Abstract
The structure of swine vesicular disease virus (SVDV) was solved and refined at a 3.0-Å resolution by X-ray crystallography to gain information about the role of sequence changes that occurred as this virus evolved from the parental human pathogen coxsackievirus B5 (CVB5). These amino acid substitutions can be clustered in five distinct regions: (i) the antigenic sites, (ii) the hydrophobic pocket of the VP1 β-sandwich, (iii) the putative CAR binding site, (iv) the putative heparan sulfate binding site, and (v) the fivefold axis. The VP1 pocket is occupied by a branched pocket factor, apparently different from that present in the closely related virus CVB3 and in other picornaviruses. This finding may be relevant for the design of new antiviral compounds against this site. Density consistent with the presence of ions was observed on the fivefold and threefold axes. The structure also provided an accurate description of the putative receptor binding sites.
Swine vesicular disease virus (SVDV) is a member of the Enterovirus genus, within the Picornaviridae family. It causes a highly contagious disease of pigs that spreads rapidly by direct contact among infected animals and by environmental contamination (12). It is not a severe disease, although the similarity between the lesions it causes and those produced by foot-and-mouth disease (FMD) has led to it being considered economically important, and routine surveillance is maintained in European countries (reviewed in reference 13). The disease was first described in Italy in 1966 (38), and numerous outbreaks have since occurred in Europe and Asia.
SVDV is a porcine variant of the human pathogen coxsackievirus B5 (CVB5) (7, 20, 47, 23, 30, 54). The antigenic and molecular relationships between these two viruses suggest that SVDV is a genetic sublineage of CVB5 that evolved between 1945 and 1965, after the transfer of CVB5 from humans to pigs (55).
Despite consisting of a single serotype, SVDV shows a high genetic and antigenic variability. More than 100 SVDV isolates from Europe and Japan have been characterized by analysis of nucleotide sequences of the capsid proteins and by determining their antigenic profile. These isolates are classified in four groups (6). Groups 1 and 2 are more closely related to each other than the more recent groups 3 and 4, which may represent two independent SVDV introductions in Europe from Asia, where SVDV strains have been isolated almost continually since 1970 (55). The fourth group includes viruses isolated between 1987 and 1994 from Romania, The Netherlands, Italy, and Spain.
The variability of SVDV also applies to its pathogenicity. Nonpathogenic isolates cause a nonsymptomatic disease, while virulent isolates produce typical lesions in infected pigs. Two amino acid changes, located in positions 1132, within the EF-loop of VP1 (The capsid residues are numbered with the first digit indicating the viral protein) and 20 of the protease 2A, have been shown to account for the differences in virulence observed between the pathogenic J1′73 and the nonpathogenic H7/3′76 SVDV isolates. The position 2A-20 appears to be the dominant determinant (29) and acts by modifying the efficacy of 2A to cleave the translation initiation factor eIF4GI, thus affecting the cap-dependent protein synthesis (46). On the other hand, the mechanism of attenuation underlying the second determinant, located in the capsid at position 1132, remains unknown.
Picornavirus particles are composed of 60 copies each of the four capsid proteins VP1 through VP4, which enclose a single-stranded messenger positive-sense RNA genome of about 7,400 nucleotides. Whereas VP1, VP2, and VP3 are exposed at the viral surface, VP4 is in intimate contact with the RNA and thus not accessible from the outer shell surface in the intact virions. A prominent feature of the capsid surface in rhino- and enteroviruses is a deep cleft around the fivefold axis of icosahedral symmetry, called the canyon, which has been proposed to be the receptor binding site for many picornaviruses (44). This prediction was unequivocally demonstrated by cryo-electron microscopy (cryo-EM) for the major group rhinoviruses and for enteroviruses, such as poliovirus and coxsackievirus, that use receptors belonging to the immunoglobulin superfamily (reviewed in reference 45). An additional structural feature observed in all enteroviruses examined to date is a hydrophobic cavity, at the heart of VP1 beneath the canyon, that contains cellularly derived lipids, known as “pocket factors,” which would stabilize the virus during its spread from cell to cell and would always be expelled upon interaction with the receptor. Antiviral compounds have been discovered that bind tightly to the VP1 pocket replacing the pocket factor and stabilizing the capsids to such an extent that uncoating is blocked (1, 19, 48).
The main cellular receptor of coxsackie B viruses is the coxsackie- and adenovirus receptor (CAR), a 46-kDa membrane glycoprotein (3, 36, 52) whereas decay-accelerating factor (DAF; also known as CD55), a ubiquitous phosphatidylinositol anchored membrane protein regulating homologous complement lysis, is also used as cellular coreceptor for CVB1, CVB3, and CVB5 (3). SVDV retains the ability to bind CAR. The oldest SVDV isolates seem able to infect human cells in a similar way as the human homologue CVB5. The SVDV UK-72 strain infects transfected CHO cells expressing human CAR and human HeLa cells, and the infection is blocked by both anti-human CAR and DAF antibodies (36). However, the new SVDV isolate SPA/2/'93 has lost its ability to bind DAF, although it retains the ability to bind CAR (M. A. Jimenez-Clavero et al., unpublished data).
Little is known on the issue of SVDV receptor usage in swine, its natural host, since neither porcine CAR nor porcine DAF have been studied as possible receptors of SVDV. Interestingly, it has recently been reported that the human coxsackieviruses that use human DAF as receptor infect pig cells using pig CAR but do not bind pig DAF in vitro (50).
On the other hand, there is evidence (E. Escribano-Romero et al., unpublished data) that SVDV strongly interacts with heparan sulfate (HS) proteoglycan (HSPG), an abundant component of the cell surface. This interaction may represent a first step in the process of binding of SVDV to cells, which, as proposed for other HS binding viruses (17) may well involve an initial attachment with HS, facilitating a second, interaction with one receptor(s) mediating virus internalization.
The SVDV crystal structure, solved and refined to a 3.0-Å resolution, provides an accurate view of this new emerged virus, serologically and phylogenetically related to CVB5. The structure allowed mapping of the surface changes occurring during the adaptation of CVB5 to infect pigs and provided some details about the nature of an unusual lipid occupying the hydrophobic pocket. An accurate description of the putative receptor binding sites is also provided.
MATERIALS AND METHODS
Crystallization and data collection.
