Abstract
Empirical evidence and theoretical studies suggest that the phenotype, i.e., cellular- and molecular-scale dynamics, including proliferation rate and adhesiveness due to microenvironmental factors and gene expression that govern tumor growth and invasiveness, also determine gross tumor-scale morphology. It has been difficult to quantify the relative effect of these links on disease progression and prognosis using conventional clinical and experimental methods and observables. As a result, successful individualized treatment of highly malignant and invasive cancers, such as glioblastoma, via surgical resection and chemotherapy cannot be offered and outcomes are generally poor. What is needed is a deterministic, quantifiable method to enable understanding of the connections between phenotype and tumor morphology. Here, we critically review advantages and disadvantages of recent computational modeling efforts (e.g., continuum, discrete, and cellular automata models) that have pursued this understanding. Based on this assessment, we propose and discuss a multi-scale, i.e., from the molecular to the gross tumor scale, mathematical and computational “first-principle” approach based on mass conservation and other physical laws, such as employed in reaction-diffusion systems. Model variables describe known characteristics of tumor behavior, and parameters and functional relationships across scales are informed from in vitro, in vivo and ex vivo biology. We demonstrate that this methodology, once coupled to tumor imaging and tumor biopsy or cell culture data, should enable prediction of tumor growth and therapy outcome through quantification of the relation between the underlying dynamics and morphological characteristics. In particular, morphologic stability analysis of this mathematical model reveals that tumor cell patterning at the tumor-host interface is regulated by cell proliferation, adhesion and other phenotypic characteristics: histopathology information of tumor boundary can be inputted to the mathematical model and used as phenotype-diagnostic tool and thus to predict collective and individual tumor cell invasion of surrounding host. This approach further provides a means to deterministically test effects of novel and hypothetical therapy strategies on tumor behavior.
THE ROLE OF PREDICTIVE SCIENTIFIC COMPUTATION AS “IN‐SILICO” CANCER MODELING
Cancer progression and invasion: current understanding
A wealth of empirical evidence links disease progression with tumor morphology, invasion, and associated molecular phenomena. However, not only is there a lack of quantitative understanding of the underlying physiological processes driving tumor-scale behavior, in particular, morphology at the tumor-host interface, but the qualitative explanations themselves may be indecisive or inconsistent. For example, a positive correlation of cell adhesion molecules (integrins) and cancer cell migration was observed in glioma cells (Tysnes et al., 1996), yet integrins can also serve as negative effectors that impede invasion and progression (Zutter et al., 1995). Similarly, conflicting data on the function of proteases in tumor invasion and metastasis (Friedl & Wolf, 2003) is illustrated by variable results from clinical trials of potential anti-invasive therapies (Lah et al., 2006). While the primary role of angiogenesis in promoting tumor growth and invasion has been well demonstrated, the results of clinical trials using various drugs to suppress neovascularization have yielded mixed results. Despite encouraging signs of tumor regression following anti-angiogenic therapy, in some cases length of survival remains the same (Kuiper et al., 1998, Bernsen et al., 1999, Bloemendal et al., 1999). In addition, experimental observations indicate that anti-angiogenic treatments may exacerbate hypoxia (Steeg, 2003), and paradoxically promote tumor fragmentation, cancer cell migration, and host tissue invasion (Page et al., 1987, Kunkel et al., 2001, Seftor et al., 2002, Lamszus et al., 2003, Bello et al., 2004).
Links between cellular‐ and tumor‐scale
In spite of abundant experimental and clinical data surrounding molecular and cellular phenomena, it is difficult to quantify their aggregate effect on gross tumor-scale behavior using conventional methods that, for the most part, investigate isolated mechanisms. Prognosis of cancer patients suffering from highly invasive tumors, such as glioblastoma, is grim despite advances in surgical and chemotherapeutic treatment, since not all tumor cells can be removed or treated because of limited delineation between healthy and tumor tissue at the tumor border, which may lead to fatal recurrences (Kansal et al., 2000a). In particular, mechanisms governing glioma invasion likely include intrinsic properties of cell proliferation, migration, and adhesion. Glioma cells have been experimentally shown to infiltrate and scatter throughout the entire central nervous system after a period of only seven days post-implantation (Chicoine & Silbergeld, 1995, Silbergeld & Chicoine, 1997, Swanson et al., 2000). This might be one reason why current treatments that focus on surgery, radiation, and chemotherapy, while perhaps having an effect on primary bulk mass characteristics, may fail to extend survival time.
A novel, in‐silico approach to cancer modeling
In this review/position paper we describe a multidisciplinary method integrating mathematical models with experimental (in vitro and in vivo) and clinical data. This methodology reflects an “engineering” approach that views tumor lesions as complex micro-structured materials, where three-dimensional tissue architecture (“morphology”) and dynamics are coupled in intricate, complex ways to cell phenotype, which in turn is influenced by factors in the microenvironment. Cellular and microenvironmental factors act both as tumor morphology regulators and as determinants of invasion potential by controlling the mechanisms of cancer cell proliferation and migration (Friedl & Wolf, 2003, Sierra, 2005, van Kempen et al., 2003). In particular, recent experimental results demonstrate that interactions between cellular proliferation, adhesion, and other phenotypic properties are reflected in both tumor-host interface morphology and invasive characteristics of tumors (Tysnes et al., 1996, Zutter et al., 1995, Lah et al., 2006, Kuiper et al., 1998, Bernsen et al., 1999, Bloemendal et al., 1999, Steeg, 2003, Page et al., 1987, Kunkel et al., 2001, Seftor et al., 2002, Lamszus et al., 2003, Bello et al., 2004, Friedl & Wolf, 2003). The goal is then to create computational (in silico) multiscale tools capable of predicting the complexity of cancer at multiple temporal and spatial resolutions, with the aim of supplementing diagnosis and treatment by helping plan more focused and effective therapy via surgical resection, standard chemotherapy, novel treatments (e.g., angiogenic, anti-invasive), or some combination of them. The tools would quantitatively examine the effect of tumor morphology regulators, which include tissue rigidity, density, adhesiveness, microenvironment gradients (e.g., oxygen, nutrient, growth factors), and the combinatorial effects of oncogenes (controlling cell proliferation, motility, and nutrient consumption) and tumor suppressor genes (controlling cell apoptosis and motility) on gross morphology. They would also define the degree of diffuse invasion of tumor cells peripheral to the tumor mass that may be beyond the detection of current non-invasive medical imaging techniques (Swanson et al., 2000), or extrapolate tumor invasiveness and metastatic potential from its morphology in fixed tissue. In-silico model development is built upon continuum, discrete, and in particular cellular automata models (e.g., see Hogea et al., 2006, Ayati et al., 2006, Chaplain & Lolas, 2005, Garner et al., 2005, Bru et al., 2003, Jackson, 2004, Castro et al., 2005, Painter, 2000, Khain et al., 2005, Khain & Sander, 2006, Sander & Deisboeck, 2002, Boushaba et al., 2006, Stein et al., 2007; see also reviews Adam, 1996, Bellomo & Preziosi, 2000, Chaplain & Anderson, 2003, Friedman, 2004, Araujo & McElwain, 2003, Byrne et al., 2006, Swanson et al., 2003, Sanga et al., 2006, Quaranta et al., 2005, Nagy, 2005, Hatzikirou et al., 2004, Frieboes et al., 2006b, Sinek et al., 2006, Sanga et al., 2007, Macklin & Lowengrub, 2007).
Incorporation of patient data: predictive modeling
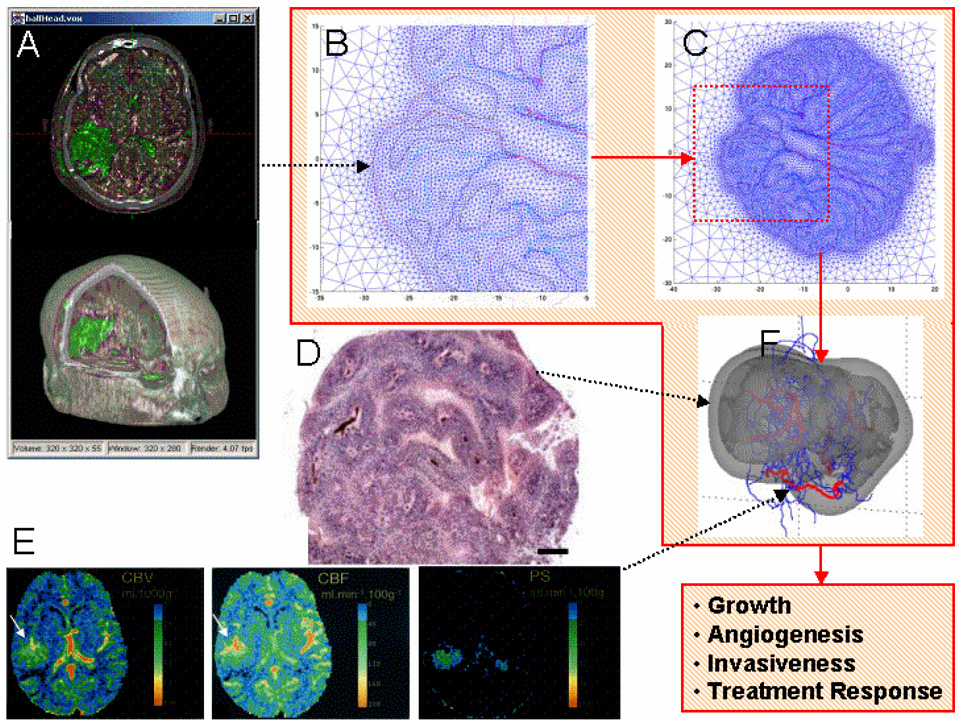
Multi-scale modeling quantifies the time- and space-dependent physics and chemistry (e.g., diffusion of substrates, mechanical forces exchanged among cells and with the matrix, molecular transport, receptor-ligand interactions, pharmacokinetics determinants) underlying tumor biological behavior. We envision that future simulators, building on the developments described herein, will operate as described in Figure 1. Initial conditions are obtained from patient data, such as MR (magnetic resonance) and CT (computed tomography) images and histopathology, which are used to set phenotypic and other model input parameters (e.g., proliferation rate). For example, CT image information would be translated voxel by voxel (using a computer program) to the coordinate system of the multi-scale model, e.g., a finite-element computational mesh discretizing the space occupied by a tumor and surrounding host tissue (Cristini et al., 2001; Zheng et al., 2005a,b; Anderson et al., 2005; Cristini and Tan, 2004). This input data would also include physical information about viable regions and cell density therein, necrosis, vasculature, blood flow, and other specific details from histopathology (Bearer and Cristini, MS submitted, Frieboes et al., 2007). The computer model then calculates local tumor growth, angiogenesis, and response to treatment under various conditions (Zheng et al., 2005a, Wise et al., MS submitted, Frieboes et al., MS submitted, Frieboes et al., 2007) by solving in time and space conservation (e.g., diffusion equation) and other laws at the tissue scale. These laws are linked to the cell molecular biology by functional relationships and parameters informed by the biopsy data. Computational solutions are obtained using finite elements and other numerical techniques (e.g., Zheng et al., 2005a; Frieboes et al., 2007). Additional patient data obtained from tissue culture, gene arrays, proteomic profiling, and other means would sharpen these parameter estimations (Frieboes et al., 2006a) in order to enable accurate prediction of behavior. Further details on the parameter estimation procedure are described below and in references cited.
