Abstract
More often than not, analyses of virus evolution have considered that virus populations are so large that evolution can be explained by purely deterministic models. However, virus populations could have much smaller effective numbers than the huge reported census numbers, and random genetic drift could be important in virus evolution. A reason for this would be population bottlenecks during the virus life cycle. Here we report a quantitative estimate of population bottlenecks during the systemic colonization of tobacco leaves by Tobacco mosaic virus (TMV). Our analysis is based on the experimental estimation of the frequency of different genotypes of TMV in the inoculated leaf, and in systemically infected leaves, of tobacco plants coinoculated with two TMV genotypes. A simple model, based on the probability that a leaf in coinoculated plants is infected by just one genotype and on the frequency of each genotype in the source, was used to estimate the effective number of founders for the populations in each leaf. Results from the analysis of three leaves per plant in plants inoculated with different combinations of three TMV genotypes yielded highly consistent estimates. Founder numbers for each leaf were small, in the order of units. This would result in effective population numbers much smaller than the census numbers and indicates that random effects due to genetic drift should be considered for understanding virus evolution within an infected plant.
Changes in the genetic structure of virus populations often lead to the failure of measures to control virus-caused diseases, and this has been a major reason for the study of virus evolution. For the last 20 years a considerable amount of research has been published on the evolution of many viruses that are important pathogens of humans, animals, and plants or that are used as experimental systems, and much effort has been done to understand the factors that drive virus evolution (for reviews, see references 7 and 14). Because virus fecundity and the number of virus particles in the infected host can be very high, it is often assumed that viruses have large populations (see, for example, reference 5), and purely deterministic models have been mostly used to explain virus evolution (cf. reference 6). As a consequence, selection is often invoked as the main force in virus evolution, and random genetic drift is seldom considered at all (see reference 13). Drift depends on the effective size of the population (i.e., the number of individuals that pass their genes to the next generation) and not on the census size (i.e., the total number of individuals in the population). Knowledge of the effective population number, Ne, is fundamental in understanding population structure and evolution because at small values of Ne random processes will predominate over deterministic ones (2). However, reliable estimates of Ne from natural populations are notoriously difficult to obtain (10) and, to our knowledge, have not been reported for any virus.
Viral populations are fragmented in demes in different organs within a host, in different host individuals, in different host populations, etc., and population bottlenecks could occur when initiating a new deme. A major cause of population differentiation could be genetic drift as a result of population bottlenecks during (i) colonization of new organs within a host, (ii) host-to-host transmission within a population, or (iii) migration to new host populations. Bottlenecks severely reduce Ne, even though census population size may recover to the former sizes. For plant viruses it has been shown that a small number of particles is involved in mechanical or vector mediated infection of a new host (12, 26). Because the number of transmission events will be related to the census size of the vector population and to the fraction of susceptible hosts infected, it has been speculated that the effective number of the viral population will be on the order of magnitude of the census number for the plant host or insect vector population (17, 24). Population bottlenecks during the colonization of different organs within an infected host individual would also result in differentiation of the within host population. Hence, the effective population number of a virus population might be much smaller than the census, and genetic drift could be important in virus evolution. This has been recognized recently: interest in understanding the evolution of human immunodeficiency virus type 1 in patients receiving antiviral therapy has led to several analyses of the virus effective population size within a patient. Most estimates indicate that the effective number can be several orders of magnitude smaller than the census (19, 25, 28, 30), with one cause being the compartmentation of the virus population in different organs (11).
For plant viruses, it is often assumed that systemic movement through the vascular system, which is necessary for the infection of new organs, results in population bottlenecks. Indeed, analyses of the genetic structure of within-plant virus populations shows population differentiation between different organs (15). However, to our knowledge, no quantitative analysis of the possible bottlenecks during organ colonization has been published for plant viruses. We present here such an analysis based on the experimental estimation of the frequency of different genotypes of Tobacco mosaic virus (TMV) in the inoculated leaf and on the upper systemically infected leaves, colonized after viral movement, of coinfected tobacco plants. The data show that severe population bottlenecks occur during systemic colonization of new organs, which should result in effective population numbers orders of magnitude smaller than the total or census population size.
MATERIALS AND METHODS
Virus genotypes and inoculations.
