Abstract
Phosphorylation of eukaryotic translation initiation factor 2α (eIF2α) is a common cellular mechanism to limit protein synthesis in stress conditions. Baculovirus PK2, which resembles the C-terminal half of a protein kinase domain, was found to inhibit both human and yeast eIF2α kinases. Insect cells infected with wild-type, but not pk2-deleted, baculovirus exhibited reduced eIF2α phosphorylation and increased translational activity. The negative regulatory effect of human protein kinase RNA-regulated (PKR), an eIF2α kinase, on virus production was counteracted by PK2, indicating that baculoviruses have evolved a unique strategy for disrupting a host stress response. PK2 was found in complex with PKR and blocked kinase autophosphorylation in vivo, suggesting a mechanism of kinase inhibition mediated by interaction between truncated and intact kinase domains.
In response to stress conditions, cells alter the level and pattern of gene expression. A common mechanism used to regulate gene expression involves activation of protein kinases. A large superfamily of structurally related serine/threonine and tyrosine protein kinases has been identified in eukaryotic organisms, and the catalytic core of these kinases is composed of 12 subdomains containing conserved residues and sequence motifs (1, 2). Three protein kinases, mammalian protein kinase RNA-regulated (PKR) and heme-regulated inhibitor (HRI) and yeast GCN2, regulate cellular protein synthesis by specifically phosphorylating Ser-51 on the α subunit of the eukaryotic translation initiation factor 2 (eIF2) (3, 4). Composed of three nonidentical subunits, eIF2 facilitates binding of the initiator Met-tRNAiMet to the 40S ribosomal subunit during translation initiation. eIF2 is a GTP-binding protein and phosphorylation of eIF2α on Ser-51 converts eIF2 into a competitive inhibitor of its guanine nucleotide exchange factor eIF2B; this results in the inhibition of general cellular protein synthesis. Phosphorylation of eIF2α can also stimulate gene-specific translation, as observed for the GCN4 mRNA in yeast. The yeast kinase GCN2 is activated by amino acid starvation and phosphorylates eIF2α, facilitating GCN4 expression by enabling ribosomes to bypass inhibitory ORFs in the leader of the GCN4 mRNA (5, 6).
In mammalian cells, the interferon-induced eIF2α kinase PKR is activated upon binding double-stranded RNA (dsRNA) and functions in the cellular antiviral defense mechanism (3, 4, 7). Viruses have developed a number of strategies to down-regulate PKR (for review, see refs. 4 and 7–10). Whereas HIV is thought to limit PKR expression, poliovirus infection of cells causes the degradation of PKR. Specific inhibitors of PKR are expressed by a number of viruses, including adenovirus, which expresses a small RNA, VAI, that binds to PKR and prevents activation of the kinase by dsRNA, and vaccinia virus, which expresses the E3L protein that is thought to sequester the dsRNA activators of PKR. The hepatitis C virus NS5A protein binds to PKR and inhibits the kinase (11), and influenza virus activates a latent cellular protein inhibitor of PKR termed P58IPK that similarly binds to and inhibits PKR kinase activity. Finally, the vaccinia virus K3L protein resembles eIF2α and acts as a pseudosubstrate inhibitor of PKR. The fact that many, if not all, viruses have developed means to inhibit PKR suggests that phosphorylation of eIF2α is an important cellular mechanism to limit viral growth and replication. In addition, expression of dominant negative alleles of PKR causes transformation of mammalian cells, suggesting that PKR plays an important role in cellular growth control (12).
The DNA sequence of the baculovirus Autographa californica multiply-embedded nuclear polyhedrosis virus (AcMNPV) revealed an ORF (ORF123) encoding a truncated protein kinase termed PK2 (13, 14). This 25-kDa protein, which could be detected in extracts of AcMNPV-infected cells (15), resembled the C-terminal half of a protein kinase domain, lacking subdomains I–IV (Fig. 1A). Crystallographic analyses have revealed that protein kinase domains fold into a two-lobe structure (2, 16), and PK2 corresponds to the larger C-terminal lobe of the kinase domain. This truncated structure with the lack of an apparent ATP-binding site and the alteration of several key residues that are usually conserved in the kinase motifs present in the truncated protein suggested that PK2 would not be a functional protein kinase. PK2 shows greatest amino acid sequence homology to the eIF2α kinase family (15); the level of sequence similarity between PK2 and each of the three eIF2α kinases is comparable to that observed in pairwise comparisons among the three eIF2α kinases themselves. Deletion of pk2, the gene encoding PK2, had no detectable effect on AcMNPV replication in cell cultures of the common host Spodoptera frugiperda (15).
Figure 1.
Reduced eIF2α phosphorylation in insect cells infected with wild-type but not pk2-deleted baculovirus. (A) Comparison of the PK2, PKR, and GCN2 proteins. Kinase subdomains (1, 2) are indicated, and the ATP-binding site is indicated as a box. The regulatory domains of PKR [dsRNA-binding domain (dsRNA-BD)] and GCN2 [histidyl-tRNA synthetase-like domain (HisRS)] are indicated as lightly shaded boxes. GCN2-TK, a truncated version of GCN2, corresponds in size to PK2 and contains only kinase subdomains V–XI (amino acids 779–996) of GCN2; the corresponding region of PKR extends from amino acid residue 357 to 551. (B) Analysis of eIF2α phosphorylation in insect cells. Twenty (lanes 1–3) or 50 (lanes 4–6) μg of crude protein extracts from SF-9 cells infected with wild-type (L1) or pk2-deleted (vKINdel) baculovirus or mock-infected (uninfected) were analyzed by IEF gel electrophoresis followed by immunoblotting with eIF2α monoclonal antibodies. The migration positions of phosphorylated and unphosphorylated eIF2α are indicated, and the percentage of eIF2α that is phosphorylated was determined by quantitative densitometry and is indicated below lanes 1–3.