Figure 1.
Patient data can be inputted into an in-silico model of cancer. The red line encloses sample in-silico representations of tumor morphology. Tumor morphology at a given time is inputted from 3-D voxel fusion of CT and MR data (A) (reprinted with permission from Xie et al. (c) 2004 IEEE) that is translated voxel by voxel (using a computer program) into the in-silico coordinate system (B) (in this example, an unstructured adaptive mesh by Cristini et al., 2001) to build a tumor representation in virtual computational space (C). Spatial information of viable cell regions and vasculature structure is obtained from histophathology (D) (reprinted from NeuroImage, Frieboes et al., in press, Copyright 2007, with permission from Elsevier; bar, 200 µm). Vasculature specific information is defined from dynamic contrast enhanced CT (E), yielding parameters such as blood volume (left), blood flow (middle), and microvascular permeability (right) (reprinted with permission from Roberts et al., 2002). Other input data to the model include cell-scale parameters such as proliferation rates obtained, for example, from in vitro cultures. The model then predicts (F) tumor growth (viable cells: light gray; necrosis: darker gray), angiogenesis (red: mature; blue: immature capillaries), invasiveness, and response to treatment for this patient by solving time- and space-dependent conservation of mass and momentum and other physical laws (reprinted from NeuroImage, Frieboes et al., in press, Copyright 2007, with permission from Elsevier). These laws contain parameters that characterize the phenotypes and are linked to the underlying molecular biology by functional mathematical relationships founded on these and other experimental data. Incorporation of models of this biology (e.g., evolution of genotype, cell signaling pathways) guarantees that the in-silico predictions of tumor behavior are realistic and accurate. Computational solutions of this multi-scale model are obtained using finite elements and other numerical techniques.
MULTI‐SCALE MODELING AND SIMULATION OF TUMOR MORPHOLOGY AND INVASION
Model development goals and choices
The progression of tumor lesions cannot be completely characterized by studying effects in isolated cells since it is known that the forces and mechanisms regulating the motion of individual cells converge and synchronize into the collective, organized, structural motion of a whole body or cluster of cells (“Functional Collective Cell-Migration Units,” FCCMU) that often precedes the onset of epithelial-mesenchymal and other phenotypic transitions leading to individual cell shedding from a tumor and eventually to metastasis (Friedl & Wolf, 2003). Both individual and collective migration modes are regulated (and complicated) by multifaceted interactions among tumor cells, stroma and tumor microenvironment (Sierra, 2005, van Kempen et al., 2003).
While “discrete” in-silico models (e.g., DiMilla et al., 1991, Dickinson & Tranquillo et al., 1993, Kansal et al., 2000a, 2000b, Patel et al., 2001, Ferreira et al., 2002, Turner & Sherratt, 2002, Leyrat et al., 2003, dos Reis et al., 2003, Anderson, 2005) are able to capture individual cell migration and easily incorporate biological rules, such as cell-cell & cell-medium interactions and motion due to chemotaxis and haptotaxis, they are limited to relatively small numbers of cells due to computational cost, among the other deficiencies and over-simplifications introduced by the discrete approach. In contrast, “continuum” models (e.g., Byrne & Chaplain, 1995a, 1995b, 1996a, 1996b, 1997, Bellomo & Preziosi, 2000, Cristini et al., 2003, 2005, Macklin & Lowengrub, 2005, Frieboes et al., 2006a, Li et al., 2007, Macklin & Lowengrub, 2007), describing tissue matter as a continuum medium rather that discrete individual cells, capture the collective motion of FCCMUs with less computational expense.
The fact that collective migration is often associated with relatively higher degrees of cell differentiation (Friedl & Wolf, 2003) than for the case of single-cell migration suggests that molecular mechanisms are relatively more robust across a tumor cell population. Thus, the multitude of cells can be averaged out and re-described as a single multi-cellular FCCMU unit obeying deterministic dynamics laws, while still employing mathematical models of single-cell migration when needed, e.g., to describe epithelial-mesenchymal transitions (Friedl & Wolf, 2003). Moreover, the domain size of realistic discrete simulations is limited to sub-millimeter-size in-vitro tumor spheroids or in-vivo patches of tumor tissue. We propose that discrete models of cell proliferation and migration should be coupled to continuum models of FCCMU to extend the computational capability to realistic, cm-size three-dimensional tumor lesions as defined and described in the following. A hybrid, multi-scale modeling methodology (Cristini et al., 2006) that links continuum (i.e., tissue-scale) with discrete (i.e., cellular-scale) formulations with appropriate functional relationships of cell adhesion and migration due to environmental conditions should provide, over the next decade, a more comprehensive understanding of the molecular basis of diversity and adaptation of cell migration, thus more efficiently and accurately predicting invasion potential from real-time tumor morphology.
This approach has the advantage that well-established engineering methods and analyses of morphology can be applied (e.g., based on continuum methods when possible). Experimental measurements, computer simulations and morphologic stability analyses can be used to study, in detail, microenvironment transport processes (e.g., of oxygen, nutrients, chemokines, growth factors), cell motion and proliferation, signaling pathways and molecular phenomena regulating cell cycling, cell-cell communications and expression of cell adhesion molecules and matrix degrading enzymes. For example, the link between hypoxic gradients and invasion, and between normoxic conditions and compact non-infiltrative tumor morphologies, can thus be explained “by exploiting the ability of mathematics to model physical and biological systems in ways that enable prediction and control” (quoting John Lowengrub, Chair, Mathematics, UC Irvine).
Significance of in‐silico modeling: a novel hypothesis‐generation tool
The computational models described in this paper represent important steps in generating hypotheses that postulate functional relationships linking the effect of molecular/cellular changes to tumor-scale morphology and invasiveness. By directly solving the mathematical equations describing underlying physical and chemical processes occurring within tumors, the complex biology of tumor behavior and the often hidden mechanisms of growth and invasion automatically are unveiled and can be accurately quantified in virtual, in-silico simulation space. Examples of novel hypotheses generated from simulations studies and tested in experiments will be provided in the following. Although these types of models are not multi-scale per se, parameters characterizing cell response to substrate concentration can be interpreted as representing underlying biochemistry and molecular biology driving tumor-scale dynamics, specifically an invasive phenotype. However, modeling of tumor behavior and cell microenvironment remains a challenge. Existing mathematical models are only capable, in general, of recapitulating a posteriori the highly variable empirical observations of morphology, once appropriate phenomenological parameters that do not incorporate direct molecular-scale description have been “fitted” to the experiments.
Here we propose that the next decade of investigation should focus on the task of developing predictive multi-scale models (e.g., see Figure 2) that incorporate new, functional relationships among macro-scale parameters characterizing differences in, and transitions among, cellular patterns, and variations in the molecular repertoire used by tumor cells to regulate proliferation, adhesion and other phenotypic properties.
Figure 2.
Main component modules of a sample predictive model based on first principles (e.g., conservation laws of mass and momentum) linking cellular/molecular scale processes to tumor growth and morphology, assuming a tumor with two clones for simplicity. Each component (Vasculature, Tumor, Genotype) lists biological processes that are implemented through a set of equations (e.g., the diffusion-reaction equation determining the local concentration n of a cell substrate within the tumor), as well as suggested experiments for validation of these functional relationships. Additional biological processes, clones, and properties of the stroma can be incorporated by augmenting the number of variables and equations. Functional relationships to be determined experimentally describe mathematically the dependence of the listed phenotypic parameters (e.g., cell proliferation rates λM) on the array M that contains genetic information. Note that several of these parameters are phenomenological, i.e., they do not correspond to direct measurables (e.g., the “strengths” α of haptotaxis and chemotaxis, which are related to the expression of integrins and the mechanical forces exchanged with molecules in the ECM). Data obtained from in vitro experiments, in vivo / ex vivo models, and clinically (e.g., tumor size, morphology) is thus input to the various modules. This data will also help refine the model’s functional relationships through an iterative exercise of multidisciplinary research that will progressively lead, over the next decade, to less phenomenological and more “exact” models, in which the parameters are directly measurable in experiments. For further mathematical details and definitions of variables and parameters, and for demonstrations of a “prototype” in-silico model see Frieboes et al. (2007), Bearer and Cristini, MS submitted, and references therein.
This methodology is expected to improve current modeling efforts because a multi-scale approach connects previous work focused on specific scales (e.g., molecular) and processes (e.g., gene transformation), affording the possibility to go beyond the current reductionist picture of tumor invasion and migration (Friedl & Wolf, 2003, Keller et al., 2006, Sierra, 2005, van Kempen et al., 2003, Wolf & Friedl, 2006, Kopfstein & Christofori, 2006, Yamaguchi et al., 2005, Elvin et al., 2005, Sahai, 2005, Friedl et al., 2004, Friedl, 2004, Condeelis et al., 2005, Ridley et al., 2003, Jones et al., 2000) by providing a platform to study cancer as a system. Next, we describe biologically founded, in-silico modeling efforts of tumor progression (e.g., Zheng et al., 2005a, Sanga et al., 2006, Bearer and Cristini, MS submitted; Frieboes et al., 2007; Wise et al., MS submitted; Frieboes et al., MS submitted) relying on known characteristics of tumor behavior (Cristini et al., 2003, Zheng et al., 2005a, Anderson et al., 2000, Cristini et al., 2005, Sinek et al., 2004, Frieboes et al., 2006a, Macklin & Lowengrub, 2007) to predict the combination of variables most likely driving progression towards invasiveness.
This effort builds on an approach (e.g., Cristini et al., 2003, Zheng et al., 2005a, Cristini et al., 2005, Frieboes et al., 2006a, Li et al., 2007) that includes reformulations and generalizations of mathematical models (Greenspan, 1976, Byrne & Chaplain, 1996a, 1996b, Adam, 1996, Chaplain, 1996, Lowengrub & Truskinovsky, 1998, Leo et al., 1998, Lee et al., 2002, Macklin & Lowengrub, 2005 & 2007, Garcke et al., 2004, Jacqmin, 1999, Anderson et al., 1998, Bellomo & Preziosi, 2000, Ambrosi & Preziosi, 2002, Byrne & Preziosi, 2003, Chaplain et al., 2006, Ambrosi & Guana, 2006, Chaplain & Anderson, 2003, see also Jackson & Byrne, 2002), solved numerically using state-of-the-art algorithms and techniques (Zheng et al., 2005a and 2005b, Wise et al., 2005, Kim et al., 2004a, Kim et al., 2004b, Wise et al., 2004, Cristini et al., 2001, Berger & Colella, 1989, Brandt, 1977; Wise et al., manuscript submitted). Figure 2 describes the main component modules (Vasculature, Tumor, and Genotype) of this model along with equations that represent mathematically the relevant biological parameters.