SVDV (isolate SPA/2/'93) was prepared and crystallized as described elsewhere (26). Three crystal forms were obtained depending on the crystallization conditions. Orthorhombic crystals, with space group I222 and unit cell parameters of a = 318.3 Å, b = 349.9 Å, and c = 371.7 Å, diffracted beyond a 3.0-Å resolution, were used for the structure determination presented in this work. A data set, with 70% completeness, was collected using synchrotron radiation (ESRF Grenoble; beamlines ID14.4 and ID29) from three crystals flash cooled to liquid-nitrogen temperature (Table 1). Crystals were cryoprotected by soaking them for 1 min in a solution containing 30% (vol/vol) glycerol in the crystallization buffer (1.2 M ammonium sulfate and 0.1 M sodium or potassium phosphate, pH 6.5 [26]). Diffraction images were processed using the DENZO package (41).
TABLE 1.
Data collection and refinement statistics of SVDV
| Parameter | Value |
|---|---|
| Data collection | |
| Dmin (Å) | 3.0 |
| No. of crystals | 3 |
| No. of reflections | 459,326 |
| No. of unique reflections | 288,614 |
| Rmerge (%) | 10.0 (19.1)a |
| Completeness (%) | 70 (51.0)a |
| Refinement | |
| Data range (Å) | 30-3.0 |
| No. of reflections [F > 2σ (F)] | 266,789 |
| Rfactor (%) | 23.5 |
| Protein model | |
| Residues | 763 |
| Atoms | 5,905 |
| Nonprotein molecules | |
| Water | 50 |
| Pocket factor | 1 |
| Ions | 6 |
| Myristic acid | 1 |
| Rmsd from ideality for bonds (Å) | 0.012 |
| Rmsd from ideality for angles (°) | 1.9 |
| Avg temp factors (Å2) | |
| All protein atoms | 28.6 |
| Main chain | 28.5 |
| Side chain | 28.7 |
| Solvent molecules | 38.8 |
| Ions | 35.0 |
| Pocket factor | 48.6 |
| Myristic acid | 66.7 |
In brackets last resolution shell (3.1 to 3.0 Å).
Structure determination.
Packing considerations indicated that the crystal cell contained two SVDV particles. Hence, the centers of each particle occupied a special 222 position, where three icosahedral twofold axes coincided with three crystallographic axes. Therefore, the particles should be positioned in one of only two possible orientations, which are related by 90° rotation around any of the three orthogonal twofold axes. The proper positioning of the coordinates of human CVB3 (37), used as a molecular model, gave an R-factor value of 38.4% in the resolution range of 15 to 3.5 Å.
The correctly oriented and positioned CVB3 model was used to calculate a set of phases to a 3.5-Å resolution that were used as starting point for extensive cycles of 15-fold averaging and phase extension with program DM (11). The quality of the electron density map, at a 3.0-Å resolution, allowed the determination of most of the sequence and structural differences between SVDV and CVB3 (Fig. 1). An atomic model was built into the 3.0-Å averaged density from the SVDV amino acid sequence (GenBank accession no. AY157625) using the graphic program O (27). The preliminary SVDV model was refined with the programs X-PLOR (version 3.0; Yale University, New Haven, Conn.) and CNS (8) using data from 20- to 3.0-Å resolution. The refined structure was used to determine phases that were combined with the observed amplitudes to calculate a new averaged electron density map. The last cycle of averaging had a correlation coefficient of 93% and an R factor of 20%. In addition, 15-fold-averaged 2Fo-Fc and Fo-Fc difference maps were calculated at this stage using the program GAP (J. M. Grimes and D. Stuart, unpublished data). The final maps showed new structural features, in particular well-defined solvent molecules and the accurate conformation of a number of side chains. Iterative positional and B-factor refinement using CNS with noncrystallographic symmetry constraints and bulk solvent correction was combined with manual model rebuilding. The final R factor for the model was 23.5%, with 88% of the residues inside the most favored region in the Ramachandran plot (Table 1). The program PROCHECK (33) was used to monitor the geometry of the model at various stages in refinement. An elongated electron density, bifurcated at one end, was observed in the hydrophobic pocket of VP1 and modeled as a ceramide (Fig. 2). Fifty well-ordered water molecules and six putative ions, five in the fivefold axis and one in the threefold axis, were included in the final model (Table 1). The solvent molecules were incorporated only when hydrogen bonded and the corresponding electron density had the same level as neighbor protein residues. Coordinates have been deposited in the Protein Data Bank (PDB code: 1MQT).
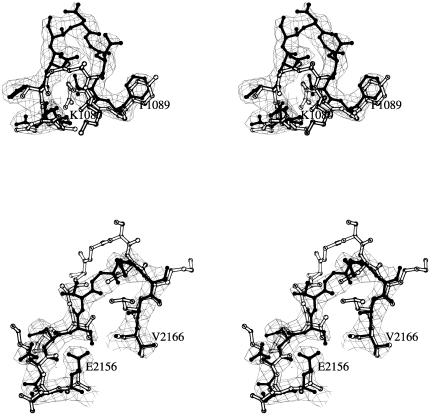
FIG. 1.
Stereo views comparing two external loops from SVDV (black) with the corresponding regions in CVB3 (white). (A) The BC loop of VP1 that contributes to antigenic site 1 in SVDV (residues 1080 to 1090) is three residues longer and more exposed than the corresponding segment of CVB3. (B) The EF loop of VP2 (residues 2156 to 2166) that contributes to antigenic site 2 is two residues shorter in SVDV than the corresponding loop of CVB3. The two averaged 2Fo-Fc maps covering the BC loop and β-knob regions are shown as chicken boxes.
FIG. 2.
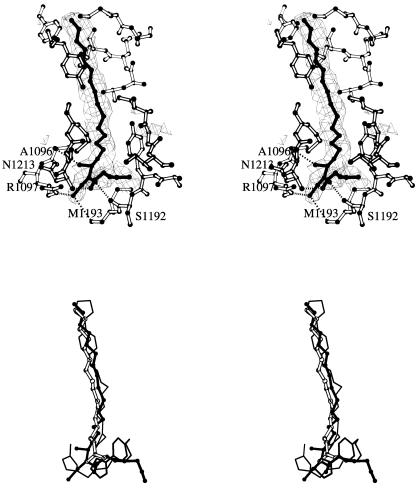
Stereo views of the hydrophobic pocket of VP1 and the pocket factor. (A) Averaged Fo-Fc map showing the branched electron density corresponding to a putative pocket factor that was interpreted as a ceramide. VP1 residues in the vicinity of the pocket are explicitly shown. (B) Superimposition of the ceramide molecule identified in the structures of SVDV (solid balls and sticks), the palmitic acid of the CVB3 structure (open balls and sticks) and the two WIN51711 molecules found in the structure of CVA9 (thin lines). The sphingosine moiety of the ceramide superimposes on the palmitic acid of CVB3 and one of the WIN51711 molecules. The fatty acid linked to the sphingosine moiety is partially coincident with the second WIN51711 molecule.