From the common identification of eIF2α kinase inhibitors in viruses that infect mammalian cells, we suspected that PK2 may function as an eIF2α kinase inhibitor. However, eIF2α phosphorylation has not been reported in S. frugiperda. A GCN2 homolog was recently identified in Drosophila melanogaster (17), suggesting that eIF2α phosphorylation does occur in insects. We present evidence that PK2 inhibits eIF2α phosphorylation in insect cells and that PK2 can bind to PKR and inhibit kinase activity. Our results suggest a mechanism for kinase inhibition mediated by direct interaction between the truncated kinase homolog PK2 and intact eIF2α kinases.
MATERIALS AND METHODS
Plasmid Construction.
The pk2 coding region was amplified by PCR using linearized baculovirus DNA (PharMingen) as a template. The primers for PCR were designed so that the pk2 coding region could be inserted between the SacI and XbaI sites of the yeast expression vector pEMBLyex4 (18). In the resulting plasmid pC201, pk2 is expressed from a hybrid GAL-CYC1 promotor. Similarly, by using the plasmid pPstI-L (15) as a template, the pk2 coding region was amplified by PCR using primers designed to add a c-myc epitope tag (-EQKLISEEDLL) to the C terminus of PK2. The resulting pk2-tag coding region was inserted between the BamHI and HindIII sites of pEMBLyex4 creating pC450, and then the GAL-CYC1-pk2-tag construct was transferred to the yeast LEU2 integrating vector pRS305 (19), creating pC451. To transform yeast cells, the plasmid pC451 was linearized by digestion with ClaI to direct the plasmid to integrate at the LEU2 locus. Finally, the region of GCN2 encoding amino acids Lys-779 to Ser-996 (of the full-length 1,659-residue protein) was amplified by using PCR, and the product was cloned between the SacI and XbaI sites in pEMBLyex4, generating the GAL-CYC1-gcn2-tk plasmid pC203. The plasmid p1548 carries a GAL-CYC1-PKR-K296R construct in the high-copy-number TRP1 vector pRS424 (20) and was a gift of P. Romano (National Institutes of Health, Bethesda, MD).
To express PKR in insect cells, the PKR coding region was isolated as a HindIII (converted to blunt-ended with mung bean nuclease)–NotI fragment and then inserted into the vector pHSP70PLVI+CAT (21) that had been digested with BglII (converted to blunt-ended with mung bean nuclease) and NotI. Similarly, the pk2 coding region from the plasmid pC201 was isolated on a SacI (made blunt with mung bean nuclease)–PstI fragment and ligated with pHSP70PLVI+CAT that had been digested with BglII (converted to blunt-ended with mung bean nuclease) and PstI. In the resulting plasmids pC452 (pk2) and pC453 (PKR), the genes are expressed under the control of the Drosophila hsp70 promotor.
Analysis of eIF2α Phosphorylation.
Crude protein extracts from uninfected or baculovirus-infected SF-9 cells were subjected to isoelectric focusing (IEF) using the same reagents and protocols established for analyzing yeast eIF2α (22), and then S. frugiperda eIF2α was detected by immunoblotting with eIF2α monoclonal antibodies (23).
Vector (pEMBLyex4) and pk2 expression plasmid (pC201) transformants of yeast strain H2544 (Mata ura3–52 trp1–63 leu2–3,-112 〈GAL-CYC1-PKR, LEU2〉) or transformants of the GCN2 (H1402), gcn2Δ (H1895) (24), and GCN2c (H1613) strains were grown overnight in synthetic medium containing 10% galactose and 2% raffinose. Cells were harvested, and crude protein extracts were subjected to IEF and then immunoblot analysis with antiserum specific for yeast eIF2α, as described (22).
Assay of eIF2B Activity.
Infected or uninfected SF-9 cells were harvested, suspended in lysis buffer [45 mM Hepes, pH 7.4/0.375 mM magnesium acetate/0.075 mM EDTA/95 mM potassium acetate/10% (vol/vol) glycerol/digitonin (2.5 mg/ml)], and lysed by two 10-s pulses of vigorous mixing using a vortex mixer. The lysate was cleared by centrifugation, and the supernatant was assayed for the exchange of [3H]GDP bound to eIF2 for nonradiolabeled GDP as described (25). Briefly, a binary complex of rat liver eIF2 (26) and [3H]GDP (eIF2⋅[3H]GDP) was combined with the SF-9 cell supernatants and a 200-fold molar excess of nonradiolabeled GDP. The exchange of [3H]GDP bound to eIF2 for free GDP was measured as a decrease in the amount of eIF2⋅[3H]GDP retained on nitrocellulose filters at various times. The results presented are the averages from four experiments.
Viral Infection Assay.