Three genotypes of TMV were used in this work: TMV wild type (wt) and two coat protein mutants. Mutant P20L has the transition C5656U, resulting in the amino acid replacement P→L at position 20 of the coat protein. Mutant 20/72 is a double mutant that, in addition to mutation C5656U, had the transversion A5932U, resulting in the amino acid replacement Y→F at position 72. These mutants were derived from biologically active cDNA clones that have been described elsewhere (3, 4) and were the gift of W. O. Dawson and J. N. Culver. Infectious RNA was transcribed from these clones with T7 (for wt) or with SP6 (for the two mutants) RNA polymerases as described previously (3, 4) and inoculated into tobacco cv. Samsun plants in 0.1 M Na2HPO4. Virus particles were purified from plants infected with RNA transcripts as described previously (1). Virus suspensions in 10 mM sodium phosphate buffer (pH 7.2) were used for further inoculations. Plants were kept in a greenhouse at 20 to 25°C.
Analysis of virus accumulation in infected tobacco plants.
Virus accumulation was quantified as viral RNA accumulation. Total RNA was extracted from 0.2 g (fresh weight) tobacco leaves at different times postinoculation (23) and resuspended in 100 μl of distilled water. RNA was quantified by dot blot hybridization with 5′ 32P-labeled oligonucleotide probes specific for each TMV genotype and densitometry analysis of the hybridization signal, as described earlier (8, 9). In each blot, internal standards for each genotype were included as a twofold dilution series of purified RNA (2 to 0.015 μg) in nucleic acid extracts from noninoculated tobacco plants. Different amounts of nucleic acid extracts from each sample to be analyzed were blotted to ensure that the hybridization signal was in the linear portion of the curve RNA concentration-hybridization signal. The genotype-specific oligonucleotide probes used for these analyses were as follows: (i) for TMV wt, 5′-TCTATTGGGTCGGCC-3′; (ii) for mutant P20L in a single infection or in a mixed infection with the wt, 5′-TCTATTAGGTCGGCC3′; (iii) for mutant P20L in mixed infection with 20/72, 5′-CGCATTGTACCTGTA-3′; and (iv) for mutant 20/72, 5′-CGCATTGAACCTGTA-3′. (Nucleotides complementary to those mutated with respect to the wt sequence are underlined.) The hybridization temperatures were 43, 37, 35, and 38°C for the four probes, respectively. All hybridizations were done overnight in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt mixture, 0.1% sodium dodecyl sulfate, and yeast tRNA at 250 μg/ml. After exposure of the X-ray film, densitometry was done by using the 1.52 version of NIH Image package (W. Rasband, NIH Research Service Branch, NIMH, Bethesda, Md.) (20). Detection of virus accumulation with this system had a lower limit of 0.001 mg of viral RNA per g (fresh weight) of leaf.
Comparisons of virus infectivity and accumulation were done by using the nonparametric Wilcoxon signed rank test (33).
RESULTS
Colonization of tobacco plants by wt and mutant TMV in single and double infections.
To analyze the progress of infection in Samsun tobacco of three TMV genotypes, plants were inoculated at the five-to-six leaf stage by gently rubbing a suspension of virus particles on the younger expanded leaf. The inoculum was 300 ng of virus particles per half leaf. This inoculum dose was chosen on the basis of single-lesion infectivity experiments in which it was shown that the number of necrotic local lesions in Xanthi-nc tobacco plants did not differ significantly for the three TMV genotypes at 250 ng/half leaf (the mean values ± the standard errors of four replicates were 201 ± 103, 162 ± 101, and 174 ± 46 for wt, P20L, and 20/72, respectively; P > 0.1 for all comparisons as determined by a Wilcoxon signed-rank test) when it reached saturation. Hence, at 300 ng per half leaf all three genotypes should be similarly infectious.
