrometry.
Surface Plasmon Resonance Binding Studies
All binding studies were performed using a BIAcore 3000 instrument (BIAcore AB). To measure the binding affinities between Siglec-5 and sialyllactoses, the recombinant Siglec-5 protein was immobilized between 2600-3300 response units (RU) in 10 mM sodium acetate (pH 5.0) onto a carboxymethylated dextran (CM5) surface using standard amine coupling. Soluble monomeric sialoglycans, α(2,3) sialyllactose, α (2,6) sialyllactose and α(2,8) disialic acid, were used as analytes. Serial dilutions of analytes were done in the range of 0.625 to 20 mM. Injections were performed at a flow rate of 20 μl/min using buffer containing 10 mM Tris-HCl (pH 8.0) and 50 mM NaCl. Surfaces were regenerated by dissociation in buffer. Dissociation constants (KD) were determined from equilibrium fitting of Req versus Analyte concentration using a simple 1:1 two state binding model. For the binding between Siglec-5 and polyacrylamide (PAA) conjugated carbohydrate polymers, the recombinant Siglec-5-Fc fusion protein (R&D Systems, Inc.) were immobilized through a protein A based capture immobilization, in which protein A was directly immobilized between 2000-4000 RU onto a carboxymethylated dextran (CM5) surface using a standard amine-coupling. The analyte consisting of PAA-conjugated α(2,3)-, α(2,6)- sialyllactose and α(2,8)-disialic acid were injected onto the protein A captured Siglec-5 sensor chips at a 10μg/ml concentration in HBS-P buffers (BIAcore, Inc.). All injections were performed at a flow rate of 20 μl/min. Sensor chip surfaces were regenerated by a brief injection of 10 mM NaOH. Dissociation constants were determined from kinetic fitting of the binding chromatograms using a 1:1 two state Langmuir binding model with the BIAcore evaluation software.
Crystallization and X-ray Data Collection
Purified soluble Siglec-5 was concentrated to 5–11 mg/ml in a buffer containing 10-50 mM NaCl, 2–10 mM imidazole, and 2–10 mM Tris-HCl (pH 9.0). The search for crystallization conditions was conducted at 4°C using the hanging drop vapor diffusion method with both commercially available sparse-matrix screens and a home-designed kit40. Crystals appeared in both a high-salt (1.4 M sodium acetate, 100 mM cacodylate pH 6.5) and a PEG (20% MPEG 550, 100 mM Tris pH 8.0) conditions. Diffraction-size native crystals (~100–350 μm) were obtained using 4 μl of protein solution and 2 μl of reservoir solution containing 20% MPEG 550 and 100 mM Tris pH 8.0. The ligand-complexed crystals were obtained by gradually soaking of 20–35 mM α(2,3)- sialyllactose and α(2,6)-sialyllactose (Sigma-Aldrich) individually into the PEG-grown Siglec-5 crystals over a period of 5–7 days.
X-ray diffraction data were collected using the SER-CAT beamline at Advanced Photon Source at Argonne National Laboratory and processed using HKL200041. Prior to flash cooling in a liquid nitrogen stream, the crystals were cryoprotected by soaking in either the mother liquor solution containing 20% glycerol for the PEG-grown crystals or 3.4 M sodium malonate (pH 6.5) for the salt-grown crystals. The native crystals belonged to a rhombohedral space group R32 with unit-cell dimensions of a=b=93.2 Å, c=205.8 Å, and one molecule in the asymmetric unit. The soaking of sialylated oligosaccharides resulted in a slight increase (~2–3%) in the crystallographic c axis (Table 1).
Structure Determination
Initial attempts to solve the structure by molecular replacement (MR) using AMoRe42, PHASER43, and EPMR44 with the V-domain only Siglec-7 as a search model (54% identity, Protein Data Band entry code: 1O7V) yielded reproducible solutions for the V-set domain of Siglec-5. No interpretable density could be attributed to the C2-type domain using the phases derived from the V-domain only MR solution. Efforts to locate the C2-domain using a C2-domain from VCAM-1, as a search model (30% identity, 1VCA), were not successful. To acquire additional phasing information, a series of heavy-atom compounds were screened for potential derivatives using mass-spectrometry45. A platinum derivative was identified with a single binding site and co-crystallized in the presence of 1 mM platinum ethylenediamine dichloride. The heavy-atom position was located next to Met 115 by the direct methods using SHELXD. The phases obtained from the platinum derivative alone, however, were insufficient to yield a traceable electron density map (Table 1). Subsequently, the heavy-atom derived phases were used in the MR with phased translation function using the program BRUTEPTF46. Both the V-domain and C2-domain search models yielded solutions with correlation coefficients of 0.44 and 0.22, respectively. When the partial models were combined and inspected, about 50% of the residues in the C2-set domain had to be removed due to either steric clash with the V-set domain or the lack of experimental density. The molecular replacement phases from the remaining model were combined with those from the Pt derivative using SIGMAA47, followed by manual model building using the program O48. The structural refinements were carried out using the program CNS 1.249. The refinement statistics for the native and complex Siglec-5 structures are given in Table 1. The refined native Siglec-5 model consisted of residues 24-238, with only two residues, 224-225, missing from the FG loop of the C2-set domain. The structural superpositions were performed using LSQMAN50 and structural drawings were created using PyMOL51.