On day 1 of the assays, SF-21 cells (1.0 × 106 cells per plate) were transfected with the PKR plasmid (Plasmid 1, 2.5 μg) by using Lipofectin reagent (GIBCO/BRL), 3 h after transfection TC-100 complete medium was added, and cells were incubated at 27°C. After a 2-h incubation, cells were heat-shocked at 42%°C for 30 min and then incubated at 27°C overnight. On day 2, cells were transfected with the pk2 plasmid (Plasmid 2, 2.5 μg), and 3 h after transfection complete medium was added. After a 1-h incubation at 27°C, cells were infected (10 plaque-forming units per cell) with either wild-type (L1) (27) or pk2-deleted (vKINdel) (15) virus, incubated at 27°C for 1 h, and then heat-shocked at 42°C for 30 min. Cells were then incubated at 27°C and screened for occluded virus 48 h after infection.
Coimmunoprecipitation Assay and Immunoblot Analysis of PKR Expression.
Polyclonal PKR antibodies were prebound to protein A-Sepharose beads (1 h at room temperature), then washed, and incubated with 300 μg of crude extracts from the yeast strain H2557 (Matα urα3–52 leu2–3,-112 trp1-Δ63 gcn2Δ GAL2+) containing either the pk2-myc construct pC451 or vector pRS305 integrated at the LEU2 locus. In addition, the strains contained the PKR-K296R expression plasmid p1548 or vector pRS424. After a 1-h incubation at 4°C, immunocomplexes were isolated by centrifugation and then washed in cell lysis buffer [20 mM Tris⋅HCl, pH 7.5/50 mM NaCl/0.2% Triton X-100/0.5 mM EDTA/1 mM phenylmethylsulfonyl fluoride/pepstatin A (7 μg/ml)]. One-sixth of the starting material or the supernatant fractions from the immunoprecipitations, and the entire pellet fraction were dissolved in SDS/PAGE sample buffer, boiled, and then subjected to SDS/PAGE on 12% gels. The samples were immunoblotted with anti-PKR (28) and anti-myc (Oncogene Research Products, Cambridge, MA) monoclonal antibodies.
For the analysis of PKR autophosphorylation, crude protein extracts were prepared from yeast strain J82 (Mata ura3–52 leu2–3,-112 trp1–63 gcn2Δ sui2Δ p[SUI2-S51A, LEU2]) cotransformed with the PKR expression vector p1545 and either vector pEMBLyex4 or pk2 expression vector pC201 or prepared from yeast strain H1816 transformed with the PKR-K296R expression vector p1421 (29) after growth under inducing conditions in medium containing 10% galactose and 2% raffinose. Crude extract (100 μg) was subjected to SDS/PAGE on 7.5% gels and immunoblotted with anti-PKR monoclonal antibodies.
RESULTS AND DISCUSSION
PK2 Inhibits eIF2α Phosphorylation and Stimulates AcMNPV Replication in Insect Cells.
To determine whether PK2 alters eIF2α phosphorylation during virus infection of insect cells, phosphorylation of S. frugiperda eIF2α was monitored by IEF gels and immunoblot analysis. As shown in Fig. 1B, SF-9 cells infected with wild-type AcMNPV (L1 strain) showed significantly reduced levels of eIF2α phosphorylation compared with uninfected cells. Strikingly, this reduction in eIF2α phosphorylation was dependent on PK2 as cells infected with the vKINdel virus lacking pk2 showed higher levels of eIF2α phosphorylation than uninfected cells. This result is consistent with the model that PK2 functions as an eIF2α kinase inhibitor in virus-infected cells.
The phosphorylated form of eIF2 is a competitive inhibitor of the eIF2 guanine nucleotide exchange factor eIF2B. From the marked difference in eIF2α phosphorylation levels between wild-type virus-infected cells and uninfected cells or cells infected with the mutant virus lacking pk2, we suspected that eIF2B activities would be similarly altered. When the eIF2B-catalyzed displacement of GDP from eIF2 was assayed in crude extracts from SF-9 cells, it was found that uninfected cells and cells infected with the pk2-deleted virus had similar activities (2.39 ± 0.23 and 2.74 ± 0.20 %GDP exchanged per min, respectively). However, the eIF2B activity in extracts from cells infected with wild-type AcMNPV was approximately 2-fold higher (5.31 ± 0.50), consistent with the lower levels of eIF2α phosphorylation observed in these cells. The 50% reduction in eIF2B activity accompanying the 13-fold increase in eIF2α phosphorylation observed in these experiments suggests that S. frugiperda eIF2B is less sensitive to regulation by phosphorylated eIF2 than the eIF2B from mammalian systems, where modest changes in eIF2α phosphorylation significantly impact eIF2B activity (3).