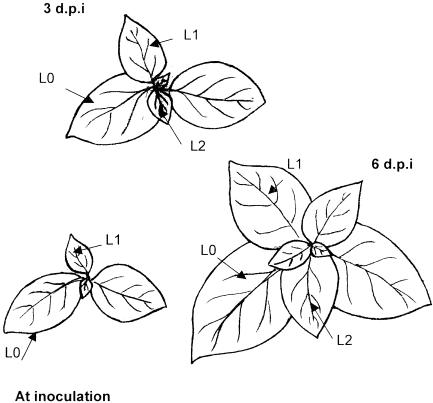
At 3 days postinoculation (dpi) no symptoms were detected in plants inoculated by any of the three TMV genotypes. Samples for analysis of virus accumulation were taken from the inoculated leaf (L0); from the second leaf above L0, which was starting to expand when plants were inoculated (L1); and from the third leaf above L0, which was starting expansion at 3 dpi (L2) (Fig. 1). Virus accumulation was estimated for each genotype separately by the quantitative dot blot hybridization analysis of virus RNA in nucleic acid extracts from leaves (Fig. 2). At 3 dpi, virus accumulation was detected only in L0 for all three genotypes. TMV-wt and P20L accumulated to similar levels (P > 0.1), whereas 20/72 accumulated to lower levels than wt and P20L (P < 0.05) (Fig. 1 and Table 1). Symptoms of TMV infection at 6 dpi were apparent in the basal portion of the leaf lamina in L1 and in the whole lamina of L2 and all younger leaves. Hence, L1 and L2 are the first and second leaves, respectively, to become systemically infected, and the leaf immediately above the inoculated one does not become infected. Analysis of TMV accumulation in systemically infected leaves indicated that all three genotypes had become systemic, but virus accumulation differed significantly for each of them (P ≤ 0.01), being highest for TMV wt and lowest for the double mutant 20/72 (Table 1).
FIG. 1.
Schematic representation of a tobacco plant at inoculation, at 3 dpi, and at harvest time (6 dpi). Leaves L0, L1, and L2 are indicated.
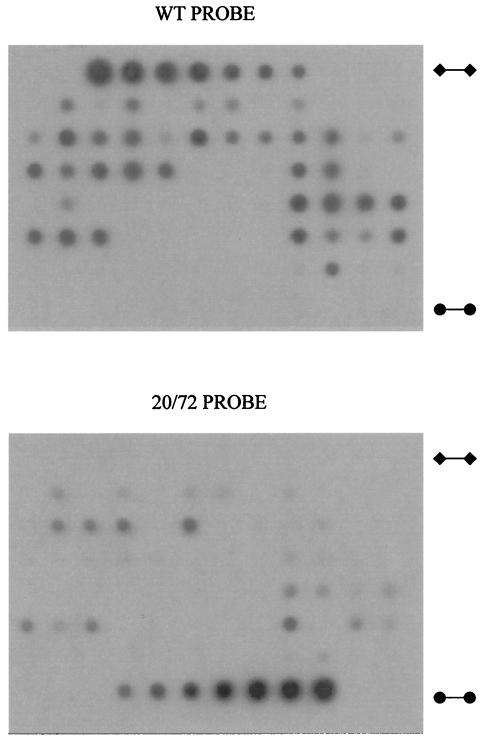
FIG. 2.
Quantification of TMV genotypes by dot blot hybridization. RNA extracts from 72 leaves were loaded together with TMV-wt (⧫—⧫) and 20/72 (•—•) standards (2 to 0.015 μg in a series of 12 dilutions) and hybridized with 32P-labeled oligonucleotide probes specific for wt (WT PROBE) and 20/72 (20/72 PROBE). The absorbances were determined by densitometry of blots and plotted against the standards.
TABLE 1.
Accumulation of three TMV genotypes in Samsun tobacco plants
| TMV genotype | Mean virus RNA accumulationa ± SE at:
|
||
|---|---|---|---|
| 3 dpi
|
6 dpi (upper leaf) | ||
| Inoculated leaf | Upper leaf | ||
| wt | 0.010 ± 0.003 | 0.000 ± 0.000 | 0.293 ± 0.085 |
| P20L | 0.014 ± 0.003 | 0.000 ± 0.000 | 0.032 ± 0.014 |
| 20/72 | 0.005 ± 0.001 | 0.000 ± 0.000 | 0.017 ± 0.009 |
Expressed as milligrams of viral RNA per gram (fresh weight) of leaf. Data are means ± standard error from five replicate plants. The youngest expanded leaf (upper leaf) was analyzed where indicated: L1 at 3 dpi and L2 at 6 dpi.