Determination of functional relationships and parameter values
The specific process of multi-scale model “training” relies on conducting experiments (Figure 2) in which molecular factors are measured in the cell and the environment, and outcome of tumor growth (e.g., morphology, shape, extent of vascularization and invasion) is correlated with expression of these factors. This data allows estimation of the mathematical model parameters and functional relationships by perturbing these parameters and comparing the resulting simulation predictions of morphology against direct measurements, thus leading, through an iterative process that reveals deficiencies in modeling choices and triggers refinements in the relationships introduced, to a validated mathematical model with calibrated constitutive parameters. By virtue of its predictive power, this approach (Cristini et al., 2006) can help plan new experiments by identifying parameter regimes of noteworthy behavior–regimes that might otherwise be time-consuming and costly to discover by systematic experimentation.
Theoretical (e.g., Frieboes et al., 2006a) and experimental work (e.g., Gatenby et al., 2006, Frieboes et al., 2006a, and reviews by Chomyak & Sidorenko, 2001, Kim, 2005, Mueller-Klieser, 2000 and references therein) can be used to develop and test functional relationships, and to estimate the microphysical parameter values of a multi-scale in silico model. Examples (Cristini et al., 2006) of these functional relationships include those between expression of membrane transport proteins (e.g., glucose transporter-1 and Na/H exchanger) and hypoxia/proliferation; between extracellular matrix macromolecules (e.g. tubulin, actin), haptotaxis and chemotaxis; and between cell-cell adhesion parameters as an increasing function of oxygenation, e.g., from recent measurements by Robert Gatenby (personal communication) showing a gradient of cell-adhesion molecules (E-cadherins) opposed to hypoxia.
In vivo animal models (e.g., dorsal wound chamber by Gatenby et al., 2006) can supply detailed measurements of angiogenesis and blood flow, which provide additional constraints to the in silico model to determine parameter values associated with a developing neovasculature. Computational models of angiogenesis (Levine et al., 2002, Plank & Sleeman, 2003, 2004, Sun et al., 2005, Stephanou et al., 2005, McDougall et al., 2006) (Figure 2) can account for endothelial cell chemotactic and haptotactic movement, proliferation, development and remodeling of capillaries and the flow of blood through the local pressure and other constraints. Under in vivo conditions, additional measurements can be performed to determine pH and pO2 gradients that provide further functional constraints on the parameters relating to proliferation and cellular adaptation to hypoxia (Cristini et al., 2006). Finally, in vivo measurements of matrix degradation at the tumor-host boundary due to acidosis and proteases can provide parameter values for the invasion component of the in silico model (Cristini et al., 2006).
COMPUTATIONAL MODELING: A FRAMEWORK FOR LINKING PHENOTYPE, MORPHOLOGY, AND CANCER INVASION
Extensive mathematical modeling has produced preliminary quantifications of the links between invasive, malignant cell phenotypes and tumor-scale morphologies. These involve cell-cell and cell-extracellular matrix (ECM) interactions, cell motility, micro vessel density and acidosis, and local concentration of cell substrates (Mareel & Leroy, 2003). We illustrate representative discrete models in each of these areas. We then review recent mathematical and computational studies of a continuum FCMU model developed in our group. Both discrete and continuum models are based on conservation formulations such as described in Figure 2.
Effects of cell‐cell and cell‐matrix interactions
The effects of tumor cell and environment heterogeneity on the overall tumor morphology were recently studied (Anderson, 2005) using a model that captured spatial distribution of oxygen, matrix-degrading enzymes, and matrix molecules in the tumor microenvironment, as well as tumor cell properties, e.g., migration and proliferation. Results support the notion that tumor cell-matrix interactions ultimately control tumor shape by driving tumor cell migration via haptotaxis and chemotaxis towards fingering, invasive tumor morphologies (Figure 3 A and B).
Figure 3.
Effects of tumor cell and environment heterogeneity on overall tumor morphology predicted by a number of “discrete” (each tumor cell’s position is “tracked” during a simulation) computational models recently introduced. Simulation results from Anderson (2005) show the importance of tumor cell-matrix interactions in aiding or hindering migration of individual cells thus defining the overall tumor-scale geometry (A and B). Tumor types I-IV correspond to cell phenotypes displaying increasing levels of aggressiveness, i.e., combinations of cell-cell adhesiveness, proliferation, degradation, and migration rates. Both simulations use the same parameter values with the exception of differing initial ECM conditions (i.e., different distributions of matrix molecules). In (A), the matrix environment is initially described as homogeneous, whereas it is heterogeneous in (B). Consequently, (B) depicts invasive, fingering morphology. Cells of phenotype IV are on the tumor-host boundary, promoting invasion into host tissue, emphasizing that more aggressive cells drive fingering morphologies. These aggressive cells have minimal cell-cell adhesion, thus they do not tend to form compact structures. Simulations are carried out on a 400 × 400 grid representing 1 cm² of a virtual, 2-D tumor. Adapted from Anderson, A.R.A, A hybrid mathematical model of solid tumour invasion: the importance of cell adhesion, Mathematical Medicine and Biology, 2005, vol. 22, issue 2, pages 175–176, by permission of Oxford University Press. Simulation results from dos Reis et al. (2003) showing how tumors growing in host tissue environments of low (C) and high (D) “rigidity” can influence compact, non-invasive (C) and fractal, fingering, invasive (D) morphologies. Simulations are carried out to approximately 5000 cells, where cells are represented as interacting particles in a 2-D continuous space with periodic boundary conditions. Reprinted from Physica A, vol. 322, dos Reis et al., The interplay between cell adhesion and environment rigidity in the morphology of tumors, page 550, Copyright 2003, with permission from Elsevier. Cell patterns simulated using the model of Ferreira et al. (2002) (E) suggest how parameters characterizing cancer cells’ response to nutrient concentrations and embodying complex genetic and metabolic processes can influence the formation of fractal, fingering tumor morphologies. For comparison, a real fractal pattern observed in trichoblastoma (F). Reprinted figure with permission from Ferreira et al., Physical Review E, 65, page 021907-6, 2002. Copyright 2002 by the American Physical Society.
However, degradation of ECM, specifically surrounding the tumor boundary, may have a stronger influence on invasion than cell-cell adhesion. Using a derivation of a Potts model (Wu et al., 1982) that incorporates homotypic and heterotypic adhesion as well as secretion of proteolytic enzymes that drive haptotaxis along their gradient, a more quantitative perspective into the role of cell adhesion and proliferation in promoting an invasive phenotype was obtained (Turner & Sherratt, 2002). The model assumes genetic mutations affect cellular adhesiveness, secretion of matrix degrading enzymes, the ability to undergo taxis along gradients, and proliferation rate (Turner and Sherratt, 2002, Stetler-Stevenson et al., 1993). Using the maximum host tissue penetration as an index of invasiveness, simulation results predict that increases in the secretion of matrix degrading enzymes in synergy with increases in cell proliferation and haptotaxis can produce fingering morphologies at the tumor-host interface as cells adhere to the ECM and spread into host tissue. The model hypothesizes a functional relationship between proliferation rates and changes in adhesiveness based on experimental evidence (Huang & Ingber, 1999).
The notion that formation of fingering protuberances at the tumor-host boundary is primarily due to an intrinsic physical property termed rigidity of the host environment to resist tumor growth has also been computationally examined (dos Reis et al., 2003). Low rigidity allows a tumor to expand through the host environment resulting in a well-defined tumor-parenchyma interface, whereas higher rigidity forces a tumor to grow by invading the host tissue resulting in a fingering morphology (Figure 3, C and D), as predicted by simulation results. In addition, cell adhesion changes growth patterns from fractal morphologies at the tumor-host interface to compact shapes.
Effects of cell motility
Computational investigations of the invasiveness of high, medium, and low-grade gliomas illustrate that the ratio of tumor growth rate and cell motility can quantify the invasive nature of a tumor (Swanson et al., 2000). Specifically, this ratio might be useful in predicting a tumor’s invasive and metastatic potential; high proliferation rates and low motility correspond to lower grade tumors with less invasive potential whereas low proliferation rates and high motility correspond to higher-grade tumors with more invasive potential.
In contrast, a 3D cellular automaton model of glioblastoma capable of predicting tumor growth according to four microscopic parameters (probability of division, necrotic thickness, proliferative thickness, and maximum tumor extent) successfully predicted tumor-scale dynamics of a test case for untreated glioblastoma progression compiled from medical literature; simulations reproduced data such as lesion radius, cell number, growth fraction, necrotic fraction, and volume doubling time at particular time points (Kansal et al., 2000a, 2000b). Human glioblastoma patients have a median survival time of 8 months from diagnosis (Kansal et al., 2000a), which these models (Kansal et al., 2000a, 2000b) accurately predict using primary tumor volume as an indicator for survival.
Effects of micro vessel density and acidosis
The potential importance of micro vessel (MV) density and acidosis in promoting tumor growth and invasion has been demonstrated through recent computational models (Patel et al., 2001, Gatenby & Gawlinski, 1996, Gatenby & Gawlinski, 2003, Gatenby et al., 2006). Simulations show that the production rate of H+ ions by cancer cells, due to their increased dependence on anaerobic glucose metabolism, is linked to an optimal micro vessel (MV) density such that the microenvironment favors tumor cells over normal cells, hence promoting growth and invasion (Patel et al., 2001). MV density below this optimal value produces an environment too acidic even for cancer cells, while MV density above the optimum reduces or even completely negates the advantage enjoyed by cancer cells over normal cells in an acidic environment, thus inhibiting overall tumor growth and invasion by promoting nutrient competition.
Depending on the metabolic phenotype, various tumor morphologies can be predicted including invasive, fingering protrusions seen in experiments and with other in silico models. This and other modeling and experimental work further supports the acid-mediated tumor invasion hypothesis (Patel et al., 2001, Gatenby & Gawlinski, 1996, Gatenby & Gawlinski, 2003, Gatenby et al., 2006), illustrating the potential importance of MV density in driving pH gradients in the microenvironment and associated tumor-scale behavior. Such microenvironmental factors, in addition to cellular dynamics, are thus quantitatively linked to tumor-scale morphology.
Effects of cell substrate concentration
Competition for nutrient and oxygen amongst normal and cancer cells, in addition to cell proliferation, motility, death, and secretion of matrix degrading enzymes, may be an important factor driving tumor invasion. Using a cellular-automaton model (Ferreira et al., 2002), cell dynamics were described where at each time step, a cell (of type normal, cancer, or necrotic tumor) has equal probability of dividing, migrating, or undergoing necrosis; each action is governed by the local substrate concentration. Cells are modeled to release a series of enzymes that progressively degrade the ECM, thus providing more space for tumor cells to invade. In Figure 3, E and F, fingering morphologies are predicted by the model (and observed) as a result of high proliferation rates demanding large amounts of substrates. Predicted tumor morphology remains compact in situations of high nutrient supply and low cell consumption, while cell clusters expressing a phenotype that increases nutrient consumption exhibit thinner “fingers.”