From the obvious differences in eIF2α phosphorylation and eIF2B activities in SF-9 cells infected with the pk2-deleted versus wild-type AcMNPV, we suspected that the presence of pk2 would better enable the virus to withstand activated eIF2α kinases. To examine this possibility, we expressed a cDNA clone of human PKR in SF-21 cells (the parent cell line of SF-9 cells) and then challenged these cells by infecting with wild-type AcMNPV or the pk2-deleted AcMNPV mutant (vKINdel). In the absence of PKR, more than 97% of the cells infected with the wild-type (L1) or vKINdel virus produced occluded virus (Table 1), confirming the previous results that showed that PK2 was not essential for virus replication in S. frugiperda cells or larvae (15). Expression of human PKR in SF-21 cells reduced the percent of wild-type virus-infected cells producing occluded virus by 60% (Table 1), indicating that PKR expression has an adverse effect on AcMNPV-occluded virus production. Consistent with the idea that PK2 can help the virus by limiting eIF2α kinase activity, occluded virus production by vKINdel, the baculovirus mutant lacking pk2, was limited to only 16% of the cells that were transfected with the PKR expression plasmid (Table 1). Thus, vKINdel was significantly more sensitive than wild-type virus to the expression of PKR. This impairment in occluded virus production by the pk2-deleted virus could be complemented by expressing pk2 in trans in SF-21 cells (Table 1). These results further support the model that PK2 functions as an eIF2α kinase inhibitor in AcMNPV-infected insect cells. Baculovirus infection of S. frugiperda cells does not appear to activate eIF2α kinases as in mammalian cells; otherwise, the wild-type virus would replicate better than vKINdel. Thus, eIF2α phosphorylation is probably not an antiviral defense mechanism in insect cells. However, it is anticipated that in their natural environments, S. frugiperda larvae will be subjected to a number of stress conditions that activate cellular eIF2α kinases, perhaps like the GCN2 kinase identified in Drosophila. Under these conditions, the expression of pk2 would be expected to increase the yield of progeny occluded virus.
Table 1.
Reduced occluded virus production in SF-21 cells expressing human PKR and infected with a baculovirus mutant lacking pk2
| Plasmid 1 | Plasmid 2 | Virus | Cells with occluded virus, % |
|---|---|---|---|
| — | — | L1 | 97.0 ± 1.0 |
| — | — | vKINdel | 98.0 ± 0.5 |
| PKR | — | L1 | 39.9 ± 0.3 |
| PKR | pk2 | L1 | 42.3 ± 0.7 |
| PKR | — | vKINdel | 15.6 ± 1.2 |
| PKR | pk2 | vKINdel | 37.3 ± 1.1 |
SF-21 cells were serially transfected with plasmids 1 and 2 expressing PKR and pk2, respectively, and then infected with either wild-type AcMNPV (L1 strain) or pk2-deleted (vKINdel) virus. At 48 h after infection, photographs were taken of two or three fields on the plate. For each field, the total number of cells and the number of cells containing occluded virus were counted. Results are expressed as the percentage of cells expressing occluded virus (mean ± SEM).
PK2 Inhibits eIF2α Phosphorylation by the Human PKR and Yeast GCN2 Protein Kinases.
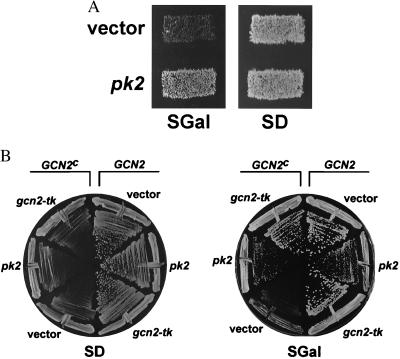
Our results from AcMNPV-infected insect cells are consistent with the idea that PK2 functions as an eIF2α kinase inhibitor. Previously, it has been shown that the vaccinia virus K3L protein (24) and the NS5A protein from hepatitis C virus (11) can suppress the toxicity caused by high-level expression of human PKR in the yeast Saccharomyces cerevisiae. To further examine the ability of PK2 to function as an eIF2α kinase inhibitor, we used the yeast model system to test the ability of PK2 to inhibit human PKR and yeast GCN2. Human PKR was expressed under the control of the galactose-inducible GAL1-CYC1 promoter in yeast cells lacking GCN2. High level expression of PKR on medium containing galactose is lethal in yeast due to extensive phosphorylation of eIF2α on Ser-51 and inhibition of protein synthesis (29, 30). When pk2 and PKR were coexpressed in yeast cells by using the same galactose-inducible promoter, PK2 was able to suppress PKR lethality (Fig. 2A). These results support the idea that PK2 is an eIF2α kinase inhibitor. To address the specificity of PK2, we tested whether PK2 could also inhibit the yeast eIF2α kinase GCN2. Hyperactive GCN2c alleles cause slow-growth phenotypes in yeast due to eIF2α phosphorylation and inhibition of cellular protein synthesis (31). As shown in Fig. 2B, induction of pk2 expression on galactose medium partially suppressed the slow growth phenotype of a yeast strain expressing the GCN2c kinase, suggesting that PK2 can also inhibit GCN2 kinase activity.
Figure 2.
Expression of baculovirus pk2 alleviates the toxic effects of human PKR and yeast GCN2 kinases on yeast cell growth. (A) PK2 inhibition of PKR. A plasmid expressing pk2 under the control of a yeast GAL-CYC1 hybrid promotor (pC201, pk2) or the vector pEMBLyex4 alone (vector) were introduced into strain H2544 (Mata ura3–52 leu2–3,-112 trp1–63 〈GAL-CYC1-PKR/LEU2〉) (24) and transformants were replica-plated to glucose (SD) medium, where PKR and pk2 expression is repressed, or galactose (SGal) medium, where PKR and pk2 expression is induced. Plates were incubated at 30°C for 2 (SD) or 3 (SGal) days. (B) Suppression of a hyperactive GCN2c allele. Plasmids expressing pk2 (pC201, pk2) or gcn2-tk (pC203, gcn2-tk) under the control of the GAL-CYC1 hybrid promotor or the vector pEMBLyex4 alone (vector) were introduced into the isogenic Matα ura3–52 leu2–3,-112 ino1 HIS4-lacZ strains expressing wild-type GCN2 (H1402) (40) or hyperactive GCN2c-E532K-E1522K (H1613) (31). Transformants were streaked on glucose (SD) or galactose (SGal) medium and the plates were incubated at 30°C for 2 (SD) or 4 (SGal) days.