Based on these results, double inoculations were carried out by applying equal amounts (600 ng/leaf) of each TMV genotype to the same leaf in the three combinations wt-P20L, wt-20/72, and P20L-20/72. According to the infectivity assays described above, at this inoculum dose the number of infection foci initiated by each genotype should be similar, and the frequency of each genotype in the infecting population should be ca. 0.5. It should be noted that infection of a leaf after mechanical inoculation is started at single epidermal cells from where virus infection spreads to adjacent cells, giving rise to infection foci (21, 22). In a hypersensitive host such as Xanthi-nc tobacco, these infection foci are apparent as necrotic local lesions. Hence, inoculations carried out at inoculum doses that result in a similar number of necrotic local lesions result in a similar number of infection foci; thus, in an initial frequency of each genotype of ca. 0.5. As infection of the inoculated leaf progresses, virus replication of the infected cells and movement of infection to neighboring cells may be more efficient for the different genotypes, resulting in higher virus accumulation, as is the case for the wt compared to either mutant (Tables 1 and 2). Inoculum doses in the double inoculations were twice those in the single-inoculation experiments described above to ensure saturation of the (finite) possible infection sites. Two hundred Samsun plants were inoculated with each virus combination. Virus accumulation was analyzed in L0, L1, and L2 at 6 dpi. Virus accumulation was always less in L1 than in L0 and L2, a finding in agreement with the partial infection of L1 observed in single infections. TMV-wt accumulated to higher levels than either of the mutants in all three leaves, and the single mutant P20L accumulated to higher levels than the double mutant 20/72 (Table 2). These results show that selection occurred in each leaf of doubly infected plants, with the wt being favored over the mutants and the single mutant being favored over the double mutant.
TABLE 2.
Average frequency and RNA accumulation of TMV genotypes in mixed infections in Samsun tobacco plants
| TMV genotype(s) | Avg frequency (mean RNA accumulation ± SE)a on leaf:
|
||
|---|---|---|---|
| L0 | L1 | L2 | |
| wt + P20L | |||
| wt | 0.80 (0.026 ± 0.003) | 0.83 (0.009 ± 0.002) | 0.94 (0.038 ± 0.004) |
| P20L | 0.20 (0.006 ± 0.001) | 0.17 (0.001 ± 0.0001) | 0.06 (0.001 ± 0.0002) |
| wt + 20/72 | |||
| wt | 0.96 (0.082 ± 0.011) | 0.85 (0.011 ± 0.002) | 0.94 (0.093 ± 0.011) |
| 20/72 | 0.04 (0.001 ± 0.0001) | 0.15 (0.00017 ± 0.00001) | 0.06 (0.00044 ± 0.0001) |
| P20L + 20/72 | |||
| P20L | 0.80 (0.005 ± 0.0003) | 0.75 (0.0006 ± 0.00001) | 0.80 (0.003 ± 0.0003) |
| 20/72 | 0.20 (0.001 ± 0.0001) | 0.25 (0.0001 ± 0.00001) | 0.20 (0.001 ± 0.0001) |
The frequency is expressed as milligrams of viral RNA per gram (fresh weight) of leaf. Data are average frequencies for each genotype and are calculated only for leaves infected by at least one of the two inoculated TMV genotypes. The mean ± the standard error of RNA accumulation at 6 dpi from 200 samples is shown in parentheses. L0, inoculated leaf; L1, second leaf above the inoculated leaf; L2, third leaf above the inoculated leaf.
For all three genotype combinations, there was a fraction of infected leaves in which only one of the two genotypes was detected (Table 3).
TABLE 3.
Frequency of leaves with single infections from Samsun tobacco plants inoculated with mixtures of different TMV genotypes
| TMV genotype(s) | Frequencya on leaf:
|
||
|---|---|---|---|
| L0 | L1 | L2 | |
| wt + P20L | |||
| wt | 0.12 (23/193) | 0.57 (83/145) | 0.38 (72/188) |
| P20L | 0.02 (4/193) | 0.12 (17/145) | 0.02 (3/188) |
| wt + 20/72 | |||
| wt | 0.30 (59/196) | 0.68 (79/114) | 0.54 (93/172) |
| 20/72 | 0.01 (2/196) | 0.12 (14/114) | 0.03 (6/172) |
| P20L + 20/72 | |||
| P20L | 0.24 (46/191) | 0.59 (58/99) | 0.40 (66/166) |
| 20/72 | 0.01 (2/191) | 0.16 (16/99) | 0.04 (7/166) |
Data are frequencies of leaves infected only by the indicated genotype over the total number of leaves infected by either of the two genotypes in each combination (these numbers are shown in parentheses). Two hundred plants were inoculated with each virus combination. L0, inoculated leaf; L1, second leaf above the inoculated leaf; L2, third leaf above the inoculated leaf.
