Abstract
We have identified disruptions of the dedicator of cytokinesis 8 gene, DOCK8, in two unrelated patients with mental retardation. In one patient, a male with MR and no speech, we mapped a genomic deletion of approximately 230 kb in subtelomeric 9p. In the second patient, a female with mental retardation and ectodermal dysplasia and a balanced translocation t(X;9)(q13.1;p24), we mapped the 9p24 breakpoint to a region overlapping with the centromeric end of the 230 kb subtelomeric deletion. We characterized the DOCK8 gene from the critical 9p deletion region and determined the longest isoform of the DOCK8 gene is truncated in both patients. Furthermore, the DOCK8 gene is expressed in several human tissues including adult and fetal brain. Recently, a role for DOCK8 in processes that affect organization of filamentous actin has been suggested. Several genes influencing actin cytoskeleton have been implicated in human cognitive function and thus a possibility exists that the rare mutations of the DOCK8 gene may contribute to some cases of autosomal dominant mental retardation.
Keywords: Subtelomere, Mental retardation, Chromosome translocation, DOCK8, Autosome
Introduction
Mental retardation (MR) is the most common developmental disability, affecting intellectual and adaptive functions in approximately 1−3% of the population. Yet, the underlying cause of MR is established in less than half the cases [1, 2]. Genetic factors in MR likely include mutations in genes distributed throughout the genome, and is already well established for the genes on the X chromosome [1, 2]. The finding of MR among almost all patients with autosomal microdeletions and the association of MR with submicroscopic alterations in the subtelomeric region of the autosomes further indicates the ubiquitous distribution of autosomal genes that influence intelligence [3-5].
Chromosomal abnormalities have been found in a significant number (10−15%) of individuals with mental retardation and developmental disabilities (MR/DD) [6]. About 5% of idiopathic MR can be explained by deletions or rearrangements of the subtelomeric regions of the autosomes [5-9]. A large number of subtelomeric deletions have been observed in patients with MR suggesting that these deletions are likely to harbor gene(s) responsible for the observed phenotypes in these patients. Furthermore, analysis of patients with submicroscopic deletions has facilitated identification of small regions of overlap linked to specific phenotypic characteristics and subsequent isolation of the causative genes. Recently, disruption of the Eu-HMTase1 gene has been shown to be associated with the 9q34 subtelomeric deletion syndrome [10, 11].
Here we characterized, at the DNA level, chromosomal rearrangements involving 9p24 in two unrelated patients with mental retardation and developmental disabilities. We have characterized the DOCK8 gene from the critical 9p subtelomeric region, determined the expression of the DOCK8 gene in fetal and adult human brains and demonstrated that the longest isoform of the gene is physically disrupted in both patients.
Results
Molecular analysis of the subtelomeric deletion in a patient with mental retardation and developmental disability
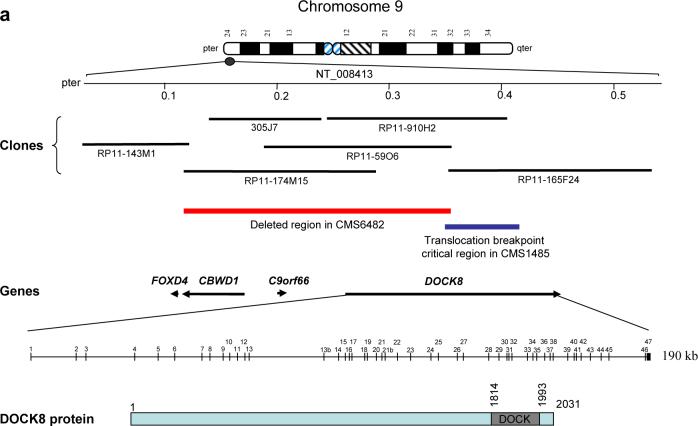