RESULTS AND DISCUSSION
Overall structure.
The SVDV crystals used in this study were orthorhombic and belonged to space group I222. The crystallographic asymmetric unit contained one quarter of a virus particle (26). The final molecular model includes 763 amino acid residues corresponding to viral proteins VP1, VP2, VP3, and VP4; a ceramide located within the hydrophobic pocket of VP1; 50 solvent molecules; and six ions in the fivefold and threefold axes (Table 1; see below). The quality of the final averaged electron density maps at a 3.0-Å resolution allowed the recognition of most of the SVDV amino acid sequence without ambiguities (Fig. 1). Three protein regions with poor density were not included in the present model: (i) residues 1001 to 1012 at the N terminus of VP1, (ii) the eight N-terminal residues of VP2, and (iii) residues 4011 to 4024 of VP4. These three protein regions were poorly defined in most picornaviral structures. Electron density was also weak for residues 1130 to 1134 on the outside of the DE loop surrounding the icosahedral fivefold axis. The root mean square deviations (RMSD) of the final model were 0.012 Å and 1.9° with respect to the ideal bond lengths and angles, respectively (Table 1). The main chain conformational angles are located within or in the close vicinity of the energetically most favored regions of the Ramachandran plot.
The SVDV structure determined is, as expected, closely related to that described for other enteroviruses. The conserved β-sandwich motif, observed in all picornaviruses, is well preserved in SVDV proteins VP1, VP2, and VP3. The most similar virus to SVDV, of known three-dimensional structure, is CVB3 (37). The RMSD for the superimposition of Cα atoms from 751 structurally equivalent residues in the two viruses is only 0.41 Å. A high structural similarity was also found between SVDV and CVA9 (22) or echovirus 1 (15), with RMSD values of only 0.57 and 0.59 Å, respectively, matching 751 residues in both superimpositions. Structural similarity is lower between SVDV and other enteroviruses such as poliovirus (14), with a RMSD for the Cα atoms from the β-sandwiches of 0.98 Å.
The structure of the external loops connecting the β-strands always shows larger differences. In particular, the BC and DE loops of VP1 and the EF loop of VP2 differ from those of the CVB3 mostly in structure (Fig. 1; see below).
Like all other picornaviruses, SVDV has a myristic acid, covalently attached to the N-terminal glycine of VP4 (10). The myristoyl groups cluster around the fivefold axis just below the N-terminal VP3 β-cylinder.
Five peaks of ion-like electron density were found along the icosahedral fivefold axis. These peaks, approximately 80% of the height of the surrounding protein density, appear to make water-mediated interactions with polar residues on the DE and EF loops of VP1. The disposition of these putative ions is essentially coincident to that found in CVA9 (22). Additional spherical density, twice the height of the surrounding protein density, was also detected on the icosahedral threefold axis. The putative cation would coordinate with the side chain oxygen Oδ1 from three symmetry-related D3203 and with three water molecules at a distance of 2.3 and 2.6 Å, respectively. The cations on the fivefold and threefold axes have been described elsewhere for other picornaviruses (22, 35, 37, 56). These ions may be involved in the regulation of capsid stability.
Mapping the structural changes in the transition CVB5 to SVDV.
Multiple sequence alignments between SVDV strains representing the four antigenic groups and the parental CVB5 revealed a number of amino acid changes on the viral surface during the evolution of CVB5 to SVDV. Isolate SPA/2/'93, which belongs to antigenic group 4 of SVDV showed 39 amino acid changes in structural proteins with respect the parental CVB5 (Table 2; Fig. 3). Thirteen of these changes were observed between SPA/2/'93 and the oldest SVDV strains (Table 2), some of which may be linked to the loss of the ability of SPA/2/'93 to bind DAF. However, most of these changes occurred in regions of considerable variation among SVDV isolates and map into an extended surface area, which complicates the localization of the DAF binding region.
TABLE 2.
Amino acid changes occurring in the capsid proteins VP1, VP2, and VP3, during the SVDV evolutiona
| Residue | Amino acid
|
Location | |||
|---|---|---|---|---|---|
| SVDVrecent | SVDVold | CVB5 | CVB3 | ||
| 1045 | S | S | A | A | N-terminus of VP1; virus interior |
| 1083 | D | D | G | G | B-C loop of VP1 (antigenic site 1) |
| 1084 | S | S | T | A | B-C loop of VP1 (antigenic site 1) |
| 1096 | A | T | T | P | Hydrophobic pocket of VP1 |
| 1131 | T | V/I | V | T | 5-fold axis; VP1 D-E loop |
| 1136 | T | S | S | A | 5-fold axis; VP1 D-E loop |
| 1175 | N | S | S | N | 5-fold axis; VP1 E-F loop |
| 1180 | M | M | I | M | Hydrophobic pocket of VP1 |
| 1186 | G | G | S | S | Hydrophobic pocket of VP1 |
| 1200 | R | R | K | E | Putative CAR binding site |
| 1210 | S | S | N | N | Putative CAR binding site |
| 1225 | G/Sb | D | D | A | 5-fold axis; H-I loop of VP1 |
| 1227 | G | G | S | S | 5-fold axis; H-I loop of VP1 |
| 1258 | K/Eb | Q | Q | E | Antigenic site IV/putatative HS binding site |
| 1266 | I/Vb | E/K | E | Q | Putative HS binding site |
| 1269 | S | G | G | G | Putative HS binding site |
| 1273 | S | S | G | T | C-terminus of VP1 (antigenic site 3A) |
| 1279 | T | T | A | T | C-terminus of VP1 (near site 3A) |
| 1281 | K/Rb | K/R | Q | T | C-terminus of VP1 (near site 3A) |
| 2040 | A | T | T | D | N-terminus of VP2. Virus interior |
| 2075 | G | S | S | T | B-C loop of VP2 |
| 2143 | P | P | Q | S | E-F loop of VP2 (near site 2B) |
| 2148 | K | K | N | G | E-F loop of VP2 (near site 2B) |
| 2151 | I | I | T | T | E-F loop of VP2 (antigenic site 2B) |
| 2153 | S | N | N | K | E-F loop of VP2 (antigenic site 2B) |
| 2160 | S | S | T | S | E-F loop of VP2 (antigenic site 2A) |
| 2163 | E | E | Q | N | E-F loop of VP2 (antigenic site 2A) |
| 2233 | A | T | T | P | H-I loop of VP2 (antigenic site 3B) |
| 3006 | A | S | L | N | N-terminus of VP3; virus interior |
| 3035 | D | D | A | N | N-terminus of VP3; virus interior |
| 3062 | E | V | V | T | β-B knob (antigenic site 3A) |
| 3063 | M | M | S | N | β-B knob (antigenic site 3A) |
| 3076 | P | P | A | E | B-C loop of VP3 (antigenic site 3B) |
| 3088 | T | T | I | Q | Putative CAR binding site |
| 3092 | N | N | S | S | Putative CAR binding site |
| 3108 | A | A | T | T | Canyon floor, protomer interphase |
| 3181 | M | M | V | S | Putative CAR binding site |
| 3204 | A | A | T | A | H-I loop of VP3 |
| 3235 | N | N | S | N | C-terminus of VP3 (antigenic site 3C) |
List of sequence changes identified from a multiple alignment computed with program ClustalW (52) of several SVDV isolates together with CVB5 and CVB3. The available crystallographic information from CVB3 (38) and SVDV SPA/2′/93 (this study) was also taken into account. Old SVDV (column three) includes isolates from 1966 to 1972. Recent SVDV (column two) includes isolate SPA/2′/93 and other post-1972 strains. Sequence differences between old and recent SVDV strains are shown in boldface type.