To further address PK2 specificity, we tested the ability of PK2 to inhibit the authentic GCN2 kinase by monitoring the GCN2-dependent regulation of GCN4 expression. In wild-type yeast strains, the expression from a GCN4-lacZ reporter is low under amino acid-sufficient or repressing conditions and increases approximately 11-fold under amino acid starvation or derepressing conditions (Table 2; for review, see ref. 5). This high-level expression of GCN4 under starvation conditions is dependent on the GCN2 kinase (Table 2) and, consistent with the idea that PK2 is an eIF2α kinase inhibitor, expression of pk2 significantly impaired derepression of GCN4-lacZ expression in the strain expressing wild-type GCN2 (Table 2). The ability of PK2 to inhibit both mammalian PKR and yeast GCN2 is consistent with our finding that PK2 can also inhibit an insect eIF2α kinase.
Table 2.
Inhibition of GCN4 expression in yeast cells overexpressing pk2 or a truncated GCN2 kinase domain
| Overexpressed protein |
GCN4-lacZ expression, units
|
|||
|---|---|---|---|---|
|
GCN2
|
gcn2Δ
|
|||
| R | DR | R | DR | |
| None (vector) | 10 | 110 | 6 | 6 |
| PK2 | 10 | 34 | 6 | 7 |
| GCN2-TK | 9 | 40 | 6 | 6 |
| PK2-tag | 9 | 34 | ND | ND |
Plasmids expressing pk2, gcn2-tk, or a c-myc epitope tagged version of pk2 were introduced into isogenic Mata ura3-52 leu2-3,-112 trpl-Δ63 〈GCN4-lacZ, TRP1〉 strains containing wild-type GCN2 (H1642) or lacking GCN2 (gcn2Δ, H1895). For repressing (R) conditions, transformants were grown for approximately 9.5 h in synthetic minimal medium containing 10% galactose and 2% raffinose. For derepressing (DR) conditions, cultures were first grown for 2 h under repressing conditions and then 3-aminotriazole, an inhibitor of histidine biosynthesis, was added to 10 mM and the cultures were incubated for another 7.5 h. Cell harvesting and β-galactosidase assays were performed as described (41, 42), and β-galactosidase activities are expressed as nanomoles of o-nitrophenyl β-d-galactopyranoside hydrolyzed per min per mg of protein. Results are the average of two or three transformants, and the individual measurements deviated from the average values shown here by 29% or less. ND, not determined.
Because PK2 structurally resembles a truncated eIF2α kinase domain and functions as an inhibitor of eIF2α kinases, we also asked whether the similar region of an authentic eIF2α kinase could act as an inhibitor. A truncated version of GCN2, composed of only subdomains V–XI (gcn2-tk, see Fig. 1A), was expressed under the control of the galactose-inducible GAL1-CYC1 promoter in yeast strains expressing a hyperactive GCN2c kinase. As observed in Fig. 2B, expression of gcn2-tk fully suppressed the slow-growth phenotype associated with the GCN2c kinase. In addition, the truncated GCN2 protein (GCN2-TK) impaired the derepression of GCN4-lacZ expression observed in wild-type GCN2 strains under amino acid starvation conditions (Table 2). GCN2-TK was at least as effective as PK2 for inhibition of GCN2 and GCN2c kinases; however, GCN2-TK was unable to suppress the toxicity associated with high-level expression of PKR in yeast (data not shown). In addition, we were unable to detect inhibitory activities with similarly truncated forms of PKR and the heme-regulated inhibitor (HRI) (data not shown). The fact that PK2 shows broad specificity inhibiting both mammalian and yeast eIF2α kinases makes it likely that it can also inhibit insect eIF2α kinases, as indicated by our results showing reduced eIF2α phosphorylation in insect cells infected with wild-type but not pk2-deleted baculovirus (Fig. 1B).