Estimation of the number of founders for populations in each infected leaf.
A simple model was developed to estimate, from the experiment above, the effective size of the population that starts an infection in a systemically infected leaf, i.e., the effective number of founders Nef. Note that in the effective size of the founder population different factors related to the infection process are taken into account, such as the number of infection events and the number of virus particles involved in each infection event. The model is based on the proportion of leaves infected with only a single genotype when the plant was inoculated with a combination of two genotypes. For each pair of genotypes A-B, the probability that one leaf is infected only by genotype A, P0B, given that the source leaf is infected by genotypes A and B with frequencies psA and psB, is equal to the Nef-th power of the frequency of variant A in the source leaf, psA
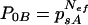 |
(1) |
Equation 1 expresses the probability that the Nef virus particles infecting a leaf are all of genotype A when drawn independently from a source in which genotype A has the frequency psA. Nef, the effective number of founders, can be estimated from this expression by estimating psA and P0B experimentally.
We should point that Nef is not the effective population number in the leaf but the number of equally fit simultaneously infecting particles that explains the distribution of singly infected leaves in Table 3. Because genotypes were not equally fit but selection for one TMV genotype had occurred in each TMV combination in coinfected leaves (Table 2), the probability of no infection by the less-fit genotype can be overestimated. This is because in coinfected leaves the less-fit genotype can be lost by selection before observation. Hence, the effective founder number should lie between that in equation (1) and that in the symmetrical expression:
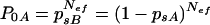 |
(2) |
where P0A is the probability that a leaf is infected only by genotype B.
Thus, considering A as the most frequent genotype, equations 1 and 2 provide upper and lower limits, respectively, for the estimated effective number of founders Nef.
The application of this procedure to estimate Nef from the experiment described above presents two difficulties. First, the inoculum source for the systemic infection of leaves L1 and L2 is not known a priori. However, because at 3dpi virus accumulation was only detected in leaf L0, whereas at 6 dpi virus accumulation was detected both in the basal portion of leaf L1 and in the whole of leaf L2 (Fig. 1 and Table 1), we assume that the contribution of L1 to the infection of L2 is negligible and that L0 is the source of inoculum for both L1 and L2. Second, the exact moment at which systemic movement from L0 to L1 and L2 occurs and how long it takes is not known; hence, the frequency of genotypes A and B in the source leaf at the moment of systemic movement is unknown. However, because systemic movement necessarily occurred between the inoculation time (time zero) and 6 dpi, the frequencies of genotypes A and B at the moment of systemic movement must be between the frequency at time zero (i.e., 0.5 for both) and that at 6 dpi. Therefore, the estimate of Nef should be between the estimates from these two extreme values of psA. In this reasoning we are assuming that genotype A is the most frequent genotype during this entire time period, which is a reasonable assumption supported by the data in Tables 1 and 2.
With these assumptions the effective number of founders for L1 and L2 was estimated from the data on the average frequency of each TMV genotype (Table 2) and on the frequency of leaves infected by only one genotype (Table 3) by using the model described above. Note that frequency data in Table 2 reflect the possibility that for individual leaves the frequency of one TMV genotype may be 1.0 (i.e., the leaf was infected only by that genotype) in spite of low virus accumulation. The estimated effective numbers of founders are shown in Table 4. The number of founders for the systemically infected leaves L1 and L2 are small, in the order of units. Note that the range of the estimates is small when either equation 1 or 2 is used and either of the psA values is estimated at time zero and at 6 dpi. As expected, this range increases for genotype combinations with stronger selection for one of the two genotypes (compare data for the pairs wt-P20L and wt-20/72). An important aspect to be stressed from the estimates in Table 4 is the similarity of the values obtained with the three pairs of genetic types, i.e., the consistency of the estimates.
TABLE 4.
Estimated effective sizes of the founder populations infecting the leaves of Samsun tobacco plants
| TMV genotypes | p̄sAa | Effective founder sizeb on leaf:
|
||
|---|---|---|---|---|
| L0 | L1 | L2 | ||
| wt + P20L | 0.50 | 3.1-5.6 | 0.8-3.1 | 1.4-6.0 |
| 0.80 | 1.3-2.4 | 2.6-4.2 | ||
| wt + 20/72 | 0.50 | 1.7-6.6 | 0.5-3.0 | 0.9-4.8 |
| 0.96 | 0.6-9.8 | 1.0-15.9 | ||
| P20L + 20/72 | 0.50 | 2.1-6.6 | 0.8-2.6 | 1.3-4.6 |
| 0.80 | 1.1-2.4 | 2.0-4.1 | ||
That is, the estimated frequency of the prevalent genotype in the source. The sources are leaf L0 for leaves L1 and L2 and inoculum for leaf L0.