Acknowledgments
We thank Dr. Z. Dauter for help with X-ray data collection, Dr. S. Radaev and Dr. M. G. Joyce for useful discussions, Dr. C. Hammer for TOF Mass-spectrometry experiments and for useful discussions, Dr. Minoru Fukuda (Burnham Institute for Medical Research, La Jolla, CA) for providing α(2,8) disialic acid. We acknowledge the use of beamline ID-22 of the Southeast Regional Collaborative Access Team at APS. This work is supported by intramural research funding from the National Institute of Allergy and Infectious Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pusztai A, Bardocz S. Lectins: biomedical perspectives. Taylor & Francis; London; Bristol, PA: 1995. [Google Scholar]
- 2.Wong Simon YC, Arsequell G. Immunobiology of carbohydrates. Landes Bioscience/Eurekah.com; Georgetown, Texas: Kluwer Academic/Plenum; New York: 2003. [Google Scholar]
- 3.Caron Michel, Sève Anne-Pierre. Lectins and pathology. Harwood Academic; Amsterdam; Marston, Abingdon: 2000. [Google Scholar]
- 4.Gabius H-J, Gabius S. Lectins and cancer. Springer-Verlag; Berlin; New York: 1991. [Google Scholar]
- 5.Sharon N, Lis H. History of lectins: from hemagglutinins to biological recognition molecules. Glycobiology. 2004;14:53R–62R. doi: 10.1093/glycob/cwh122. [DOI] [PubMed] [Google Scholar]
- 6.Sharon N, Lis H. Lectins. Chapman and Hall; London, New York: 1989. [Google Scholar]
- 7.Crocker PR, Varki A. Siglecs, sialic acids and innate immunity. Trends Immunol. 2001;22:337–342. doi: 10.1016/s1471-4906(01)01930-5. [DOI] [PubMed] [Google Scholar]
- 8.Varki A, Angata T. Siglecs -the major subfamily of I-type lectins. Glycobiology. 2006;16:1R–27R. doi: 10.1093/glycob/cwj008. [DOI] [PubMed] [Google Scholar]
- 9.Nitschke L, Crocker PR. The Sialic Acid-Binding Siglec Family. In: Wong SYC, Arsequell G, editors. Immunobiology of carbohydrates. Landes Bioscience/Eurekah.com; Georgetown, Texas: Kluwer Academic/Plenum; New York: 2003. pp. 119–127. [Google Scholar]
- 10.Crocker PR, Paulson JC, Varki A. Siglecs and Their Roles in the Immune System. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 11.Angata Takashi, Tabuchi Yukako, Nakamura Kazunori, Nakamura Mitsuru. Siglec-15: An immune system Siglec that is conserved throughout vertebrate evolution. Glycobiology. 2007;17(8):838–846. doi: 10.1093/glycob/cwm049. [DOI] [PubMed] [Google Scholar]
- 12.Angata T, Hayakawa T, Yamanaka M, Varki A, Nakamura M. Discovery of Siglec-14, a novel sialic acid receptor undergoing concerted evolution with Siglec-5 in primates. The FASEB Journal. 2006;20:1964–1973. doi: 10.1096/fj.06-5800com. [DOI] [PubMed] [Google Scholar]
- 13.Avril T, Freeman SD, Attrill H, Clarke RG, Crocker PR. Siglec-5 (CD170) can mediate inhibitory signaling in the absence of immunoreceptor tyrosine-based inhibitory motif phosphorylation. J Biol Chem. 2005;280:19843–19851. doi: 10.1074/jbc.M502041200. [DOI] [PubMed] [Google Scholar]
- 14.Crocker PR. Siglecs in innate immunity. Curr Opin Pharmacol. 2005;5:1–7. doi: 10.1016/j.coph.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Crocker PR. Siglecs: sialic-acid-binding immunoglobulin-like lectins in cell-cell interactions and signaling. Curr Opin Struct Biol. 2002;12:609–615. doi: 10.1016/s0959-440x(02)00375-5. [DOI] [PubMed] [Google Scholar]
- 16.von Gunten S, Simon H-U. Sialic acid binding immunoglobulin-like lectins may regulate innate immune responses by modulating the life span of granulocytes. The FASEB Journal. 2006;20:601–605. doi: 10.1096/fj.05-5401hyp. [DOI] [PubMed] [Google Scholar]
- 17.Nicoll G, Avril T, Lock K, Furukawa K, Bovin N, Crocker PR. Ganglioside GD3 expression on target cells can modulate NK cell cytotoxicity via siglec-7-dependent and -independent mechanisms. Eur J Immunol. 2003;33:1642–1648. doi: 10.1002/eji.200323693. [DOI] [PubMed] [Google Scholar]