Strain that differs from the Spanish strain.
FIG. 3.
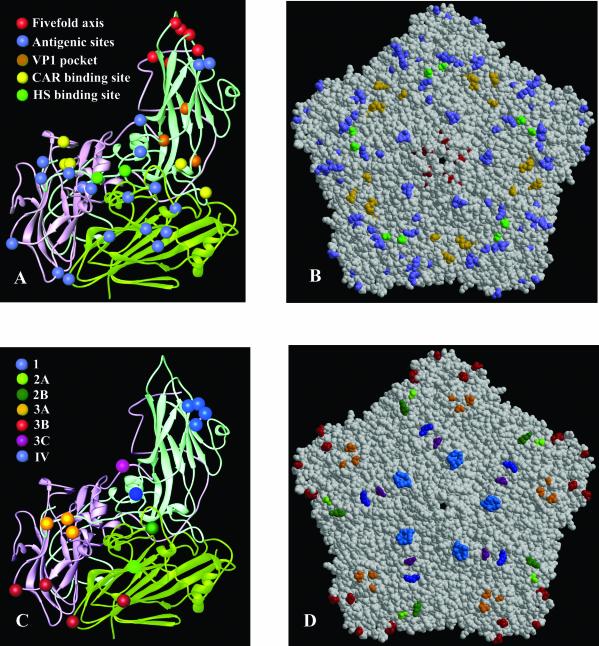
(A) Ribbon diagram showing the secondary structural elements of a viral protomer of SVDV (VP1, light blue; VP2, light green; VP3, pink). The location of the amino acid changes between CVB5 and SVDV (SPA/2/'93) is shown as differently colored spheres and explicitly indicated. (B) Space-filling model of an SVDV pentamer subunit showing the disposition of substitutions on the external part of the capsid colored as in A. (C) Ribbon diagram of an SVDV protomer highlighting the amino acids contributing to the seven antigenic sites residues as different colored spheres and explicitly indicated. (D) Space-filling model of an SVDV pentamer subunit showing the disposition of the antigenic sites colored as in C.
The structure shows that with the exception of three changes (at positions 1046, 2040, 3006, and 3035) which are located at the capsid interior, the position of all of the amino acid substitutions mapped on the surface of the virion and was clustered in five well-defined regions (Fig. 3; Table 2).
(i) The antigenic sites.
The antigenic structure of SVDV has been examined by monoclonal antibody-resistant mutant analysis (5, 28, 40). Seven neutralization sites have been defined, five of which (1, 2A, 2B, 3A, 3B, and 3C) are analogous to those described for poliovirus, the type species for the enterovirus group (28). The other two are new and specific for isolates of the most recent antigenic group of SVDV (group 4) (5, 40). The seven antigenic sites of SVDV can now be positioned on the corresponding capsid structure and compared with the arrangement of epitopes found in other picornaviruses (Fig. 3). Fifteen of the 33 amino acid changes found in the CVB5-SVDV comparison (Table 2; Fig. 3) mapped at the external loops containing the antigenic sites described in the following paragraphs.
Amino acid residues D1083 and S1084 were located within the BC loop of VP1. This loop flanks the north rim of the canyon closest to the fivefold axis and is particularly variable in both length and sequence among picornaviruses. In SVDV, it is two residues longer and more exposed than the equivalent one in CVB3 (Fig. 1). The BC loop contains a relevant antigenic site for enteroviruses and rhinoviruses. In particular, residues D1083 and S1084 are frequently substituted in mutants escaping neutralization by monoclonal antibodies and contributed to define antigenic site 1 (5) (Fig. 3), which is conserved in all the four antigenic groups of SVDV, including the oldest isolate, IT66 (5, 28, 40).
Most substitutions in VP2 (Table 2) map on the largest and most variable loop of this protein, the puff or EF loop. The SVDV puff consists of two loops. The first loop (residues 2130 to 2154) lies on the rim of the canyon closest to the twofold axes and contains the antigenic site 2B in SVDV (28, 40). The substituted residues P2143 and K2148 map within this region. The second loop (amino acids 2155 to 2170; Fig. 1) points to the outer viral surface. Replaced residues S2160 and E2163 are located within this loop, which contains the antigenic site 2A (5, 28, 40).
The substituted amino acids E3062 and M3063 are located at the center of the VP3 “knob,” which is the major surface protrusion of VP3. This region, together with the VP1 C-terminal residues 1272 and 1275 define the antigenic site 3A (28, 40). The changes at residues S1273 and T1279 map close to this antigenic region (Fig. 3). The neutralizing immunogenic site 3B, consisting of residues within the BC loop of VP3 and the C-terminal end of VP2, includes residues P3076 and A2233, respectively, the latter being substituted only in the SVDV strains that appeared after 1972 (Table 2). The last VP3 residue substituted N3235 is located at the carboxyl terminus of the protein that contains the antigenic site 3C (Fig. 3).
The amino acid K1258 (Table 2), near the VP1 C termini, maps on the antigenic site IV (Fig. 3). This epitope is specific for isolates of the most recent antigenic group of SVDV (5). In addition, it may be associated with the ability of SVDV to bind HS (see below).