Continuum‐based parameter‐sensitivity studies of FCCMU
Morphologic instability as a mechanism of tumor invasion
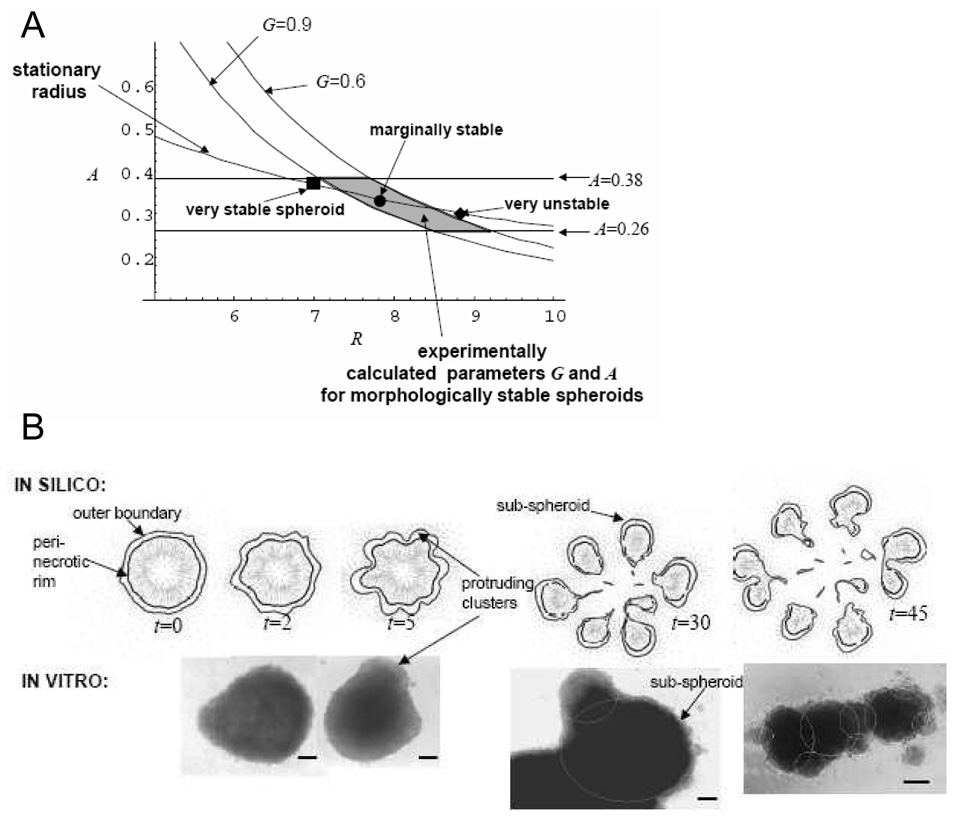
The current conceptual framework of continuum FCCMU models is based on reaction–diffusion formulations (Figure 2). Accordingly, tumor morphologic “stability” is regulated by the competition of pro- and anti-migratory/proliferative factors. When the former prevail, complex, unstable FCCMU patterns can develop (Cristini et al., 2003; Li et al. 2007). The power of this approach is that it is based on a physical mechanism that can account for the various invasive morphologies observed, and is thus potentially predictive of tumor growth. This mathematical analysis of morphologic stability has suggested that tumor tissue dynamics is regulated by two dimensionless parameters: the parameter G quantifies the competition between local tumor mass growth due to proliferation, and cell adhesion that tends to minimize tumor surface area and thus maintain compact nearly spherical tumors; the parameter A quantifies tumor mass shrinkage due to cell death (these parameters are obtained from some of those listed in Figure 2 using dimensional analysis; for the sake of simplicity, the associated cumbersome formulation is not reported here: see Cristini et al., 2003, Li et al. 2007). During glioblastoma tumor growth in vitro, cell proliferation is observed in a viable region where nutrients, oxygen, and growth factor levels are adequate, and cell death and necrosis in the inner regions where diffusion limitations prevent these substances from being present in adequate levels (Frieboes et al., 2006a). In the presence of these substrate gradients, morphology can be “unstable”, i.e., invasive, when cell adhesion is weak (large G). In contrast for small G, spheroid morphology is “stabilized” (i.e., spherical or nearly spherical) by cell adhesion (Cristini et al., 2003).
This is illustrated in a morphologic stability diagram (Cristini et al., 2003, Frieboes et al., 2006a), Figure 4, A. The G-curves divide the parameter space into stable (left) and unstable (right) regions. The G-curves are more shifted to the left as cell adhesion decreases (higher G), thus reducing the range of sizes of tumors that will be morphologically stable. The in silico model parameters A and G were calibrated using data from “stable” spheroids (shaded area) until agreement was obtained between the model calculations of morphology and growth and in vitro measures of tumor growth curves and thickness of the viable rim of cells. The model was then tested by predicting morphology stability conditions for an independent set of experiments (filled symbols) where the cell medium levels of growth factors and glucose were changed over a wider range to manipulate glioblastoma cell proliferation and adhesion (Frieboes et al., 2006a).
Figure 4.
In-silico model predictions and in-vitro measurements of locally invasive cell clusters in collective migration, using a “continuum model” (see text for a definition). Adapted from Frieboes et al. (2006a) with permission from the American Association of Cancer Research. In the morphologic stability diagram (A), obtained from a mathematical analysis of the model, “stationary radius” describes tumor dimension R (unit length = 100 µm), monotonically decreasing as death (described by the parameter A) increases. The G-curves calculated by the mathematical model divide, for each value of G (a parameter related to cell adhesion), the parameter space into morphologically stable (left) and unstable (right) tumors. Stable tumors remain roughly spherical during growth; unstable tumors are invasive and form wavy protrusions at the tumor-host boundary that further develop into sub-tumors that break-up from the parent tumor (B). The shaded region was determined from calibration of the parameter values under “stable” in vitro conditions. Representative “stable” and “unstable” spheroids (filled symbols) from different sets of experiments are shown to agree with the model predictions. This means that the mathematical model was capable of predicting invasive behavior of tumors under conditions of growth factors and substrate concentrations different from the “control” that was used to calibrate the model parameters. These results indicate that wavy patterns of cell arrangements at the tumor-host boundary could be inputted to a mathematical model to predict future invasive potential. (B) Time progression (arbitrary units) of avascular glioma predicted by simulations of the in-silico model (top) compared to observations in vitro (bottom). Tumor morphology and invasiveness are predicted to be heavily influenced by substrate gradients (e.g., nutrient) in the cellular microenvironment, driving detachment of bulbs or clusters of cells. Bar, 130 µm.
A remarkable result of this study was that the in silico model was capable of predicting growth and invasion of tumors from experiments that were not used for model training, thus successfully testing, under relatively simple highly controllable in vitro conditions, the hypothesized phenomenological relationships of adhesion and proliferation with substrate levels and their effects on tissue-scale growth and morphology (e.g., see Figure 2). Computer simulations and experimental observations of “unstable” spheroids that develop protrusions and detachment of cell clusters are shown in Figure 4, B. As described below, these infiltrative morphologies are also universally observed in tumors in vivo and in data from patients.
Clinical relevance
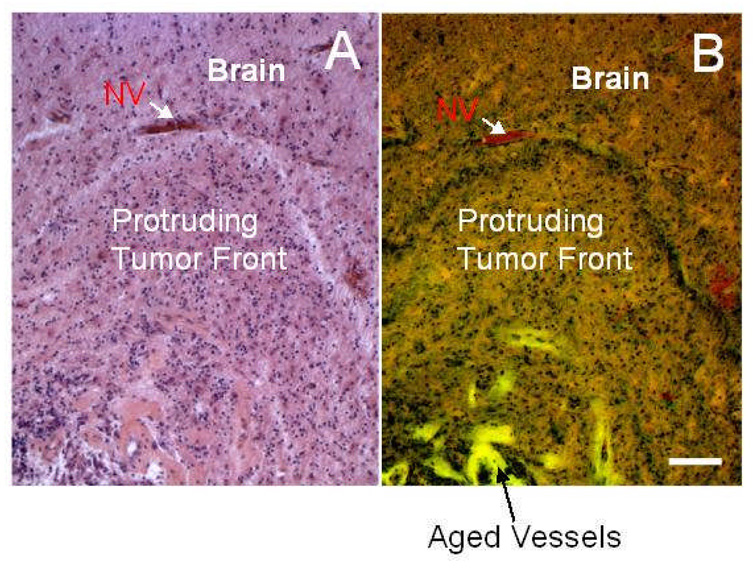
Morphologic instability during tumor growth is predicted to result from genomic changes that produce variations in sub-tumor clonal expansion, rates of mitosis and apoptosis, oxygen consumption, and diffusion gradients. This physical hypothesis is corroborated by in vitro and in vivo observations (e.g., Rubenstein et al., 2000, Kunkel et al., 2001, Lamszus et al., 2003, Bello et al., 2004, Frieboes et al., 2006a) and by patient data. In a study of several clinical samples of glioblastoma multiforme from multiple patients (Figure 5), histology reveals protruding fronts of cells in collective motion away from a necrotic area into an area of the host brain where neo-vascularization is present, thus following substrate gradients (Bearer and Cristini, MS submitted; Frieboes et al., 2007). This data strongly resembles the morphology of the tumor boundary predicted by computer simulation (Bearer and Cristini, MS submitted; Frieboes et al., 2007) and by the in vitro experiments described above (Frieboes et al., 2006a). These infiltrative shapes were consistently observed in high-grade gliomas, although their size may vary.
Figure 5.
Glioblastoma histopathological sections from one patient stained for H&E and viewed by bright field (A) and fluorescence (B) microscopy (Frieboes et al. 2007, reproduced with permission from Elsevier) showing tumor (bottom) pushing into more normal brain (top). Note demarcated margin between tumor and brain parenchyma to the middle top of the image and green fluorescent outlines of large vascular channels deeper in the tumor. Neovascularization (NV) at the tumor-brain interface can be detected by red fluorescence from the erythrocytes inside the vessels. Altogether, these data support our “morphologic instability” hypothesis. Bar, 100 µm.