That is, the effective size of the founder population as derived from equations 1 and 2 in the text for the indicated threshold values of p̄sA.
The model presented above can also be applied to estimate the effective number of founders in the inoculated leaf. In this case, because inoculations were done under conditions in which both TMV genotypes in each pair will initiate a similar number of infection foci, psA, in theory, will be 0.5. Table 4 shows that estimates of effective numbers of founders were again small, a result similar to those for systemically infected leaves and, again, consistent for the three pairs of genetic types. For inoculated leaves, Nef can be compared to the census number of the population that initiates the infection. We may reasonably assume that the number of infection foci in the systemic host tobacco cv. Samsun is similar to that in the local lesion host tobacco cv. Xanthi-nc. Because at the inoculum dose used the number of necrotic local lesions per leaf was 450 to 500 on the average, this should be an estimate of the census size of the founder population. Hence, data from the inoculated leaf shows that the estimated effective number of founders is about 2 orders of magnitude smaller than the census size of the infecting population.
DISCUSSION
Viruses are known to accumulate to very high numbers within their infected hosts. However, effective numbers within an infected host could be much smaller than the census. This has been analyzed in recent years for human immunodeficiency virus type 1 and most analyses, based on different models and assumptions, conclude that effective numbers are small (11, 19, 25, 30). A major reason for this could be strong population differentiation within and between infected organs (11). Population differentiation within the infected host could also result in small effective numbers for within-host plant virus populations. Systemic movement of virus infection through the vasculature is the key process in the colonization of plants by viruses. The mechanisms and dynamics of systemic movement are incompletely understood (31), but systemic movement has often been assumed to result in population bottlenecks (cf. references 13 and 29), although evidence for this is scant. It has been shown that in plant tissues infected with two similar genotypes of one virus species, most cells become infected by either one or the other genotype, which mutually exclude each other (15, 18). This results in within-tissue differentiation of virus populations, i.e., spatial structuring of the within-plant virus populations. The structure of virus populations within infected plants has been described, to our knowledge, only for Wheat streak mosaic virus (WSMV). In detailed analyses of the distribution of two genetic types of WSMV within coinfected wheat plants, Hall et al. (15, 16) presented strong evidence of genetic differentiation within leaves, as well as between leaves and tillers. They concluded that exclusion of genetically similar types within tissues and organs, and population bottlenecks during the virus infection cycle, were causes of genetic differentiation.
Here we report a quantitative estimate of the founding number of the TMV population involved in the colonization of new tobacco leaves. Our estimates derive from an experimental design in which the frequencies of two genotypes of TMV were estimated in three infected leaves of tobacco plants inoculated with combinations of TMV genotypes. From these data, the effective number of founders, Nef, was derived by using a simple probabilistic model. Nef estimates are only upper and lower limits because selection acted on the TMV genotypes, causing frequencies to change from the time of inoculation until it can be estimated empirically. Therefore, the actual frequency of each genotype in the source leaf at the moment of systemic infection was unknown. However, the range of variation of the estimates was small, and data for the two different systemically infected leaves and for the three pairs of variants were highly consistent, strongly supporting the validity of these estimates. Our results are evidence of strong population bottlenecks during systemic movement of TMV in tobacco plants. Once each new leaf is infected, population expansion would have to be huge to reach the very high census numbers reported of ca. 107 to 109 (21, 22), and during this expansion selection for the genotype that accumulates faster would occur again. This model for population dynamics during plant colonization could possibly be extended to other viruses, as suggested by analyses of WSMV within wheat plants (15, 16).
Quantitative estimates of the number of founders for systemic infection of leaves by viruses have not been reported previously, and the major contribution of the present study is to show that they are in the order of units and not in the order of hundreds or thousands. The comparison of the estimated number of founders in the inoculated leaf with the actual observed number of viruses initiating an infection, i.e., initiating a necrotic local lesion in Xanthi-nc tobacco plants, indicates a difference of about 2 orders of magnitude. A similar difference could occur in systemically infected leaves, because it has been shown that systemic infection is initiated from numerous foci where virus is unloaded along the major veins of the sink leaves (27, 32).