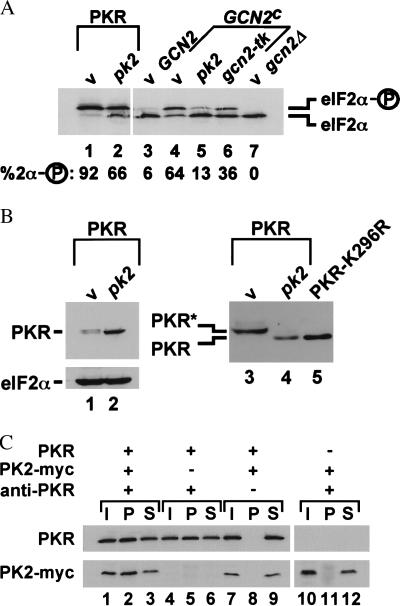
To define how expression of pk2 or gcn2-tk suppressed the toxicity associated with expression of PKR and GCN2c kinases in yeast, we monitored eIF2α phosphorylation by using IEF gels. As we have shown, eIF2α resolves into two species on IEF gels and the upper form detected in strains expressing GCN2 or human PKR is phosphorylated on Ser-51 (22, 29). Consistent with the severe inhibition of translation and lethal phenotype, practically all of the eIF2α was phosphorylated in yeast cells expressing PKR (Fig. 3A, lane 1). Coexpression of pk2 in this yeast strain expressing PKR resulted in a significant decrease in the percentage of eIF2α phosphorylated on Ser-51 (Fig. 3A, lane 2). Similarly, expression of pk2 or gcn2-tk lowered the percentage of the phosphorylated form of eIF2α in strains expressing the GCN2c kinase (Fig. 3A, lanes 4–6). The greater inhibition of the GCN2c kinase by PK2 versus GCN2-TK observed in the IEF gel results contrasts with the results of the growth-rate tests in Fig. 2B, where GCN2-TK appeared more potent than PK2. However, high-level expression of pk2 caused a slow-growth phenotype in yeast (see Fig. 2B, pk2 expressed in GCN2 strain) that partially masked the ability of PK2 to suppress the toxicity due to the GCN2c kinase. Immunoblot analysis revealed that expression of pk2 or gcn2-tk did not impair expression of PKR or GCN2c in yeast cells. PKR levels were significantly elevated in cells coexpressing pk2 (Fig. 3B, compare lanes 2 to 1). This inverse relationship between PKR toxicity and protein levels has been noted previously when studying the expression of wild-type and catalytically impaired mutants of PKR in both yeast and mammalian cells (24, 29, 32, 33), and it has been attributed to negative translational autoregulation of PKR expression resulting from localized activation of the PKR kinase in the vicinity of the PKR mRNA.
Figure 3.
Inhibition of eIF2α phosphorylation in yeast cells by expression of pk2 and coprecipitation of PK2 with PKR. (A) Analysis of eIF2α phosphorylation. Plasmids encoding pk2 or gcn2-tk or an empty vector (v) were introduced into yeast strains expressing PKR, wild-type GCN2, hyperactive GCN2c-E532K-E1522K (GCN2c), or no eIF2α kinase (gcn2), as indicated. Crude protein extracts were prepared and analyzed by IEF gel electrophoresis on a polyacrylamide slab gel followed by immunoblotting with yeast eIF2α-specific antiserum. The eIF2α phosphorylated on Ser-51 focuses above the protein lacking the Ser-51 phosphorylation, as indicated. The percentage of eIF2α that is phosphorylated on Ser-51 was determined by quantitative densitometry and is indicated below each lane. (B) Analysis of PKR expression. (Lanes 1 and 2) PKR expression in the strains shown in A, lanes 1 and 2, was analyzed by immunoblotting 50 μg of whole cell extracts and probing with antiserum specific for yeast eIF2α or with human PKR monoclonal antibodies, as indicated. (Lanes 3–5) Crude protein extracts from an eIF2α-S51A yeast strain expressing PKR, PK2, or PKR-K296R, as indicated, were subjected to SDS/PAGE on 7.5% gels and then immunoblotted with human PKR monoclonal antibodies. The phosphorylated form of PKR (PKR*) migrates slower than the unphosphorylated form (PKR), as indicated. The slower mobility of wild-type PKR, compared with inactive PKR-K296R, in SDS/PAGE is due to extensive autophosphorylation of the wild-type protein. Treatment of wild-type PKR with phosphatase increases its mobility and causes wild-type PKR to comigrate with nonphosphorylated PKR-K296R (ref. 35; P. Romano and A. Hinnebusch, personal communication). (C) Coprecipitation of PKR and PK2. Crude protein extracts from yeast strains expressing PKR-K296R (PKR) and c-myc epitope-tagged pk2 (PK2-myc), as indicated, were incubated with anti-PKR polyclonal antiserum bound to protein A-agarose beads; after precipitation, fractions of the starting materials (I), pellets (P), and supernatants (S) were subjected to SDS/PAGE followed by immunoblotting with human PKR or c-myc monoclonal antibodies, as indicated.
PK2 Stably Interacts with PKR.