Effects of phenotype on morphology and growth
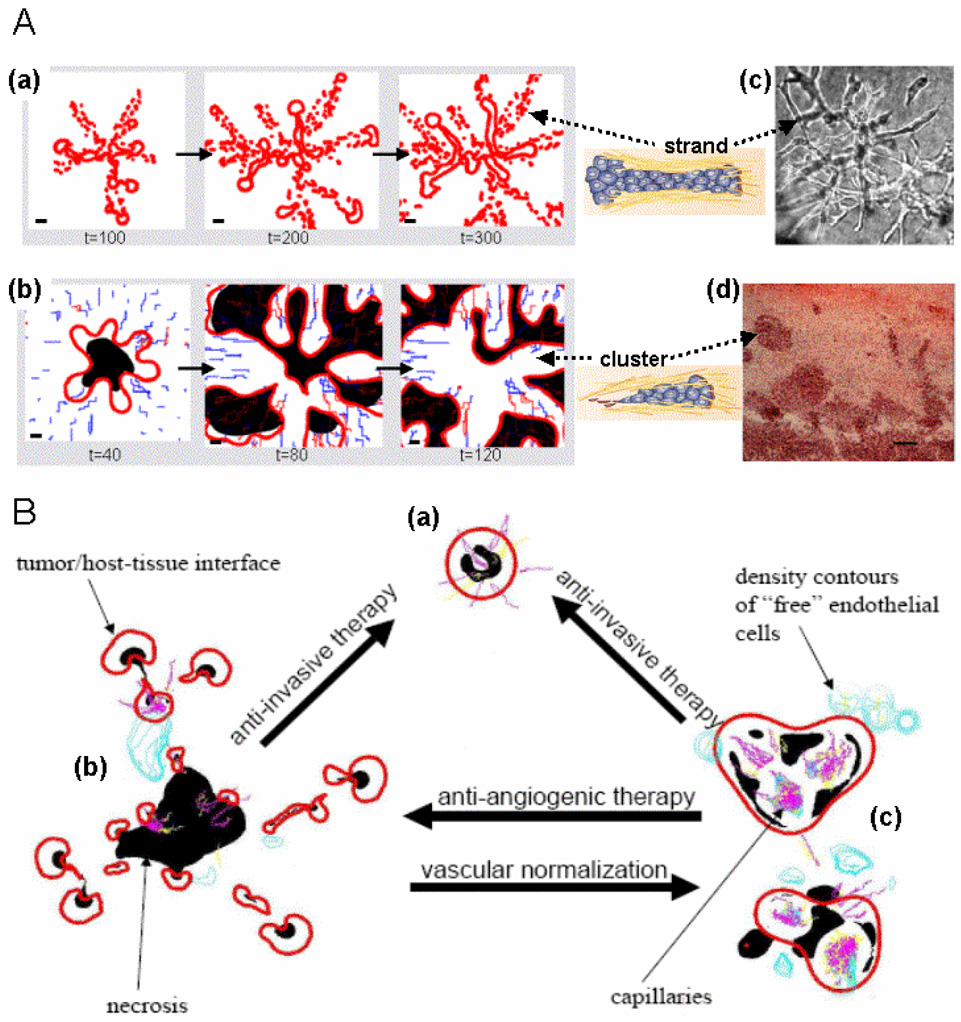
In Figure 6, A, different morphologies predicted by a continuum FCCMU model (Zheng et al., 2005a, Cristini et al., 2005) are shown, starting from the same initial condition of a spherical tumor. The model predicts that when cell taxis, but no proliferation is present (a), cells tend to align in chains or strands. When cell proliferation is significant (b), more “bulb like” protrusions form and detach into the host. These are also predicted to be more hypoxic. In all cases, these complex morphologic patterns developed because cell adhesion parameters were set very low ("morphologic instability,” Cristini et al., 2003). Corresponding structures observed after inducing hypoxia (c) in vitro (proliferation was inhibited) (Pennacchietti et al., 2003) and (d) in vivo (Rubenstein et al., 2000) are reported for comparison. The underlying molecular phenomena (Friedl & Wolf, 2003) responsible for the prevalence of one over another morphology, and for the spatial frequency of finger-like protrusions originating from a primary tumor, are captured by (phenomenological) model parameters describing proliferation and taxis and the associated convective cell fluxes on one side, and cell-adhesion forces on the other (Cristini et al., 2003).
Figure 6.
In-silico predictions of tumor morphology based on varying cellular and micro-environmental conditions in a parameter-sensitivity simulation study of the continuum model by Zheng et al. 2005 (figures are not to scale). These results extend the findings illustrated in Figure 4 and demonstrate that the in-silico model can account for the variety of invasive morphologies observed in tumors, and that the in-silico model is thus potentially predictive of tumor growth. (A) When cell taxis but not proliferation is present, cells are predicted to align in chains or strands (a). When cell proliferation is significant, more bulb- or cluster-like protrusions form and detach into the host (b). Red: tumor boundary; Black: hypoxia; Blue and Pink: neovascularization (immature and mature, respectively); time units are arbitrary. Drawings of cell strand and cluster adapted by permission from Macmillan Publishers Ltd: Nature Rev. Cancer, Vol. 3, p. 366, Friedl & Wolf, Copyright 2003. Corresponding structures observed after inducing hypoxia in vitro (proliferation was also inhibited) (c) (Pennacchietti et al., 2003) and in vivo (Rubenstein et al., 2000) through anti-angiogenic therapy (d) are shown for comparison. Bar, 80 µm. Reprinted from Cancer Cell, Vol. 3, Pennacchietti et al., page 354, Copyright (2003), with permission from Elsevier. Reprinted from Neoplasia, vol. 2, Rubenstein et al., page 311, Copyright 2000, with permission from Neoplasia Press. (B) Snapshots from three simulated tumor morphologies corresponding to different values of cell adhesion and vascularization parameters: high cell adhesion (a); low cell adhesion (b); low cell adhesion and with higher microvascular density or more efficient vascularization (c). Arrows indicate morphology transitions following different therapy strategies. Adapted from Cristini et al. (2005) with permission from the American Association of Cancer Research.
This model was further used to predict changes in the system dynamics following a range of possible perturbations of parameters related to cell-adhesion forces and to oxygen distribution in the environment, with the goal of providing suggestions for novel treatment protocols aimed at restoring normoxia and thus preventing “unstable” morphologies (Cristini et al., 2005, Frieboes et al., 2006a). Since these phenotypic and environmental parameters are also associated with invasion, this perturbation study provided preliminary quantification of the effects of anti-invasive and “vasculature-normalizing” anti-angiogenic therapeutic strategies that alter the balance of morphology-stabilizing and -destabilizing micro-environmental and molecular processes. In particular, this study helped explain the undesirable effects on morphology following current anti-angiogenic therapy due to exacerbation of micro-environmental hypoxic gradients and enhancement of cell migration (as reviewed above).
In Figure 6, B, case (a) corresponds to sufficiently high cell adhesion so that the simulated tumor growth is morphologically “stable”. Due to hypoxic gradients, necrotic regions have formed where concentrations are inadequate, leading to a diffusion-limited tumor size. Angiogenic factors (not shown) emanate from the peri-necrotic regions and diffuse outward, reaching pre-existing vessels and triggering neo-vascularization of the lesion. Even after angiogenesis, the model predicts that lesion (a) will maintain a compact shape because of high cell adhesion.
Case (b) corresponds to low cell adhesion, in which the tumor experiences morphological instability driven by hypoxic gradients as it progresses (Cristini et al., 2003, Zheng et al., 2005a, Byrne & Chaplain, 1997, Macklin & Lowengrub, 2007). Cell adhesion is insufficient to maintain proliferating cells together. The lesion breaks up into fragments, or detached cell clusters (Friedl & Wolf, 2003), moving away following outward gradients of nutrient and oxygen concentration (Zheng et al., 2005a, Macklin & Lowengrub, 2007). The model predicts that anti-invasive therapy enforces a morphology transition from (b) to (a) by increasing cell adhesion.
Case (c) corresponds to low cell adhesion (as in (b)), but with a more spatially uniform distribution of vessels. The simulation predicts that this “vascular normalization” would lead to reduced oxygen gradients, and hence to suppression of instability and to clustering of cells into a more compact tumor morphology. This result could be achieved by pruning immature and inefficient blood vessels, leading to a more normal vasculature of vessels reduced in diameter, density, and permeability (Jain, 1990, 2001, 2005). In contrast, after anti-angiogenic therapy ((c) to (b)), increased scattering of tumor cell clusters in response to hypoxia is predicted, as documented in vivo by Bello et al. (2004) and by others. Remarkably, the simulations also predict that in this case some tumor cell clusters tend to co-opt the vasculature to maximize nutrient uptake, as documented previously in vivo (Kunkel et al., 2001, Lamszus et al., 2003, Rubenstein et al., 2000).
Figure 7 shows a summary of some of the biology revealed by the predictive model presented here (Zheng et al., 2005, Sinek et al., 2004 and in press, Cristini et al., 2005, Frieboes et al., 2006 and 2007, Sanga et al., 2006) under the categories of Tumor, Microenvironment, Treatment Response, and Vasculature, including gross tumor morphology in 3-D (A), gradients of cell substrates (B), tissue fragmentation in response to chemotherapy involving large nanoparticles and adjuvant anti-angiogenic therapy (C), and tumor vasculature with both conducting and non-conducting vessels (D).
Figure 7.
Tumor biology revealed by parameter-sensitivity studies of a continuum FCCMU computer model is listed under the categories of Tumor, Microenvironment, Treatment Response, and Vasculature. (A) Gross tumor growth and morphology in 3-D are predicted based on cell-scale parameters (e.g., proliferation, cell adhesion) set from experimental values. Viable (light grey) and necrotic (dark grey) tissue as well as extensive vascularization (conducting vessels in red, non-conducting in blue) are shown. Reprinted from NeuroImage, Frieboes et al., in press, Copyright 2007, with permission from Elsevier. (B) Gradients of cell substrates (from highest (red) to lowest concentration (blue)) are predicted from the vasculature topology (dark red lines). Thin dashed line denotes tumor boundary. Reprinted from Journal of Mathematical Biology, Sinek et al., in press, Copyright 2007 Springer. With kind permission of Springer Science and Business Media. (C) Local tumor fragmentation (top) is predicted in response to chemotherapy involving large nanoparticles and adjuvant anti-angiogenesis (bottom). Boundary of tumor fragments is in red, vessels are pink (conducting) and light blue (non-conducting). Gradient of drug (red, highest, blue, lowest) is centered in middle area of tumor tissue. Adapted from Biomedical Microdevices, Vol 6, 2004, p. 307, Sinek et al., Figure 5. Copyright 2004 Kluwer Academic Publishers. With kind permission of Springer Science and Business Media. (D) Abnormal tumor vasculature architecture with conducting (red) and nonconducting vessels (blue) is predicted based on angiogenic regulators produced by tumor and host tissue. Reprinted from NeuroImage, Frieboes et al., in press, Copyright 2007, with permission from Elsevier.
CONCLUSIONS AND FUTURE WORK
The research direction we envision focuses on the development and application to tumor biology of quantitative methods traditionally confined to engineering and the physical sciences. Indeed, it is clear that such complex biological systems dominated by large numbers of processes and highly complex dynamics are very difficult to approach by experimental methods alone. They can typically be understood only by using appropriate mathematical models and sophisticated computer simulations complementary to experimental investigations. In the innovative and powerful multidisciplinary approach reviewed here, mathematical and computational modeling completes the circle of discovery: laboratory experiments provide data that, in turn, informs the construction of a mathematical model that can then predict behavior and guide the design of future experiments to test these predictions.
This multi-scale approach captures tumor progression by taking into account ongoing molecular and cellular scale events (Martinez-Zaguilan et al., 1996, Schlappack et al., 1991, Rofstad et al., 1999, Gatenby et al., 2006, Jensen, 2006). One of the key links established in a more quantitative manner is that mutations drive increased cellular uptake, which introduces perturbations in spatial gradients of oxygen and nutrient; these gradients enhance hypoxia and cause heterogeneous cell proliferation and migration leading towards diffusional shape instabilities. This supports the hypothesis that cellular and extra-cellular properties driving tumor growth and invasiveness also determine tumor morphology (Cristini et al., 2005, Frieboes et al., 2006a) and suggests that morphological characteristics including neo-vasculature and harmonic content of the tumor edge should serve as predictors of tumor growth (Cristini et al., 2006).
Predictive modeling assumes that criteria and critical microphysical conditions for tumor invasion can be formulated in terms of physical laws linking tissue architecture and morphology to cell phenotype. Future multidisciplinary investigations should exploit the power of predictive modeling that allows observable properties of the tumor, such as its morphology in general and specifically the cell spatial arrangements at the tumor boundary, to be used to both understand the underlying cellular physiology and predict subsequent invasive behavior. We envision this research taking steps towards further establishing the dependence of tumor cell motion into surrounding host tissue on the balance between cell proliferation and adhesion, as well as perturbations caused by microenvironmental factors such as oxygen, nutrient, and H+ diffusion gradients. This will include the continuing application of mathematical and empirical methods to quantify the competition between gradient-related pro-invasion phenomena and molecular forces that govern proliferation and taxis, and forces opposing invasion through cell adhesion. In addition, a more detailed description of the complex in vivo environment, which better recapitulates the conditions of tumors in patients, would be valuable.