The effective population number is a theoretical concept that indicates the size of an ideal population that will have the same genetic variance as the observed population. Our results show that the genetic variance between leaves of the TMV population within an infected plant is as if ca. 2 to 20 virus particles were simultaneously infecting each leaf. The fact that these numbers are small indicates that strong population bottlenecks occur during the systemic infection of new leaves, which will result in subdivision of the virus population within the infected plant. It has been shown, under diverse assumptions, that population subdivision leads to a decreased effective population size (reviewed in reference 34). Hence, our results indicate that virus evolution within an infected plant cannot be explained by purely deterministic models and that random effects due to genetic drift should be taken into account.
Acknowledgments
We thank William O. Dawson and James K. Cluver for providing cDNA clones of TMV wt and mutants. We also thank Michael M. Milgroom for helpful discussions on an earlier version of the manuscript. Antolín López Quirós and Fernanda Afonso provided excellent technical assistance.
This work was in part supported by grant BIO99-1121-C02-02 from Ministerio de Educación y Cultura to F.G.-A. S.S. was supported by a fellowship from the programme Formación de Personal Investigador (AP98 03463406), Ministerio de Educación y Cultura (Spain).
REFERENCES
- 1.Bruening, G., R. N. Beachy, R. Scalla, and M. Zailtin. 1976. In vitro and in vivo translation of the ribonucleic acids of a cowpea strain of tobacco mosaic virus. Virology 71:498-517. [DOI] [PubMed] [Google Scholar]
- 2.Crow, J. F., and M. Kimura. 1970. An introduction to population genetics theory. Harper & Row, Publishers, Inc., New York, N.Y.
- 3.Culver, J. N., and W. O. Dawson. 1989. Point mutations in the coat protein gene of Tobacco mosaic virus induce hypersensitivity in Nicotiana sylvestris. Mol. Plant-Microbe Interact. 2:209-213. [Google Scholar]
- 4.Dawson, W. O., D. L. Beck, D. A. Knorr, and G. L. Grantham. 1986. cDNA cloning of the complete genome of tobacco mosaic virus and production of infectious transcripts. Proc. Natl. Acad. Sci. USA 83:1832-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 6.Domingo, E., E. Escarmís, L. Menéndez-Arias, and J. J. Holland. 1999. Viral quasispecies and fitness variations, p. 141-162. In E. Domingo, R. Webster, and J. J. Holland (ed.), Origin and evolution of viruses. Academic Press, Inc., San Diego, Calif.
- 7.Domingo, E., R. Webster, and J. J. Holland. 1999. Origin and evolution of viruses. Academic Press, Inc., San Diego, Calif.
- 8.Escriu, F., A. Fraile, and F. García-Arenal. 2000. Evolution of virulence in natural populations of the satellite RNA of Cucumber mosaic virus. Phytopathology 90:480-485. [DOI] [PubMed] [Google Scholar]
- 9.Fraile, A., F. Escriu, M. A. Aranda, J. M. Malpica, A. J. Gibbs, and F. García-Arenal. 1997. A century of tobamovirus evolution in an Australian population of Nicotiana glauca. J. Virol. 71:8316-8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frankham, R. 1995. Effective population size/adult population size ratios in wildlife: a review. Genet. Res. 66:95-107. [DOI] [PubMed] [Google Scholar]
- 11.Frost, S. D. W., M.-J. Dumaurier, S. Wain-Hobson, and A. J. Leigh Brown. 2001. Genetic drift and within host metapopulation dynamics of HIV-1 infection. Proc. Natl. Acad. Sci. USA 98:6975-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furumoto, W., and R. Mickey. 1967. A mathematical model for the infectivity-dilution curve of tobacco mosaic virus: experimental tests. Virology 32:216-223. [DOI] [PubMed] [Google Scholar]
- 13.García-Arenal, F., A. Fraile, and J. M. Malpica. 2001. Variability and genetic structure of plant virus populations. Annu. Rev. Phytopathol. 39:157-186. [DOI] [PubMed] [Google Scholar]
- 14.Gibbs, A., C. H. Calisher, and F. García-Arenal. 1995. Molecular basis of virus evolution. Cambridge University Press, Cambridge, United Kingdom.