All neutralization sites identified by monoclonal antibody-resistant mutant analyses are predicted to be exposed on the surface of the capsid (5, 28, 40). However, sera from SVDV-infected pigs recognize other epitopes that are located in the capsid but not exposed on its surface (24). According to a widely accepted model, the capsids of entero- and rhinoviruses undergo conformational rearrangements upon virus binding to the cell receptor (16). In this process, the capsid transforms into a structurally and antigenically altered form, the A particle. These conformationally altered particles undergo two specific changes, namely, the externalization of the N terminus of VP1 and the loss of VP4 (2, 16). This transition is an essential event in the mechanism of infection of many picornaviruses as it was well illustrated by the fact that antiviral drugs that inhibit a broad range of entero- and rhinoviruses (1) act by stabilizing native virus capsids, thus preventing these conformational changes (19, 34, 53). The relevance of the immune response to the VP1 N terminus for host protection against poliovirus has been pointed out by in vitro studies of viral neutralization using synthetic peptides (9). In addition, this region is immunogenic in humans vaccinated with an attenuated (Sabin) poliovirus vaccine (43), in rabbits inoculated with CVA9 (42), and in SVDV infected pigs (24). The substituted residue S1045 maps to this region (Table 2). It has been shown that an epitope within this region, residues 1005 to 1018 become exposed upon viral attachment to the cell surface and that in this conformation the virus can be neutralized with antibodies directed against this epitope (25).
(ii) The VP1 hydrophobic pocket.
Amino acid residues A1096, M1180, and G1186 (Table 2) map to the hydrophobic pocket, inside the β-barrel of VP1 which is often occupied by lipids of various length depending on the pocket size. In fact, the structure of SVDV showed a portion of elongated extra density, bifurcated near the entrance filling most of the pocket (Fig. 2). This extra density was only slightly weaker than that corresponding to the surrounding protein residues, suggesting that the putative pocket factor was present with a high occupancy. The size and shape of the density in the SVDV structure largely differed from the density of the CVB3 structure, which was interpreted as palmitic acid (37). The shape of the electron density and the environment around the polar end may be consistent with the tentative identification of a branched sphyngolipid modeled as a ceramide. Sphingosine has been previously identified as binding the pocket of polioviruses 1 and 3 (14). However, a single sphingosine molecule would not account for all the density observed in the SVDV structure (Fig. 2A). The orientation of the sphingosine moiety in SVDV is the same as that observed for the palmitic acid in CVB3 and for the sphingosine in polioviruses. In turn, the aliphatic tail of the 8-carbon fatty acid linked to the sphingosine runs near to the pocket entrance, but making contacts only with the hydrophobic side chains of the protein (Fig. 2). The polar head group of the lipid forms very precise stabilizing interactions with the virus; The fatty acyl oxygen contacts the main chain nitrogen of N1212 and the amide nitrogen is hydrogen bonded to the main chain oxygen of S1192. Finally, the hydroxyl group of the sphingosine moiety interacts with the side chain of R1097 (Fig. 2).
The dimensions of the hydrophobic pocket are comparable in both the SVDV and CVB3 structures, the two major differences being an alanine at position 1096 instead of a proline and a glycine, instead of a serine, at residue 1186. Both substitutions represent a small increment in the pocket size and in the hydrophobicity in the pocket entrance, which favors the presence of a larger pocket factor in SVDV. The structure shows the first identification of a branched lipid in the hydrophobic pocket of a picornavirus and may be relevant for the design of antiviral compounds to bind here. A similar situation has been described for the structure of CVA9 complexed to the antiviral drug WIN51711 (22), where two drug molecules were found together filling the VP1 pocket and the location of the second WIN molecule was partially coincident with the second branch of the lipid found in SVDV (Fig. 2).
The fact that the pocket can be occupied by lipids of various lengths and shapes in closely related viruses suggests that these viruses show different preferences for cell-specific membrane components. The apparent selectivity of pocket factors by specific picornaviruses may be a factor, in addition to the cellular receptor, determining the tissue tropism of the virus (22, 49).
(iii) The putative CAR binding site.
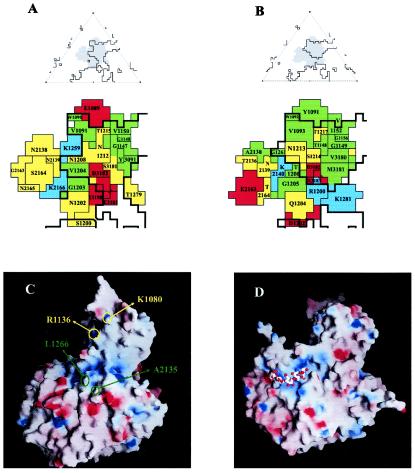
Replaced residues R1200, S1210, at the GH loop of VP1 and N3092 and M3181, located at the BC and GH loops of VP3, respectively (Table 2; Fig. 3), map at the base of the canyon which is the CAR binding site of CVB3 (21). In addition, the amino acid residues K1281 at the C termini of VP1 and E2163 within the EF loop of VP2 (Table 2) lie next to or within the CAR binding site, as revealed by the cryo-EM structure of the complex CVB3-CAR (21). The structure of this complex showed several charged residues lining the binding interface that make complementary interactions with CAR. The putative footprint of CAR on the surface of SVDV can be predicted after superimposition of the CVB3-CAR structure on the SVDV structure (Fig. 4A and B). Comparison of both footprints revealed moderate conservation of residues on the CAR recognition site in the two viruses. The distribution of hydrophobic and hydrophilic amino acids in the footprint areas is similar in both viruses, and the negatively charged area in the canyon floor (residues D3182, E3183) is mostly conserved (Fig. 4A and B). The main difference between the CVB3 and SVDV CAR footprints is the presence of a negatively charged residue at position 2163 of SVDV instead of a neutral one in CBV3. It has been reported that a variant of CVB3 with the mutation N2165-D attenuates the myocarditic phenotype (31). This mutation, adjacent to K2166 may affect the electrostatic interactions between this residue in CVB3 and two acidic residues within the BC loop of CAR (21). In addition, the parental CVB5 shows a glutamine in position 2163. In the projected SVDV CAR footprint, position 2166 is replaced by a threonine (Fig. 4B). Nevertheless, the electrostatic interaction between CVB3 and the acidic BC-loop of CAR may be maintained in SVDV by a slightly different orientation of the D1 domain of CAR onto the SVDV surface. A similar situation has been described in the interactions between the major group HRVs 14 and 16 with ICAM-1 (32). These hypotheses are consistent with the conserved CAR binding activity of SVDV strains (36).
FIG. 4.