Currently, pathologic analysis is often limited to a set of morphological features that are rarely quantitatively assessed (the main quantitative factors are mitotic rates and size of invasive tumor “fingers”), and these measures differ depending on the types of tumor. “Degree of pleiomorphism” (variable phenotypes) is also used as a prognosticator, although this has no absolute quantitative definition and is subjective. Multi-scale modeling of cancer would allow predictions of cellular and molecular perturbations that alter invasiveness and are measured through changes in tumor morphology. This opens the possibility of designing novel individualized therapeutic strategies in which the microenvironment and cellular factors are manipulated with the aim of imposing compact tumor morphology by both decreasing invasiveness and promoting defined tumor margins—an outcome that would benefit cancer therapy by improving local tumor control through surgery or chemotherapy.
Acknowledgements
We gratefully acknowledge John Lowengrub and Steven M. Wise (Mathematics, U.C. Irvine), and the reviewers for helpful comments and suggestions. We thank Ed Stopa at Rhode Island Hospital and the Columbia University Alzheimer's Brain Bank for archived human glioma specimens. Funding from NIH-NIGMS RO1 GM47368 and RO1-NS046810_(E.L.B.), from the National Science Foundation (V.C.) and National Cancer Institute (V.C.; R.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam J. General aspects of modeling tumor growth and the immune response. In: Adam J, Bellomo N, editors. A survey of models on tumor immune systems dynamics. Boston: Birkhauser; 1996. pp. 15–87. [Google Scholar]
- Ambrosi D, Guana F. Stress-modulated growth. Math. Meth. Solids. 2006 in press. [Google Scholar]
- Ambrosi D, Preziosi L. On the closure of mass balance models for tumor growth. Math. Mod. Meth. Appl. Sci. 2002;12:737–754. [Google Scholar]
- Anderson ARA, Chaplain MAJ. Continuous and discrete models of tumour-induced angiogenesis. Bull. Math. Biol. 1998;60:857–899. doi: 10.1006/bulm.1998.0042. [DOI] [PubMed] [Google Scholar]
- Anderson ARA, Chaplain MAJ, Newman EL, Steele RJC, Thompson AM. Mathematical modeling of tumour invasion and metastasis. J. Theor. Med. 2000;2:129–154. [Google Scholar]
- Anderson ARA. A hybrid mathematical model of solid tumour invasion: the importance of cell adhesion. Math. Med. Biol. 2005;22:163–186. doi: 10.1093/imammb/dqi005. [DOI] [PubMed] [Google Scholar]
- Anderson A, Zheng X, Cristini V. Adaptive unstructured volume remeshing-I: The method. J. of Computational Physics. 2005;208(2):616–625. [Google Scholar]
- Anderson DM, McFadden GB, Wheeler AA. Diffuse interface methods in fluid mechanics. Ann. Rev. Fluid Mech. 1998;30:139–165. [Google Scholar]
- Araujo RP, McElwain DLS. A history of the study of solid tumor growth: The contribution of mathematical modeling. Bull. Math. Biol. 2003;66:1039–1091. doi: 10.1016/j.bulm.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Ayati BP, Webb GF, Anderson ARA. Computational methods and results for structured multiscale models of tumor invasion. Multiscale Model. Simul. 2006;5:1–20. [Google Scholar]
- Bello L, Lucini V, Costa F, Pluderi M, Giussani C, Acerbi F, Carrabba G, Pannaci M, Caronzolo D, Grosso S, Shinkaruk S, Colleoni F, Canron X, Tomei G, Deleris G, Bikfalvi A. Combinatorial Administration of Molecules That Simultaneously Inhibit Angiogenesis and Invasion Leads to Increased Therapeutic Efficacy in Mouse Models of Malignant Glioma. Clin Cancer Res. 2004;10:4527–4537. doi: 10.1158/1078-0432.CCR-04-0194. [DOI] [PubMed] [Google Scholar]
- Bellomo N, Preziosi L. Modeling and mathematical problems related to tumor evolution and its interaction with the immune system. Math. Comp. Modell. 2000:413–452. [Google Scholar]
- Berger M, Colella P. Local adaptive mesh refinement for shock hydrodynamics. J. Comp. Phys. 1989;82:64–84. [Google Scholar]
- Bernsen HJJA, Van der Kogel AJ. Antiangiogenic therapy in brain tumor models. J. Neuro-oncology. 1999;45:247–255. doi: 10.1023/a:1006395802727. [DOI] [PubMed] [Google Scholar]
- Bloemendal HJ, Logtenberg T, Voest EE. New strategies in anti-vascular cancer therapy. Euro. J. Clinical Investig. 1999;29:802–809. doi: 10.1046/j.1365-2362.1999.00534.x. [DOI] [PubMed] [Google Scholar]
- Boushaba K, Levine HA, Nilsen-Hamilton M. A mathematical model for the regulation of tumor dormancy based on enzyme kinetics. Bull. Math. Biol. 2006;68:1495–1526. doi: 10.1007/s11538-005-9042-z. [DOI] [PubMed] [Google Scholar]
- Brandt A. Multi-level adaptive solutions to boundary-value problems. Math. Comput. 1977;31:333–390. [Google Scholar]
- Bru A, Albertos S, Subiza JL, Garcia-Asenjo JL, Bru I. The universal dynamics of tumor growth. Biophys. J. 2003;85:2948–2961. doi: 10.1016/S0006-3495(03)74715-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne HM, Alarcon T, Owen MR, Webb SD, Maini PK. Modeling aspects of cancer dynamics: A review. Phil. Trans. R. Soc. A. 2006;364:1563–1578. doi: 10.1098/rsta.2006.1786. [DOI] [PubMed] [Google Scholar]
- Byrne H, Chaplain M. Growth of nonnecrotic tumors in the presence and absence of inhibitors. Math. Biosci. 1995a;130:151–181. doi: 10.1016/0025-5564(94)00117-3. [DOI] [PubMed] [Google Scholar]
- Byrne H, Chaplain M. Free boundary value problems associated with the growth and development of multicellular spheroids. Eur. J. Appl. Math. 1995b;8:639. [Google Scholar]
- Byrne HM, Chaplain MAJ. Growth of necrotic tumors in the presence and absence of inhibitors. Math. Biosci. 1996a;135:187–216. doi: 10.1016/0025-5564(96)00023-5. [DOI] [PubMed] [Google Scholar]
- Byrne HM, Chaplain MAJ. Modeling the role of cell-cell adhesion in the growth and development of carcinomas. Math. Comput. Model. 1996b;24:1–17. [Google Scholar]
- Byrne H, Chaplain M. Free boundary value problems associated with the growth and development of multicellular spheroids. Eur. J. Appl. Math. 1997;8:639–658. [Google Scholar]
- Byrne HM, Preziosi L. Modeling solid tumor growth using the theory of mixtures. Math. Meth. Biol. 2003;20:341–366. doi: 10.1093/imammb/20.4.341. [DOI] [PubMed] [Google Scholar]
- Castro M, Molina-Paris C, Deisboeck TS. Tumor growth instability and the onset of invasion. Phys. Rev. E. 2005;72:041907.1–041907.12. doi: 10.1103/PhysRevE.72.041907. [DOI] [PubMed] [Google Scholar]
- Chaplain MAJ. Avascular growth, angiogenesis and vascular growth in solid tumours: the mathematical modelling of the stages of tumour development. Math. Comput. Model. 1996;23:47–87. [Google Scholar]
- Chaplain MAJ, Anderson ARA. In: Mathematical modeling of tissue invasion. Preziosi L, editor. CRC Press: Cancer Modeling and Simulation; 2003. pp. 269–297. [Google Scholar]
- Chaplain MAJ, Lolas G. Mathematical modeling of cancer cell invasion of tissue: The role of the urokinase plasminogen activation system. Math. Models Meth. Appl. Sci. 2005;15:1685–1734. [Google Scholar]
- Chaplain MAJ, Graziano L, Preziosi L. Mathematical modeling of the loss of tissue compression responsiveness and its role in solid tumor development. Math. Med. Biol. 2006;23:192–229. doi: 10.1093/imammb/dql009. [DOI] [PubMed] [Google Scholar]
- Chicoine MR, Silbergeld DL. Assessment of brain tumour cell motility in vivo and in vitro. J. Neurosurg. 1995;82:615–622. doi: 10.3171/jns.1995.82.4.0615. [DOI] [PubMed] [Google Scholar]
- Chomyak OG, Sidorenko MV. Multicellular spheroids model in oncology. Exp. Oncology. 2001;23:236–241. [Google Scholar]
- Condeelis J, Singer RH, Segall JE. The great escape: When cancer cells hijack the genes for chemotaxis and motility. Annu. Rev. Cell Dev. Biol. 2005;21:695–718. doi: 10.1146/annurev.cellbio.21.122303.120306. [DOI] [PubMed] [Google Scholar]
- Cristini V, Blawzdziewicz J, Loewenberg M. An adaptive mesh algorithm for evolving surfaces: simulations of drop breakup and coalescence. J Computational Physics. 2001;168:445–463. [Google Scholar]
- Cristini V, Lowengrub J, Nie Q. Nonlinear simulation of tumor growth. J. Math. Biol. 2003;46:191–224. doi: 10.1007/s00285-002-0174-6. [DOI] [PubMed] [Google Scholar]
- Cristini V, Tan Y-C. Theory and numerical simulation of droplet dynamics in complex flows-A review. Lab on a Chip. 2004;4(4):257–264. doi: 10.1039/b403226h. [DOI] [PubMed] [Google Scholar]
- Cristini V, Frieboes H, Gatenby R, Caserta M, Ferrari M, Sinek J. Morphological instability and cancer invasion. Clinical Cancer Res. 2005;11:6772–6779. doi: 10.1158/1078-0432.CCR-05-0852. [DOI] [PubMed] [Google Scholar]
- Cristini V, Gatenby R, Lowengrub J. Multidisciplinary studies of tumor invasion and the role of the microenvironment. 2006 NIH 1R01CA127769-01.