- 15.Hall, J. S., R. French, G. L. Hein, T. J. Morris, and D. C. Stenger. 2001. Three distinct mechanisms facilitate genetic isolation of sympatric Wheat streak mosaic virus lineages. Virology 282:230-236. [DOI] [PubMed] [Google Scholar]
- 16.Hall, J. S., R. French, T. J. Morris, and D. C. Stenger. 2002. Structure and temporal dynamics of populations within Wheat streak mosaic virus isolates. J. Virol. 75:10231-10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison, B. D. 1981. Plant virus ecology: ingredients, interactions and environmental influences. Ann. Appl. Biol. 99:195-209. [Google Scholar]
- 18.Hull, R., and A. Plaskitt. 1970. Electron microscopy of the behavior of two strains of alfalfa mosaic virus in mixed infections. Virology 42:773-776. [DOI] [PubMed] [Google Scholar]
- 19.Leigh-Brown, A. J. 1997. Analysis of HIV-1 env gene sequences reveals evidence for a low effective number in the viral population. Proc. Natl. Acad. Sci. USA 94:1862-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lennard, P. R. 1990. Image analysis for all. Nature 347:103-104. [DOI] [PubMed] [Google Scholar]
- 21.Malpica, J. M., A. Fraile, I. Moreno, C. I. Obies, J. W. Drake, and F. García-Arenal. 2002. The rate and character of spontaneous mutation in an RNA virus. Genetics 162:1505-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthews, R. E. F. 1970. Plant virology, p. 201-204. Academic Press, Inc., San Diego, Calif.
- 23.Moriones, E., I. Díaz, E. Rodríguez-Cerezo, A. Fraile, and F. García-Arenal. 1992. Differential interactions among strains of tomato aspermy virus and satellite RNAs of cucumber mosaic virus. Virology 186:475-480. [DOI] [PubMed] [Google Scholar]
- 24.Moya, A., E. Rodríguez-Cerezo, and F. García-Arenal. 1993. Genetic structure of natural populations of the plant RNA virus tobacco mild green mosaic virus. Mol. Biol. Evol. 10:449-456. [Google Scholar]
- 25.Nijhuis, M., C. A. B. Boucher, P. Scipper, T. Leitner, R. Schuurman, and J. Albert. 1998. Stochastic processes strongly influence HIV-1 evolution during suboptimal protease-inhibitor therapy. Proc. Natl. Acad. Sci. USA 95:14441-14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pirone, T. P., and D. W. Thornbury. 1988. Quantity of virus required for aphid transmission of a potyvirus. Phytopathology 78:104-107. [Google Scholar]
- 27.Roberts, A. G., S. Santa Cruz, I. M. Roberts, D. A. M. Prior, R. Turgeon, and K. J. Oparka. 1997. Phloem unloading in sink leaves of Nicotiana benthamiana: comparison of a fluorescent solute with a fluorescent virus. Plant Cell 9:1381-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodrigo, A., E. G. Shpaer, E. L. Delwart, A. K. N. Iversen, M. V. Gallo, J. Brojatsch, M. S. Hirsch, B. D. Walker, and J. I. Mullins. 1999. Coalescent estimates of HIV-1 generation time in vivo. Proc. Natl. Acad. Sci. USA 96:2187-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roossinck, M. J. 1997. Mechanisms of plant virus evolution. Annu. Rev. Phytopathol. 35:191-209. [DOI] [PubMed] [Google Scholar]
- 30.Rouzine, I. M., and J. M. Coffin. 1999. Linkage disequilibrium test implies a large effective population number for HIV in vivo. Proc. Natl. Acad. Sci. USA 96:10758-10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz-Medrano, R., B. Xoconostle-Cazares, and W. J. Lucas. 2001. The phloem as a conduit for inter-organ communication. Curr. Opin. Plant Biol. 4:202-209. [DOI] [PubMed] [Google Scholar]
- 32.Santa Cruz, S., A. G. Roberts, D. A. M. Prior, S. Chapman, and K. J. Oparka. 1997. Cell-to-cell and phloem-mediated transport of potato virus X: the role of virions. Plant Cell 10:495-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokal, R. R., and F. J. Rohlf. 1995. Biometry, p. 440-444. W. H. Freeman & Co., New York, N.Y.
- 34.Wang, J., and A. Caballero. 1999. Developments in predicting the effective size of subdivided populations. Heredity 82:212-226. [Google Scholar]