The putative receptor binding sites in SVDV SPA/2/'93 (A) Roadmap diagram of the CAR footprint on the surface of CVB3 (PDB code 1JEW) (22). On the top is an icosahedral asymmetric unit with the footprints shown as grey shadows and the limits of the canyon shown as black outlines. On the bottom are residues defining the CAR footprint. Basic amino acids are shown in blue, acidic amino acids are shown in red, hydrophobic amino acids are shown in green, and polar amino acids in yellow. (B) Projected footprint of CAR on the SVDV surface after superimposition of the cryo-EM structure of the CVB3-CAR complex on the crystal structure of SVDV. Coloring and top-bottom representations as in A. (C) Electrostatic accessible surface representation for the biological protomer of SVDV calculated with GRASP (39). The electrostatic potential with positive charge was colored in blue and that with negative charge in red. The position of the residues (I1266 and A2135) substituted in all SVDV mutants lacking the ability to bind heparin are indicated in green. These residues are adjacent to a region of positively electrostatic charge, the candidate to be the HS binding site in SVDV. The basic residues R1236 and K1080 which would define a second putative binding site for HS proteoglycans near the fivefold axis are indicated in yellow. (D) Electrostatic accessible surface representation for the biological protomer of FMDV type O in complex, with HS (PDB entry code 1QQP) (17) shown for comparison. The bound HS molecule (represented by sticks) is located within a slight depression of positively charged residues making contacts with all three major capsid proteins. The FMDV and SVDV capsid surfaces differ from each other both in shape and in charge distributions. This may explain why the predicted HS binding site of SVDV does not overlap the FMDV HS binding site.
(iv) The putative HS binding site.
Three amino acid substitutions (positions 1258, 1266, and 1269) located near the C terminus of VP1 were detected between the oldest SVDV strains, isolates from 1966 to 1972, and the more recently isolated viruses (Table 2). All three substitutions may be related to the ability of SVDV SPA/2/'93 to bind HS. The use of HSPGs as receptors for initial cell contact of SVDV has been recently investigated (Escribano-Romero et al., unpublished data). The binding of SVDV to glycosaminoglycans was established by heparin-affinity chromatography as well as by inhibition of the virus infection, either by treatment with soluble HS or by enzymatic digestion of cell surface glycosaminoglycans. In addition, the analysis of heparin resistant SVDV mutants allowed the localization of the putative HS binding site. The amino acid sequences of the mutants revealed the same two substitutions in all SVDV variants analyzed: VP1 I1266→K and VP2 A2155→V. The structure of SVDV shows that both substitutions are close to each other and adjacent to a region of positively charged residues, characteristic of the heparin-binding domains of proteins. Three lysines K1253, K1258, and K1259 at the C termini of VP1 and one arginine R3073 at the BC loop of VP3 contributed to this region (Fig. 4C). The crystal structure of FMD virus (FMDV) serotype O in complex with HS (17) showed the HS binding site of the virus in a shallow depression of positively electrostatic charge on the virion surface contributed by the three capsid proteins VP1, VP2, and VP3 (Fig. 4D). The predicted HS binding site of SVDV maps near, but does not overlap the FMD virus HS binding site (Fig. 4C and D).
(v) Fivefold axis.
The substituted residues T1131, T1136, N1175, S1225, and G1227 form part of three loops of VP1 (DE, FG, and HI) that cluster at the fivefold axis and the particular amino acids T1136 and N1175 participate in interactions with two of the putative cations located along this axis. The VP1 DE loop (sequence 1125TQEQSTTQGQDTP1137) is the most prominent of the exterior loops about the fivefold axis and the residues at the base of the loop make extensive interactions with its icosahedral fivefold symmetry related DE loops. Because of the extensive protein-protein and protein-ion interactions, the DE loop may contribute to the stabilization of the virus pentamers and be partially responsible for the stability of the SVDV virions. In addition to the stabilizing role of the DE loop, the residue Q1132 is involved, at least in part, in the expression of pathogenic versus nonpathogenic phenotypes of SVDV (29). This loop was partially disordered in the X-ray structure (residues from 1130 to 1134 presented poor electron density). A similar situation has been described in the equivalent loop of the closely related CVB3, where two amino acids within this loop are disordered (37). It appears that the external part of the DE loop is flexible and can adopt more than one conformation. The flexibility of the loops surrounding the fivefold pore seems to contribute, at least in part, to the stability of the virus pentamers. The higher virulence observed in SVDV strains that show the substitution Q→G at position 1132 may be due to an increment in flexibility of the DE loop and thus in the stability of SVDV virions.
The third loop running close to the fivefold axis that contains the replaced residues S1225 and G1227 is the HI-loop of VP1 (Table 2; Fig. 3) This loop, together with the DE and HI loops of a neighbor protomer within a pentamer and the C terminus of VP3 define a small depression on the SVDV surface that was partially filled by a nonidentified electron density. We failed to identify the chemical moiety of this feature. The density may be due to a tight cluster of water molecules, but its general shape together with the lack of hydrogen bond interactions in the region is not consistent with it being solvent. Or else the density may be due to the presence of sucrose molecules from the purification process that occupy the crevice. The presence of three positively charged residues (K1081, R1136 and R1221 from a neighboring protomer) flanking the cavity suggest a second binding site for HSPGs within this region (Fig. 4).
Conclusions.
The structure of SVDV strain SPA/2/'93 determined at a 3.0-Å resolution allowed us to interpret most of the amino acid changes that take place in the evolution of CVB5 to SVDV. These changes are distributed into five regions: (i) the antigenic sites, (ii) the hydrophobic pocket of the VP1 β-sandwich, (iii) the putative CAR binding site, (iv) the putative HS binding site, and (v) the fivefold axis. The last three regions may be related with the adaptation of the virus to bind the porcine receptors and to the SVDV capsid stability.
The finding of a branched lipid occupying the hydrophobic pocket of VP1, apparently different from that found in other picornaviruses, offers great potential for the design of new antiviral compounds against this site.
ADDENDUM
The structure of SVDV strain UKG/27/72 was reported while this paper was under review (18). Our structure confirms and extends the main conclusions reported there.
Acknowledgments
We thank J. M. Grimes and D. I. Stuart for the program GAP, M. G. Rossmann for the program Roadmap, and P. B. Pereira for assistance in the preparation of figures.
This study was supported by grants BIO2002-04419 and BIO2002-00517 of CICYT to the IBMB and CICYT BIO098-0833 to INIA. Data were collected at the EMBL protein crystallography beamlines at ESRF (Grenoble). Financial support was provided by the ESRF and by grant HPRI-CT-1999-00022 of the European Union.