Two of the simplest models for inhibition of kinase activity by expression of a truncated kinase domain are sequestration of the substrate or formation of inactive kinase heterodimers. Two observations support the former model. (i) The substrate binding site of the cAMP-dependent protein kinase has been localized to the C-terminal half of the protein kinase domain (34). The fact that PK2 resembles the C-terminal half of a protein kinase supports the model that PK2 inhibits eIF2α phosphorylation by sequestering the substrate. (ii) The C-terminal half of a protein kinase domain has not been reported to function in kinase dimerization. However, in favor of the dimerization model, we found that high-level expression of pk2 in yeast cells caused a slow-growth phenotype and that overexpression of catalytically inactive PKR-K296R, but not eIF2α, could alleviate this slow-growth phenotype (data not shown). To explore further the possibility of direct interaction between PK2 and PKR, we tested whether PK2 and PKR could be coimmunoprecipitated from crude yeast extracts. For these experiments a c-myc epitope tag was added to the C terminus of PK2, and as shown in Table 2, this tagged protein retained full activity. The tagged PK2 was coproduced in yeast cells along with the catalytically inactive PKR-K296R. Anti-PKR polyclonal antiserum was used to immunoprecipitate PKR, and the pellet and supernatant fractions were subjected to SDS/PAGE and then immunoblotted using monoclonal antibodies specific for PKR and c-myc. PK2 was coprecipitated along with PKR by using the PKR antibodies (Fig. 3C, lane 2), and this was dependent on both inclusion of PKR antibodies (lane 8) in the precipitation reaction and the presence of PKR in the extract (lane 11). These results demonstrate that PKR and PK2 can be found in a complex in vivo and suggest that PK2 inhibits PKR activity through heterodimer formation. Yeast two-hybrid assays provided further evidence of an interaction between PKR and PK2. By using this assay, the PK2 protein was found to interact with both full-length PKR and a portion of the PKR kinase domain containing subdomains I–V (data not shown). These genetic and biochemical data support the model that PK2 inhibits PKR (and GCN2) kinase function through formation of inactive heterodimers mediated by contacts in the kinase domains.
Upon binding dsRNA activators, PKR undergoes autophosphorylation in an intermolecular reaction (33, 35, 36), and it is the autophosphorylated form of PKR that is competent to phosphorylate eIF2α. When expressed in yeast cells, wild-type PKR, but not the inactive PKR-K296R mutant, is phosphorylated (P. Romano and A. Hinnebusch, personal communication), indicating that PKR undergoes autophosphorylation when expressed in yeast cells. The active phosphorylated forms of PKR are known to migrate slower in SDS/PAGE (33, 35). In our experiments, the PKR from cells expressing pk2 had a slightly faster mobility in SDS/PAGE than the PKR present in vector-transformed cells (Fig. 3B, compare lanes 2 to 1). By using an SDS/PAGE system with better resolving abilities and an eIF2α-S51A yeast strain to block the autoregulation of PKR expression, we found that the PKR in extracts from cells expressing pk2 comigrated with the inactive and nonphosphorylated PKR-K296R (Fig. 3B, lanes 3–5). These results demonstrate that expression of pk2 blocks PKR autophosphorylation in vivo. In addition, these results support the idea that dimerization of the eIF2α kinases, mediated by protein–protein contacts between kinase domains, may be an obligatory step in kinase activation.
Our results demonstrate that PK2 is an eIF2α kinase inhibitor, and thus the paradigm that viruses express eIF2α kinase inhibitors can be extended to insect viruses. The wide variety of strategies for inhibiting eIF2α kinases found in different viruses underscores the importance of these mechanisms in virus replication, whereas the mode of kinase inhibition used by PK2 suggests that this strategy arose independently in baculoviruses. Previously, it has been demonstrated that fragments of trans-membrane protein kinases containing the activation or ligand-binding domains can function in a dominant negative manner by forming heterodimers with the corresponding intact protein kinase (37, 38). Our identification of a truncated protein kinase domain that can inhibit kinase activity reveals a different mechanism of inhibition mediated by contacts between homologous kinase catalytic domains. The C-terminal half of many protein kinase domains contain key autophosphorylation sites, especially within a loop between kinase subdomains VII and VIII known as the activation lip (39). Although PK2 shares sequence similarity with the eIF2α kinases in subdomains VII and VIII, it lacks the phosphorylation loop and potential autophosphorylation sites. Our results demonstrate that PK2 interacts with PKR and blocks kinase autophosphorylation. We propose that PK2 prevents the intermolecular interactions required for kinase autophosphorylation, perhaps by functioning as an autophosphorylation pseudosubstrate inhibitor and competing with the activation lip for binding in the active site of the kinase. Consistent with this idea, PK2 does not appear to be phosphorylated during viral infections (15). Alternatively, PK2 may inhibit PKR function by blocking PKR dimerization. Finally, our discovery that eIF2α phosphorylation in insects interferes with viral propagation provides an additional system in which to study cellular translational regulation and antiviral defense mechanisms.
Acknowledgments
We thank T. Ung and L. Hugendubler for technical assistance; and A. Hinnebusch, G. Pavitt, P. Romano, and members of the Dever and Hinnebusch laboratories for useful discussions and comments on the manuscript. This work was supported in part by National Institutes of Health Grants AI23719 (L.K.M.) and DK13499 (S.R.K.).