- 18.Connolly NP, Jones M, Watt SM. Human Siglec-5: tissue distribution, novel isoforms and domain specificities for sialic acid-dependent ligand interactions. Br J Haematol. 2002;119:221–238. doi: 10.1046/j.1365-2141.2002.03808.x. [DOI] [PubMed] [Google Scholar]
- 19.Virgo P, Denning-Kendall PA, Erickson-Miller CL, Singha S, Evely R, Hows JM, Freeman SD. Identification of the CD33-related Siglec receptor, Siglec-5 (CD170), as a useful marker in both normal myelopoiesis and acute myeloid leukaemias. Br J Haematol. 2003;123:420–430. doi: 10.1046/j.1365-2141.2003.04625.x. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen DH, Ball ED, Varki A. Myeloid precursors and acute myeloid leukemia cells express multiple CD33-related Siglecs. Experimental Hematology. 2006;34:728–735. doi: 10.1016/j.exphem.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 21.May AP, Robinson RC, Vinson M, Crocker PR, Jones EY. Crystal Structure of the N-Terminal Domain of Sialoadhesin in Complex with 3′ Sialyllactose at 1.85 Å Resolution. Mol Cell. 1998;1:719–728. doi: 10.1016/s1097-2765(00)80071-4. [DOI] [PubMed] [Google Scholar]
- 22.Alphey MS, Attrill H, Crocker PR, Van Aalten DMF. High Resolution Crystal Structures of Siglec-7. Insights into ligand specificity in the Siglec family. J Biol Chem. 2003;278:3372–3377. doi: 10.1074/jbc.M210602200. [DOI] [PubMed] [Google Scholar]
- 23.Yamaji T, Teranishi T, Alphey MS, Crocker PR, Hashimoto Y. A Small Region of the Natural Killer Cell Receptor, Siglec-7, Is Responsible for Its Preferred Binding to α2,8-Disialyl and Branched α2,6-Sialyl Residues. A comparison with Siglec-9. J Biol Chem. 2002;277:6324–6332. doi: 10.1074/jbc.M110146200. [DOI] [PubMed] [Google Scholar]
- 24.Dimasi N, Moretta A, Moretta L, Biassoni R, Mariuzza RA. Structure of the saccharide-binding domain of the human natural killer cell inhibitory receptor p75/AIRM1. Acta Crysallogr. 2004;D60:401–403. doi: 10.1107/S0907444903028439. [DOI] [PubMed] [Google Scholar]
- 25.Attrill H, Takazawa H, Witt S, Kelm S, Isecke R, Brossmer R, Ando T, Ishida H, Kiso M, Crocker PR, van Aalten DM. The structure of siglec-7 in complex with sialosides: leads for rational structure-based inhibitor design. Biochem J. 2006;397:271–278. doi: 10.1042/BJ20060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cornish AL, Freeman S, Forbes G, Ni J, Zhang M, Cepeda M, Gentz R, Augustus M, Carter KC, Crocker PR. Characterization of Siglec-5, a novel glycoprotein expressed on myeloid cells related to CD33. Blood. 1998;92:2123–2132. [PubMed] [Google Scholar]
- 27.Brinkman-Van der Linden ECM, Varki A. New Aspects of Siglec Binding Specificities, Including the Significance of Fucosylation and of the Sialyl-Tn Epitope. J Biol Chem. 2000;275:8625–8632. doi: 10.1074/jbc.275.12.8625. [DOI] [PubMed] [Google Scholar]
- 28.Blixt O, Collins BE, van den Nieuwenhof IM, Crocker PR, Paulson JC. Sialoside specificity of the siglec family assessed using novel multivalent probes: identification of potent inhibitors of myelin-associated glycoprotein. J Biol Chem. 2003;278:31007–31019. doi: 10.1074/jbc.M304331200. [DOI] [PubMed] [Google Scholar]
- 29.Freeman SD, Kelm S, Barber EK, Crocker PR. Characterization of CD33 as a new member of the sialoadhesin family of cellular interaction molecules. Blood. 1995;85:2005–2012. [PubMed] [Google Scholar]
- 30.Angata T, Varki A. Siglec-7: a sialic acid-binding lectin of the immunoglobulin superfamily. Glycobiology. 2000;10:431–438. doi: 10.1093/glycob/10.4.431. [DOI] [PubMed] [Google Scholar]
- 31.Zhang JQ, Nicoll G, Jones C, Crocker P. Siglec-9, a Novel Sialic Acid Binding Member of the Immunoglobulin Superfamily Expressed Broadly on Human Blood Leukocytes. J Biol Chem. 2000;275:22121–22126. doi: 10.1074/jbc.M002788200. [DOI] [PubMed] [Google Scholar]