REFERENCES
- 1.Andries, K., B. Dewindt, J. Snoeks, R. Willebrords, R. Stokbroekx, and P. J. Lewi. 1991. A comparative test of fifteen compounds against all known human rhinovirus serotypes as a basis for a more rational screening program: mini-review. Antivir. Res. 16:213-225. [DOI] [PubMed] [Google Scholar]
- 2.Belnap, D. M., D. J. Filman, B. L. Trus, N. Cheng, F. P. Booy, J. F. Conway, S. Curry, C. N. Hiremath, S. K. Tsang, A. C. Steven, and J. M. Hogle. 2000. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J. Virol. 74:1342-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 4.Bergelson, J. M., M. Chan, K. R. Solomon, N. F. St John, H. Lin, and R. W. Finberg. 1994. Decay-accelerating factor (CD55), a glycosylphosphatidylinositol-anchored complement regulatory protein, is a receptor for several echoviruses. Proc. Natl. Acad. Sci. USA 91:6245-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrego, B., E. Carra, J. A. Garcia-Ranea, and E. Brocchi. 2002. Characterization of neutralization sites on the circulating variant of swine vesicular disease virus (SVDV): a new site is shared by SVDV and the related coxsackie B5 virus. J. Gen. Virol. 83:35-44. [DOI] [PubMed] [Google Scholar]
- 6.Brocchi, E., G. Zhang, N. J. Knowles, G. Wilsden, J. W. McCauley, O. Marquardt, V. F. Ohlinger, and F. De Simone. 1997. Molecular epidemiology of recent outbreaks of swine vesicular disease: two genetically and antigenically distinct variants in Europe, 1987-94. Epidemiol. Infect. 118:51-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, F., P. Talbot, and R. Burrows. 1973. Antigenic differences between isolates of swine vesicular disease virus and their relationship to coxsackie B5 virus. Nature 245:315-316. [DOI] [PubMed] [Google Scholar]
- 8.Brünger, A. T., P. D. Adams, M. G. Clore, W. L. Delano, P. Gros, W. Grose-Kunsleve, J. S. Jiang, J. Kuszewski, N. Nilges, N.-S. Pannu, R. J. Read., L. M. Rice., T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 9.Chow, M., R. Yabrov, J. Bittle, J. Hogle, and D. Baltimore. 1985. Synthetic peptides from four separate regions of the poliovirus type 1 capsid protein VP1 induce neutralizing antibodies. Proc. Natl. Acad. Sci. USA USA 82:910-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow, M., J. F. E. Newman, J. Filman, J. M. Hogle, D. Rowlands, and F. Brown. 1987. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327:482-486. [DOI] [PubMed] [Google Scholar]
- 11.Cowtan, K., and P. Main. 1998. Miscellaneous algorithms for density modification. Acta Crystallogr. D Biol. Crystallogr. 54:487-493. [DOI] [PubMed] [Google Scholar]
- 12.Dekker, A., P. Moonen, E. A. de Boer-Luijtze, and C. Terpstra. 1995. Pathogenesis of swine vesicular disease after exposure of pigs to an infected environment. Vet. Microbiol. 45:243-250. [DOI] [PubMed] [Google Scholar]
- 13.Escribano-Romero, E., M. A. Jimenez-Clavero, and V. Ley. 2000. Swine vesicular disease virus. Pathology of the disease and molecular characteristics of the virion. Anim. Health Res. Rev. 1:119-126. [DOI] [PubMed] [Google Scholar]
- 14.Filman, D. J., R. Syed, M. Chow, A. J. Macadam, P. D. Minor, and J. M. Hogle. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8:1567-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filman, D. J., M. W. Wien, J. A. Cunningham, J. M. Bergelson, and J. M. Hogle. 1998. Structure determination of echovirus 1. Acta Crystallogr. D Biol. Crystallogr. 54:1261-1272. [DOI] [PubMed] [Google Scholar]
- 16.Fricks, C. E., and J. M. Hogle. 1990. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol. 64:1934-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fry, E. E., S. M. Lea, T. Jackson, J. W. Newman, F. M. Ellard, W. E. Blakemore, R. Abu-Ghazaleh, A. Samuel, A. M. King, and D. I. Stuart. 1999. The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex. EMBO J. 18:543-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fry, E. E., N. J. Knowles, J. W. Newman, G. Wilsden, Z. Rao, A. M. King, and D. I. Stuart. 2003. Crystal structure of swine vesicular disease virus and implications for host adaptation. J. Virol. 77:5475-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant, R. A., C. N. Hiremath, D. J. Filman, R. Syed, K. Andries, and J. M. Hogle. 1994. Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design. Curr. Biol. 4:784-797. [DOI] [PubMed] [Google Scholar]
- 20.Graves, J. H. 1973. Serological relationship of swine vesicular disease virus and coxsackie B5 virus. Nature 245:314-315. [DOI] [PubMed] [Google Scholar]
- 21.He, Y., P. R. Chipman, J. Howitt, C. M. Bator, M. A. Whitt, T. S. Baker, J. R. Kuhn, C. W. Anderson, P. Freimuth, and M. G. Rossmann. 2001. Interaction of coxsackievirus B3 with the full length coxsackievirus-adenovirus receptor. Nat. Struct. Biol. 8:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendry E, H. Hatanaka, E. Fry, M. Smyth, J. Tate, G. Stanway, J. Santti, M. Maaronen, T. Hyypia, and D. Stuart. 1999. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure 15:1527-1538. [DOI] [PubMed] [Google Scholar]
- 23.Inoue, T., S. Yamaguchi, T. Kanno, S. Sugita, and T. Saeki. 1993. The complete nucleotide sequence of a pathogenic swine vesicular disease virus isolated in Japan (J1′73) and phylogenetic analysis. Nucleic Acids Res. 21:3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jimenez-Clavero, M. A., A. Douglas, T. Lavery, J. A. Garcia-Ranea, and V. Ley. 2000. Immune recognition of swine vesicular disease virus structural proteins: novel antigenic regions that are not exposed in the capsid. Virology 270:76-83. [DOI] [PubMed] [Google Scholar]
- 25.Jimenez-Clavero, M. A., E. Escribano-Romero, A. J. Douglas, and V. Ley. 2001. The N-terminal region of the VP1 protein of swine vesicular disease virus contains a neutralization site that arises upon cell attachment and is involved in viral entry. J. Virol. 75:1044-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jimenez-Clavero, M. A., V. Ley, I. Fita, and N. Verdaguer. 2003. Crystallization and preliminary X-ray analysis of swine vesicular disease virus (SVDV). Acta Crystallogr. D Biol. Crystallogr. 59:541-543. [DOI] [PubMed] [Google Scholar]