- Dickinson RB, Tranquillo RT. A stochastic model for adhesion-mediated cell random motility and haptotaxis. J. Math. Biol. 1993;31:1432–1416. doi: 10.1007/BF00161199. [DOI] [PubMed] [Google Scholar]
- DiMilla PA, Barbee K, Lauffenburger DA. Mathematical model for the effects of adhesion and mechanics on cell migration speed. Biophys. J. 1991;60:15–37. doi: 10.1016/S0006-3495(91)82027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis AN, Mombach JCM, Walter M, de Avila LF. The interplay between cell adhesion and environment rigidity in the morphology of tumors. Physica A. 2003;322:546–554. [Google Scholar]
- Elvin P, Garner AP. Tumour invasion and metastasis: challenges facing drug discovery. Curr. Opin. Pharmacol. 2005;5:374–381. doi: 10.1016/j.coph.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Ferreira SC, Martins ML, Vilela MJ. Reaction-diffusion model for the growth of avascular tumor. Physical Review E. 2002;65:021907. doi: 10.1103/PhysRevE.65.021907. [DOI] [PubMed] [Google Scholar]
- Frieboes H, Zheng X, Sun CH, Tromberg B, Gatenby R, Cristini V. An integrated computational/experimental model of tumor invasion. Cancer Res. 2006a;66:1597–1604. doi: 10.1158/0008-5472.CAN-05-3166. [DOI] [PubMed] [Google Scholar]
- Frieboes HB, Sinek JP, Nalcioglu O, Fruehauf JP, Cristini V. BioMEMS and Biomedical Nanotechnology. Volume I. Springer-Verlag: Biological and Biomedical Nanotechnology; 2006b. Nanotechnology in cancer drug therapy: a biocomputational approach; pp. 441–466. [Google Scholar]
- Frieboes HB, Lowengrub J, Wise S, Zheng X, Macklin P, Bearer E, Cristini V. Computer simulation of glioma growth and morphology. NeuroImage. 2007 doi: 10.1016/j.neuroimage.2007.03.008. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Wolf A. Tumor cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- Friedl P. Prespecification and plasticity: shifting mechanisms of cell migration. Curr. Opin. Cell Biol. 2004;16:14–23. doi: 10.1016/j.ceb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Friedl P, Hegerfeldt Y, Tilisch M. Collective cell migration in morphogenesis and cancer. Int. J. Dev. Biol. 2004;48:441–449. doi: 10.1387/ijdb.041821pf. [DOI] [PubMed] [Google Scholar]
- Friedman A. A hierarchy of cancer models and their mathematical challenges. Discrete Cont. Dyn. Systems Ser. B. 2004;4:147–159. [Google Scholar]
- Garcke H, Nestler B, Stinner B. A diffuse interface model for alloys with multiple components and phases. SIAM J. Appl. Math. 2004;64:775–799. [Google Scholar]
- Garner AL, Lau YY, Jackson TL, Uhler MD, Jordan DW, Gilgenbach RM. Incorporating spatial dependence into a multicellular tumor spheroid growth model. J. App. Phys. 2005;98:124701.1–124701-8. [Google Scholar]
- Gatenby RA, Gawlinski ET. A reaction-diffusion model of acid-mediated invasion of normal tissue by neoplastic tissue. Cancer Res. 1996;56:5745–5753. [PubMed] [Google Scholar]
- Gatenby RA, Gawlinski ET. The glycotic phenotype in carcinogenesis and tumor invasion: insights through mathematical models. Cancer Res. 2003;63:3847–3854. [PubMed] [Google Scholar]
- Gatenby RA, Gawlinski ET, Gmitro AF, Kaylor B, Gillies RJ. Acid-mediated tumor invasion: a multidisciplinary study. Cancer Res. 2006;66:5216–5223. doi: 10.1158/0008-5472.CAN-05-4193. [DOI] [PubMed] [Google Scholar]
- Greenspan HP. On the growth and stability of cell cultures and solid tumors. J. Theor. Biol. 1976;56:229–242. doi: 10.1016/s0022-5193(76)80054-9. [DOI] [PubMed] [Google Scholar]
- Hatzikirou H, Deutsch A, Schaller C, Simon M, Swanson K. Mathematical modeling of glioblastoma tumour development: A review. Math. Models Meth. Appl. Sci. 2005;15:1779–1794. [Google Scholar]
- Hogea CS, Murray BT, Sethian JA. Simulating complex tumor dynamics from avascular to vascular growth using a general level set method. J. Math. Biol. 2006;53:86–134. doi: 10.1007/s00285-006-0378-2. [DOI] [PubMed] [Google Scholar]
- Huang S, Ingber DE. The structural and mechanical complexity of cell-growth control. Nat. Cell. Biol. 1999;1:E131–E138. doi: 10.1038/13043. [DOI] [PubMed] [Google Scholar]
- Jacqmin D. Calculation of two-phase Navier-Stokes flows using phase-field modeling. J. Comp. Phys. 1999;155:96–127. [Google Scholar]
- Jackson TL, Byrne HM. A mechanical model of tumor encapsulation and transcapsular spread. Math. Biosci. 2002;180:307–328. doi: 10.1016/s0025-5564(02)00118-9. [DOI] [PubMed] [Google Scholar]
- Jackson TL. A mathematical model of prostate tumor growth and androgen-independent relapse. Cont. Discr. Dyn. Syst. B. 2004;4:187–202. [Google Scholar]
- Jain RK. Physiological barriers to delivery of monoclonal antibodies and other macromolecules in tumors. Cancer Res. 1990;50 Suppl:814s–819s. [PubMed] [Google Scholar]
- Jain RK. Delivery of molecular medicine to solid tumors: lessons from in vivo imaging of gene expression and function. J. Controlled Release. 2001;74:7–25. doi: 10.1016/s0168-3659(01)00306-6. [DOI] [PubMed] [Google Scholar]
- Jain RK. Normalization of tumor vasculature: An emerging concept in antiangiogenic therapy. Science. 2005;307:58–62. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- Jensen RL. Hypoxia in the tumorigenesis of gliomas and as a potential target for therapeutic measures. Neurosurg. Focus. 2006;20:E24. doi: 10.3171/foc.2006.20.4.16. [DOI] [PubMed] [Google Scholar]
- Jones AF, Byrne HM, Gibson JS, Dold JW. Mathematical model of the stress induced during avascular tumour growth. J. Math. Biol. 2000;40:473–499. doi: 10.1007/s002850000033. [DOI] [PubMed] [Google Scholar]
- Kansal AR, Torquato S, Harsh GR, Chiocca EA, Diesboeck TS. Cellular automaton of idealized brain tumor growth dynamics. Biosystems. 2000a;55:119–127. doi: 10.1016/s0303-2647(99)00089-1. [DOI] [PubMed] [Google Scholar]
- Kansal AR, Torquato S, Harsh GR, Chiocca EA, Deisboeck TS. Simulated brain tumor growth dynamics using a three-dimensional cellular automaton. J. Theor. Biol. 2000b;203:367–382. doi: 10.1006/jtbi.2000.2000. [DOI] [PubMed] [Google Scholar]
- Keller PJ, Pampaloni F, Stelzer EHK. Life sciences require the third dimension. Curr. Opin. Cell. Biol. 2006;18:117–124. doi: 10.1016/j.ceb.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Khain E, Sander LM, Stein AM. A model for glioma growth. Complexity. 2005;11:53–57. [Google Scholar]
- Khain E, Sander LM. Dynamics and pattern formation in invasive tumor growth. Phys. Rev. Lett. 2006;96:188103.1–188103.4. doi: 10.1103/PhysRevLett.96.188103. [DOI] [PubMed] [Google Scholar]
- Kim JB. Three-dimensional tissue culture models in cancer biology. Seminars Cancer Biol. 2005;15:236–241. doi: 10.1016/j.semcancer.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Kim JS, Kang K, Lowengrub JS. Conservative multigrid methods for Cahn-Hilliard fluids. J. Comp. Phys. 2004a;193:511–543. [Google Scholar]
- Kim JS, Kang K, Lowengrub JS. Conservative multigrid methods for ternary Cahn-Hilliard systems. Comm. Math. Sci. 2004b;12:53–77. [Google Scholar]
- Kim JS, Lowengrub JS. Phase field modeling and simulation of three-phase flows. Int. Free Bound. 2005;7:435. [Google Scholar]
- Kopfstein L, Christofori G. Metastasis: cell-autonomous mechanisms versus contributions by the tumor microenvironment. Cell. Mol. Life Sci. 2006;63:449–468. doi: 10.1007/s00018-005-5296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper RAJ, Schellens JHM, Blijham GH, Beijnen JH, Voest EE. Clinical research on antiangiogenic therapy. Pharmacol Res. 1998;37:1–16. doi: 10.1006/phrs.1997.0268. [DOI] [PubMed] [Google Scholar]
- Kunkel P, Ulbricht U, Bohlen P, Brockmann MA, Fillbrandt R, Stavrou D, Westphal M, Lamszus K. Inhibition of glioma angiogenesis and growth in vivo by systemic treatment with a monoclonal antibody against vascular endothelial growth factor receptor-2. Cancer Res. 2001;61:6624–6628. [PubMed] [Google Scholar]
- Lah TT, Alonso MBD, Van Noorden CJF. Antiprotease therapy in cancer: hot or not? Expert Opin. Biol. Ther. 2006;6:257–279. doi: 10.1517/14712598.6.3.257. [DOI] [PubMed] [Google Scholar]
- Lamszus K, Kunkel P, Westphal M. Invasion as limitation to anti-angiogenic glioma therapy. Acta Neurochir Suppl. 2003;88:69–77. doi: 10.1007/978-3-7091-6090-9_23. [DOI] [PubMed] [Google Scholar]
- Lee H, Lowengrub JS, Goodman J. Modeling pinchoff and reconnection in a Hele-Shaw cell I. The models and their calibration. Phys. Fluids. 2002;14:492–513. [Google Scholar]
- Leo PH, Lowengrub JS, Jou H-J. A diffuse interface model for elastically stressed solids. Acta Metall. 1998;46:2113–2130. [Google Scholar]
- Levine HA, Pamuk S, Sleeman BD, Nilsen-Hamilton M. Mathematical modeling of capillary formation and development in tumor angiogenesis: penetration into the stroma. Bull. Math. Biol. 2002;54:423. doi: 10.1006/bulm.2001.0240. [DOI] [PubMed] [Google Scholar]
- Leyrat A, Duperray A, Verdier C. “Adhesion mechanisms in cancer metastasis” Chap. 8. In: Preziosi L, editor. Cancer Modelling and Simulation. CRC Press; 2003. pp. 221–242. [Google Scholar]
- Li X, Cristini V, Nie Q, Lowengrub J. Nonlinear three-dimensional simulation of solid tumor growth. Discrete and Continuous Dynamical Systems B. 2007 In press. [Google Scholar]
- Lowengrub JS, Truskinovsky L. Quasi-incompressible Cahn-Hilliard fluids and topological transitions. Proc. R. Soc. London A. 1998;454:2617–2654. [Google Scholar]
- Macklin P, Lowengrub JS. Evolving interfaces via gradients of geometry-dependent interior Poisson problems: Application to tumor growth. J. Comp. Phys. 2005;203:191–220. [Google Scholar]
- Macklin P, Lowengrub JS. Nonlinear simulation of the effect of microenvironment on tumor growth. J. Theor. Biol. 2007 doi: 10.1016/j.jtbi.2006.12.004. in press. [DOI] [PubMed] [Google Scholar]
- Mareel M, Leroy A. Clinical, cellular, and molecular aspects of cancer invasion. Physiol. Rev. 2003;83:337–376. doi: 10.1152/physrev.00024.2002. [DOI] [PubMed] [Google Scholar]
- Martinez-Zaguilan R, Seftor EA, Seftor REB, Chu YW, Gillies RJ, Hendrix MJC. Acidic pH enhances the invasive behavior of human melanoma cells. Clin. Exp. Metastasis. 1996;14:176–186. doi: 10.1007/BF00121214. [DOI] [PubMed] [Google Scholar]