ABBREVIATIONS
- eIF2
eukaryotic translation initiation factor 2
- PKR
protein kinase RNA-regulated
- dsRNA
double-stranded RNA
- AcMNPV
Autographa californica multiply-embedded nuclear polyhedrosis virus
- IEF
isoelectric focusing
References
- 1.Hanks S K, Quinn A M, Hunter T. Science. 1988;241:41–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 2.Hanks S K, Hunter T. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 3.Clemens M J. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 139–172. [Google Scholar]
- 4.Hinnebusch A G. Sem Cell Biol. 1994;5:417–426. doi: 10.1006/scel.1994.1049. [DOI] [PubMed] [Google Scholar]
- 5.Hinnebusch A G. In: Translational Control. Hershey J W B, Matthews M B, Sonenberg N, editors. Plainview: Cold Spring Harbor Lab. Press; 1996. pp. 199–244. [Google Scholar]
- 6.Hinnebusch A G. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- 7.Mathews M B. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 505–548. [Google Scholar]
- 8.Katze M G. J Interferon Res. 1992;12:241–248. doi: 10.1089/jir.1992.12.241. [DOI] [PubMed] [Google Scholar]
- 9.Katze M G. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 607–630. [Google Scholar]
- 10.Schneider R J. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 575–605. [Google Scholar]
- 11.Gale M J, Jr, Korth M J, Tang N M, Tan S-L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 12.Koromilas A E, Roy S, Barber G N, Katze M G, Sonenberg N. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 13.Morris T D, Todd J W, Fisher B, Miller L K. Virology. 1994;200:360–369. doi: 10.1006/viro.1994.1200. [DOI] [PubMed] [Google Scholar]
- 14.Ayres M D, Howard S C, Kuzio J, Lopez-Ferber M, Possee R D. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Miller L K. Virology. 1995;206:314–323. doi: 10.1016/s0042-6822(95)80047-6. [DOI] [PubMed] [Google Scholar]
- 16.Knighton D R, Zheng J, Ten Eyck L F, Xuong N H, Taylor S S, Sowadski J M. Science. 1991;253:407–414. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- 17.Santoyo J, Alcalde J, Mendez R, Pulido D, de Haro C. J Biol Chem. 1997;272:12544–12550. doi: 10.1074/jbc.272.19.12544. [DOI] [PubMed] [Google Scholar]
- 18.Cesareni G, Murray J A H. In: Genetic Engineering: Principals and Methods. Setlow J K, Hollaender A, editors. Vol. 9. New York: Plenum; 1987. pp. 135–154. [Google Scholar]
- 19.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 21.Clem R J, Miller L K. Mol Cell Biol. 1994;14:5212–5222. doi: 10.1128/mcb.14.8.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dever T E, Feng L, Wek R C, Cigan A M, Donahue T D, Hinnebusch A G. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- 23.Scorsone K A, Panniers R, Rowlands A G, Henshaw E C. J Biol Chem. 1987;262:14538–14543. [PubMed] [Google Scholar]
- 24.Kawagishi-Kobayashi M, Silverman J B, Ung T L, Dever T E. Mol Cell Biol. 1997;17:4146–4158. doi: 10.1128/mcb.17.7.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimball S R, Everson W V, Flaim K E, Jefferson L S. Am J Physiol. 1989;256:C28–C34. doi: 10.1152/ajpcell.1989.256.1.C28. [DOI] [PubMed] [Google Scholar]
- 26.Kimball S R, Everson W V, Myers L M, Jefferson L S. J Biol Chem. 1987;262:2220–2227. [PubMed] [Google Scholar]
- 27.Lee H H, Miller L K. J Virol. 1978;27:754–767. doi: 10.1128/jvi.27.3.754-767.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurent A G, Krust B, Galabru J, Svab J, Hovanessian A G. Proc Natl Acad Sci USA. 1985;82:4341–4345. doi: 10.1073/pnas.82.13.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dever T E, Chen J-J, Barber G N, Cigan A M, Feng L, Donahue T F, London I M, Katze M G, Hinnebusch A G. Proc Natl Acad Sci USA. 1993;90:4616–4620. doi: 10.1073/pnas.90.10.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chong K L, Feng L, Schappert K, Meurs E, Donahue T F, Friesen J D, Hovanessian A G, Williams B R G. EMBO J. 1992;11:1553–1562. doi: 10.1002/j.1460-2075.1992.tb05200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramirez M, Wek R C, Vazquez de Aldana C R, Jackson B M, Freeman B, Hinnebusch A G. Mol Cell Biol. 1992;12:5801–5815. doi: 10.1128/mcb.12.12.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barber G N, Wambach M, Wong M L, Dever T E, Hinnebusch A G, Katze M G. Proc Natl Acad Sci USA. 1993;90:4621–4625. doi: 10.1073/pnas.90.10.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romano P R, Green S R, Barber G N, Mathews M B, Hinnebusch A G. Mol Cell Biol. 1995;15:365–378. doi: 10.1128/mcb.15.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knighton D R, Zheng J, Eyck L F T, Xuong N H, Taylor S S, Sowadski J M. Science. 1991;253:414–420. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- 35.Thomis D C, Samuel C E. J Virol. 1993;67:7695–7700. doi: 10.1128/jvi.67.12.7695-7700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kostura M, Mathews M B. Mol Cell Biol. 1989;9:1576–1586. doi: 10.1128/mcb.9.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kashles O, Yarden Y, Fischer R, Ullrich A, Schlessinger J. Mol Cell Biol. 1991;11:1454–1463. doi: 10.1128/mcb.11.3.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ueno H, Colbert H, Escobedo J A, Williams L T. Science. 1991;252:844–848. doi: 10.1126/science.1851331. [DOI] [PubMed] [Google Scholar]
- 39.Johnson L N, Noble M E M, Owen D J. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 40.Hannig E H, Williams N P, Wek R C, Hinnebusch A G. Genetics. 1990;126:549–562. doi: 10.1093/genetics/126.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moehle C M, Hinnebusch A G. Mol Cell Biol. 1991;11:2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lucchini G, Hinnebusch A G, Chen C, Fink G R. Mol Cell Biol. 1984;4:1326–1333. doi: 10.1128/mcb.4.7.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]