- 27.Jones, T., Y. Zou, S. Cowan, and M. Kjeldgaard. 1991. Improved methosd for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 28.Kanno, T., T. Inoue, Y. Wang, A. Sarai, and S. Yamaguchi. 1995. Identification of the location of antigenic sites of swine vesicular disease virus with neutralization-resistant mutants. J. Gen. Virol. 76:3099-3106. [DOI] [PubMed] [Google Scholar]
- 29.Kanno, T., D. Mackay, T. Inoue, G. Wilsden, M. Yamakawa, R. Yamazoe, S. Yamaguchi, J. Shirai, P. Kitching, and Y. Murakami. 1999. Mapping the genetic determinants of pathogenicity and plaque phenotype in swine vesicular disease virus. J. Virol. 73:2710-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knowles, N. J., and J. W. McCauley. 1997. Coxsackievirus B5 and the relationship to swine vesicular disease virus. Curr. Top. Microbiol. Immunol. 223:153-167. [DOI] [PubMed] [Google Scholar]
- 31.Knowlton, K. U., E. S. Jeon, N. Berkley, R. Wessely, and S. Huber. 1996. A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J. Virol. 70:7811-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolatkar, P. R., J. Bella, N. H. Olson, C. M. Bator, T. S. Baker, and M. G. Rossmann. 1999. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 18:6249-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laskowski, R. A., J. A. Rullmannn, M. W. MacArthur, R. Kaptein, and J. M. Thornton. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477-486. [DOI] [PubMed] [Google Scholar]
- 34.Lewis, J. K., B. Bothner, T. J. Smith, and G. Siuzdak. 1998. Antiviral agent blocks breathing of the common cold virus. Proc. Natl. Acad. Sci. USA 95:6774-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo, M., G. Vriend, G. Kamer, I. Minor, E. Arnold, M. G. Rossmann, U. Boege, D. G. Scraba, G. M. Duke, and A. C. Palmenberg. 1987. The atomic structure of Mengo virus at 3.0 A resolution. Science 235:182-191. [DOI] [PubMed] [Google Scholar]
- 36.Martino, T. A., M. Petric, H. Weingart, J. M. Bergelson, M. A. Opavsky, C. D. Richardson, J. F. Modlin, R. W. Finberg, K. C. Kain, N. Willis, N., C. J. Gauntt, and P. P. Liu. 2000. The coxsackie-adenovirus receptor (CAR) is used by reference strains and clinical isolates representing all six serotypes of coxsackievirus group B and by swine vesicular disease virus. Virology 271:99-108. [DOI] [PubMed] [Google Scholar]
- 37.Muckelbauer, J. K., M. Kremer, I. Minor, G. Diana, F. J. Dutko, J. Groarke, D. C. Pevear, and M. G. Rossmann. 1995. The structure of coxsackievirus B3 at 3.5 angstrom resolution. Structure 3:653-667. [DOI] [PubMed] [Google Scholar]
- 38.Nardelli, L., E. Lodetti, G. L. Gualandi, R. Burrows, D. Goodridge, F. Brown, and B. Cartwright. 1968. A foot and mouth disease syndrome in pigs caused by an enterovirus. Nature 219:1275-1276. [DOI] [PubMed] [Google Scholar]
- 39.Nichols, A., K. Sharp, and B. Honing. 1991. GRASP, graphical representation and analysis of structural properties of proteins. Proteins Struct. Funct. Genet. 11:281-296.1758883 [Google Scholar]
- 40.Nijhar, S. K., D. K. Mackay, E. Brocchi, N. P. Ferris, R. P. Kitching, and N. J. Knowles. 1999. Identification of neutralizing epitopes on a European strain of swine vesicular disease virus. J. Gen. Virol. 80:277-282. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski, Z., and I. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 42.Pulli, T., M. Roivainen, T. Hovi, and T. Hyypia. 1998. Induction of neutralizing antibodies by synthetic peptides representing the C terminus of coxsackievirus A9 capsid protein VP1. J. Gen. Virol. 79:2249-2253. [DOI] [PubMed] [Google Scholar]
- 43.Roivainen, M., A. Narvanen, M. Korkolainen, M. L. Huhtala, and T. Hovi. 1991. Antigenic regions of poliovirus type 3/Sabin capsid proteins recognized by human sera in the peptide scanning technique. Virology 180:99-107. [DOI] [PubMed] [Google Scholar]
- 44.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, and A. G. Mosser. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145-153. [DOI] [PubMed] [Google Scholar]
- 45.Rossmann, M. G., Y. He, and J. Kuhn. 2002. Picornavirus-receptor interactions. Trends Microbiol. 10:324-331. [DOI] [PubMed] [Google Scholar]
- 46.Sakoda, Y., N. Ross-Smith, T. Inoue, and G. J. Belsham. 2001. An attenuating mutation in the 2A protease of swine vesicular disease virus, a picornavirus, regulates cap- and internal ribosome entry site-dependent protein synthesis. J. Virol. 75:10643-10650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seechurn, P., N. J. Knowles, and J. W. McCauley. 1990. The complete nucleotide sequence of a pathogenic swine vesicular disease virus. Virus Res. 16:255-274. [DOI] [PubMed] [Google Scholar]
- 48.Smith, T. J., M. J. Kremer, M. Luo, G. Vriend, E. Arnold, G. Kamer, M. G. Rossmann, M. A. McKinlay, G. D. Diana, and M. J. Otto. 1986. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 233:1286-1293. [DOI] [PubMed] [Google Scholar]
- 49.Smyth, M., J. Tate, E. Hoey, C. Lyons, S. Martin, and D. Stuart. 1995. Implications for viral uncoating from the structure of bovine enterovirus. Nat. Struct. Biol. 2:224-231. [DOI] [PubMed] [Google Scholar]
- 50.Spiller, O. B., I. G. Goodfellow, D. J. Evans, S. J. Hinchliffe, and B. P. Morgan. 2002. Coxsackie B viruses that use human DAF as a receptor infect pig cells via pig CAR and do not use pig DAF. J. Gen. Virol. 83:45-52. [DOI] [PubMed] [Google Scholar]
- 51.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomko, R. P., R. Xu, and L. Philipson. 1997. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. USA 94:3352-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsang, S. K., P. Danthi, M. Chow, and J. M. Hogle. 2000. Stabilization of poliovirus by capsid-binding antiviral drugs is due to entropic effects. J. Mol. Biol. 296:335-340. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, G., G. Wilsden, N. J. Knowles, and J. W. McCauley. 1993. Complete nucleotide sequence of a coxsackie B5 virus and its relationship to swine vesicular disease virus. J. Gen. Virol. 74:845-853. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, G., D. T. Haydon, N. J. Knowles, and J. W. McCauley. 1999. Molecular evolution of swine vesicular disease virus. J. Gen. Virol. 1999. 80:639-651. [DOI] [PubMed] [Google Scholar]
- 56.Zhao, R., D. C. Pevear, M. J. Kremer, V. L. Giranda, J. A. Kofron, R. J. Kuhn, and M. G. Rossmann. 1996. Human rhinovirus 3 at 3.0 angstrom resolution. Structure 4:1205-1220. [DOI] [PubMed] [Google Scholar]