- McDougall SR, Anderson ARA, Chaplain MAJ. Mathematical modelling of dynamic adaptive tumour-induced angiogenesis: clinical applications and therapeutic targeting strategies. J. Theor. Biol. 2006;241:564–589. doi: 10.1016/j.jtbi.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Mueller-Klieser W. Tumor biology and experimental therapeutics. Crit. Rev. Oncol. Hemat. 2000;36:159–178. doi: 10.1016/s1040-8428(00)00082-2. [DOI] [PubMed] [Google Scholar]
- Nagy JD. The ecology and evolutionary biology of cancer: A review of mathematical models of necrosis and tumor cell diversity. Math. Biosci. Eng. 2005;2:381–418. doi: 10.3934/mbe.2005.2.381. [DOI] [PubMed] [Google Scholar]
- Page DL, Anderson TJ, Sakamoto G. Diagnostic Histopathology of the Breast Churchill Livingstone. New York: 1987. pp. 219–222. [Google Scholar]
- Painter KJ. Development and applications of a model for cellular response to multiple chemotactic cues. J. Math Biol. 2000;41:285–314. doi: 10.1007/s002850000035. [DOI] [PubMed] [Google Scholar]
- Patel AA, Gawlinski ET, Lemieux SK, Gatenby RA. A cellular automaton model of early tumor growth and invasion: the effects of native tissue vascularity and increase anaerobic tumor metabolism. J. Theor. Biol. 2001;213:315–331. doi: 10.1006/jtbi.2001.2385. [DOI] [PubMed] [Google Scholar]
- Pennacchietti S, Michieli P, Galluzzo M, Mazzone M, Giordano S, Comoglio PM. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell. 2003;3:347–361. doi: 10.1016/s1535-6108(03)00085-0. [DOI] [PubMed] [Google Scholar]
- Plank MJ, Sleeman BD. A reinforced random walk model of tumour angiogenesis and anti-angiogenic strategies. Math. Med. Biol. 2003:20135–20181. doi: 10.1093/imammb/20.2.135. [DOI] [PubMed] [Google Scholar]
- Plank MJ, Sleeman BD. Lattice and non-lattice models of tumour angiogenesis. Bull. Math. Biol. 2004;66:1785–1819. doi: 10.1016/j.bulm.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Quaranta V, Weaver AM, Cummings PT, Anderson ARA. Mathematical modeling of cancer: The future of prognosis and treatment. Clinica Chimica Acta. 2005;357:173–179. doi: 10.1016/j.cccn.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: Integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Roberts HC, Roberts TPI, Lee T-Y, Dillon WP. Dynamic, contrast-enhanced CT of human brain tumors: quantitative assessment of blood volume, blood flow, and microvascular permeability: report of two cases. Am. J. Neuroradial. 2002;23:828–832. [PMC free article] [PubMed] [Google Scholar]
- Rofstad EK, Danielsen T. Hypoxia-induced metastasis of human melanoma cells: involvement of vascular endothelial growth factor-mediated angiogenesis. Br. J. Cancer. 1999;80:1697–1707. doi: 10.1038/sj.bjc.6690586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Kim J, Ozawa T, Zhang M, Westphal M, Deen DF, Shuman MA. Anti-VEGF antibody treatment of glioblastoma prolongs survival but results in increased vascular cooption. Neoplasia. 2000;2:306–314. doi: 10.1038/sj.neo.7900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E. Mechanisms of cancer cell invasion. Curr. Opin. Genet. Dev. 2005;15:87–96. doi: 10.1016/j.gde.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Sander LM, Deisboeck TS. Growth patterns of microscopic brain tumors. Phys. Rev. E. 2002;66:051901.1–051901.7. doi: 10.1103/PhysRevE.66.051901. [DOI] [PubMed] [Google Scholar]
- Sanga S, Sinek JP, Frieboes HB, Ferrari M, Fruehauf JP, Cristini V. Mathematical modeling of cancer progression and response to chemotherapy. Expert Rev. Anticancer Ther. 2006;6:1361–1376. doi: 10.1586/14737140.6.10.1361. [DOI] [PubMed] [Google Scholar]
- Sanga S, Frieboes HB, Sinek JP, Cristini V. Cancer Nanotechnology (American Scientific) 2007. A Multiscale Approach for Computational Modeling of Biobarriers to Cancer Chemotherapy via Nanotechnology Ch. 10; pp. 1–21. [Google Scholar]
- Schlappack OK, Zimmermann A, Hill RP. Glucose starvation and acidosis: effect on experimental metastasic potential, DNA content and MTX resistance of murine tumour cells. Br. J. Cancer. 1991;64:663–670. doi: 10.1038/bjc.1991.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seftor EA, Meltzer PS, Kirschmann DA, Pe’er J, Maniotis AJ, Trent JM, Folberg R, Hendrix MJC. Molecular determinants of human uveal melanoma invasion and metastasis. Clin. Exp. Metastasis. 2002;19:233–246. doi: 10.1023/a:1015591624171. [DOI] [PubMed] [Google Scholar]
- Sierra A. Metastases and their microenvironments: linking pathogenesis and therapy. Drug Resist. Updat. 2005;8:247–257. doi: 10.1016/j.drup.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Silbergeld DL, Chicoine MR. Isolation and characterization of human malignant glioma cells from histologically normal brain. J. Neurosurg. 1997;86:525–531. doi: 10.3171/jns.1997.86.3.0525. [DOI] [PubMed] [Google Scholar]
- Sinek J, Frieboes HB, Zheng X, Cristini V. Two-dimensional Chemotherapy Simulations Demonstrate Fundamental Transport and Tumor Response Limitations Involving Nanoparticles. Biomed Microdev. 2004;6:297–309. doi: 10.1023/B:BMMD.0000048562.29657.64. [DOI] [PubMed] [Google Scholar]
- Sinek JP, Frieboes HB, Sivaraman B, Sanga S, Cristini V. Nanotechnologies for the Life Sciences. Vol. 4. Wiley-VCH: Nanodevices for the Life Sciences; 2006. Mathematical and Computational Modeling: Towards the development and application of nanodevices for drug delivery; pp. 29–66. [Google Scholar]
- Sinek JP, Sanga S, Zheng X, Cristini V. Predicting drug pharmacokinetics and effect in vascularized tumors using computer simulation. J. Math. Biol. 2007 doi: 10.1007/s00285-008-0214-y. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeg PS. Angiogenesis inhibitors: motivators of metastasis? Nature Medicine. 2003;9:822–823. doi: 10.1038/nm0703-822. [DOI] [PubMed] [Google Scholar]
- Stein AM, Demuth T, Mobley D, Berens M, Sander LM. A mathematical model of glioblastoma tumor spheroid invasion in a three-dimensional in vitro experiment. Biophys. J. 2007;92:356–365. doi: 10.1529/biophysj.106.093468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanou A, McDougall SR, Anderson ARA, Chaplain MAJ. Mathematical modeling of flow in 2D and 3D vascular networks: applications to anti-angiogenic and chemotherapeutic drug strategies. Math. Comp. Modeling. 2005;41:1137–1156. [Google Scholar]
- Stetler-Stevenson WG, Aznavoorian S, Liotta LA. Tumor cell interactions with the extracellular matrix during invasion and metastasis. Ann. Rev. Cell. Biol. 1993;9:541–573. doi: 10.1146/annurev.cb.09.110193.002545. [DOI] [PubMed] [Google Scholar]
- Sun S, Wheeler MF, Obeyesekere M, Patrick CW. A deterministic model of growth factor-induced angiogenesis. Bull. Math. Biol. 2005;67:313–337. doi: 10.1016/j.bulm.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Swanson KR, Alvord EC, Murray JD. A quantitative model for differential motility of gliomas in grey and white matter. Cell Prolif. 2000;33:317–329. doi: 10.1046/j.1365-2184.2000.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson KR, Bridge C, Murray JD, Alvord EC., jr Virtual and real brain tumors: Using mathematical modeling to quantify glioma growth and invasion. J. Neuro. Sci. 2003;216:1–10. doi: 10.1016/j.jns.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Turner S, Sherratt JA. Intercellular adhesion and cancer invasion: a discrete simulation using the extended Potts model. J. Theor. Biol. 2002;216:85–100. doi: 10.1006/jtbi.2001.2522. [DOI] [PubMed] [Google Scholar]
- Tysnes BB, Larsen LF, Ness GO, Mahesparan R, Edvardsen K, Garcia-Cabrera I, Bjerkvig R. Stimulation of glioma-cell migration by laminin and inhibition by anti-alpha3 and anti-beta 1 integrin antibodies. Int. J. Cancer. 1996;67:777–784. doi: 10.1002/(SICI)1097-0215(19960917)67:6<777::AID-IJC5>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- van Kempen LCLT, Ruiter DJ, van Muijen GNP, Coussens LM. The tumor microenvironment: a critical determinant of neoplastic evolution. Eur. J. Cell. Biol. 2003;82:539–548. doi: 10.1078/0171-9335-00346. [DOI] [PubMed] [Google Scholar]
- Wise SM, Lowengrub JS, Kim JS, Johnson WC. Efficient phase-field simulation of quantum dot formation in a strained heteroepitaxial film. Superlattices and Microstructures. 2004;36:293–304. [Google Scholar]
- Wise SM, Lowengrub JS, Kim JS, Thornton K, Voorhees PW, Johnson WC. Quantum dot formation on a strain-patterned epitaxial thin film. Appl. Phys. Lett. 2005;87:133102. [Google Scholar]
- Wolf K, Friedl P. Molecular mechanisms of cancer cell invasion and plasticity. Br. J. Dermatol. 2006;154 Suppl:11–15. doi: 10.1111/j.1365-2133.2006.07231.x. [DOI] [PubMed] [Google Scholar]
- Wu FY. The Potts model. Rev. Mod. Phys. 1982;54:235–268. [Google Scholar]
- Xie H, Li G, Ning H, Menard C, Coleman CN, Miller RW. 3D voxel fusion of multi-modality medical images in a clinical treatment planning system; Proceedings of the 17th IEEE Symposium on Computer-Based Medical Systems (CBMS’04); 2004. [Google Scholar]
- Yamaguchi H, Wyckoff J, Condeelis J. Cell migration in tumors. Curr. Opin. Cell Biol. 2005;17:559–564. doi: 10.1016/j.ceb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Zheng X, Wise SM, Cristini V. Nonlinear simulation of tumor necrosis, neo-vascularization and tissue invasion via an adaptive finite-element/level-set method. Bull. Math. Biol. 2005a;67:211–259. doi: 10.1016/j.bulm.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Zheng X, Lowengrub JS, Anderson A, Cristini V. Adaptive unstructured volume remeshing-II: Application to two- and three-dimensional level-set simulations of multiphase flow. J. Comp. Phys. 2005b;208:626–650. [Google Scholar]
- Zutter MM, Santoro SA, Staatz WD, Tsung YL. Re-expression of the alpha 2 beta 1 integrin abrogates the malignant phenotype of breast carcinoma cells. Proc. Natl Acad. Sci. USA. 1995;92:7411–7415. doi: 10.1073/pnas.92.16.7411. [DOI] [PMC free article] [PubMed] [Google Scholar]