- 32.Powell LD, Varki A. 14246 I-type Lectins. J Biol Chem. 1995;270:14243–14246. doi: 10.1074/jbc.270.24.14243. [DOI] [PubMed] [Google Scholar]
- 33.Kelm S, Pelz A, Schauer R, Filbin MT, Tang S, Bellard ME, Schnaar RL, Mahoney JA, Hartnell A, Bradfield P, Crocker P. Sialoadhesin, myelin-associated glycoprotein and CD22 define a new family of sialic acid-dependent adhesion molecules of the immunoglobulin superfamily. Curr Biology. 1994;4:965–972. doi: 10.1016/s0960-9822(00)00220-7. [DOI] [PubMed] [Google Scholar]
- 34.Freeman SD, Kelm S, Barber EK, Crocker PR. Characterization of CD33 as a new member of the sialoadhesin family of cellular interaction molecules. Blood. 1995;85:2005–2012. [PubMed] [Google Scholar]
- 35.Attrill H, Imamura A, Sharma RS, Kiso M, Crocker PR, van Aalten DMF. Siglec-7 undergoes a major conformational change when complexed with the α2,8-disialylganglioside GT1b. J Biol Chem. 2006;281:32774–32783. doi: 10.1074/jbc.M601714200. [DOI] [PubMed] [Google Scholar]
- 36.Wang J-H, Stehle T, Pepinsky B, Liu J-H, Karpusas M, Osborn L. Structure of a Functional Fragment of VCAM-1 Refined at 1.9 Å Resolution. Acta Crystallogr. 1996;D52:369–379. doi: 10.1107/S0907444995012352. [DOI] [PubMed] [Google Scholar]
- 37.Snyder GA, Brooks AG, Sun PD. Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2. Proc Natl Acad Sci USA. 1999;96:3864–3869. doi: 10.1073/pnas.96.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cantoni C, Ponassi M, Biassoni R, Conte R, Spallarossa A, Moretta A, Moretta L, Bolognesi M, Bordo D. The Three-Dimensional Structure of the Human NK Cell Receptor NKp44, a Triggering Partner in Natural Cytotoxicity. Structure. 2003;11:725–734. doi: 10.1016/s0969-2126(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 39.Radaev S, Rostro B, Brooks AG, Colonna M, Sun PD. Conformational Plasticity Revealed by the Cocrystal Structure of NKG2D and Its Class I MHC-like Ligand ULBP3. Immunity. 2001;15:1039–1049. doi: 10.1016/s1074-7613(01)00241-2. [DOI] [PubMed] [Google Scholar]
- 40.Radaev S, Sun PD. Crystallization of protein-protein complexes. Acta Crystallogr. 2002;D35:674–676. [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- 43.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr. 2005;D61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 44.Kissinger CR, Gehlhaar DK, Smith BA, Bouzida D. Molecular replacement by evolutionary search. Acta Crystallogr. 2001;D57:1474–1479. doi: 10.1107/s0907444901012458. [DOI] [PubMed] [Google Scholar]
- 45.Sun PD, Hammer CH. Mass-spectrometry assisted heavy-atom derivative screening of human FcγRIII crystals. Acta Crystollgr. 2000;D56:161–168. doi: 10.1107/s0907444999015188. [DOI] [PubMed] [Google Scholar]
- 46.Strokopytov BV, Fedorov A, Mahoney NM, Kessels M, Drubin DG, Almo SC. Phased translation function revisited: structure solution of the cofilin-homology domain from yeast actin-binding protein 1 using six-dimensional searches. Acta Crystalogr. 2005;D61:285–293. doi: 10.1107/S0907444904033037. [DOI] [PubMed] [Google Scholar]
- 47.Collaborative Computational Project, Number 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 48.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 49.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: A New Software Suite for Macromolecular Structure Determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 50.Kleywegt GJ, Jones TA. Detecting folding motifs and similarities in protein structures. Methods in Enzymology. 1997;277:525–545. doi: 10.1016/s0076-6879(97)77029-0. [DOI] [PubMed] [Google Scholar]
- 51.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]