Subtelomeric FISH analysis revealed deletion of a probe from the 9p subtelomeric region (ish 9pter (305J7-T7 ×1) in patient CMS6482. (Fig. 1a, b). This probe was found to map within a genomic contig (NT_008413; NCBI) in 9p24.3 (Fig. 1a). We identified physically mapped genomic clones from the contig and used them systematically in FISH analyses to determine the extent of the subtelomeric 9p deletions in this patient (Fig. 1a and Table 1). A part of the 9p subtelomeric region has previously been shown to share sequence homology with other regions of chromosome 9 [12]. Consistent with that report, probes RP11−143M1 and RP11−174M15 gave three FISH signals on a normal chromosome 9: one at the 9p subtelomere and two others in 9p12 and 9q21.2 regions. However, the 9p subtelomere-specific FISH signal with clone RP11−174M15 was absent in CMS6482, indicating a deletion of this region in the patient (Supplementary Fig. 1 and Table 1). Probe RP11−59O6 gave a signal on the normal chromosome 9 but not on the derivative chromosome 9 (Fig. 1c). Probe RP11−910H2 gave signals on the normal chromosome 9, as well as a reduced signal on the on the derivative chromosome 9 suggesting this probe is partially deleted (Fig. 1d). All FISH probes gave expected signals on the normal chromosome 9 in a control sample (not shown). These results determined the deletion in patient CMS6482 to be between genomic clones RP11−143M15 (telomeric) and RP11−910H2 (centromeric) (Fig. 1a).
Fig. 1.
Mapping of the subtelomeric 9p chromosomal rearrangements. (a) Ideogram of chromosome 9 with a partial physical map of the subtelomeric 9p24.3 region, BAC clones used in FISH analysis, and known genes in italics are shown. Black arrows indicate the direction of the transcription. FISH analyses determined a critical deletion region (red bar) in patient CMS6482 and a region (blue bar) harboring the 9p24 translocation breakpoint in CMS1495. The genomic structure of the DOCK8 gene and a schematic of the 2031 amino acids DOCK8 variant are shown. The predicted DOCK domain in the C-terminal region is indicated. (b-d) FISH analyses in CMS6482. (b) Chromosome 9 subtelomeric probes (9p, green; 9q red) produced a green signal only on one chromosome 9 but not the other, indicating a 9p subtelomeric deletion in CMS6482. Red signal was present on both the normal and the derivative chromosomes 9. (c) RP11−59O6 (red) gave a signal (dark red) only on the normal chromosome 9 and not on the derivative chromosome 9 (arrow) indicating deletion of this probe in CMS6482. The chromosome 9 centromeric probe is in light red color. (d) Probe RP11−910H2 (green, arrowheads) is partially deleted on the derivative chromosome 9 and gave signals on both the normal and the derivative chromosome 9. The chromosome 9 centromeric probe is in light red color. (e) FISH analysis in CMS 1485. Probe RP11−910H2 (green) is distal to the 9p24.3 translocation breakpoint in CMS1485. The probe is present on the normal chromosome 9 as well as the derivative X chromosome. (f) PFGE analysis with a probe located in the 3’ portion of DOCK8 reveals an abnormally migrating fragment in CMS1485 but not in the control sample. The chromosome 9 centromeric probe is in light red color. (g) Fine mapping of the 9p critical region with quantitative genomic PCR in CMS6482, CMS1485 and in three controls (CMS11195, CMS5865 and CMS6265) reveals the 9p subtelomeric deletion in CMS6482 extends at least through exon 23 of the DOCK8 gene. CMS1485 has a deletion presumably associated with the translocation located between exon 23 and 34 with exon 29 deleted. The values represent the average of three replicates used in each case.
Table 1.
Chromosome 9p BAC/PAC FISH and subtelomeric analysis
| FISH Probe | CMS6482 9p24 del | CMS1485 t(X;9) |
|---|---|---|
| RP11−143M1 | Normal | Distal |
| RP11−174M15 | Deleted | Distal |
| #Subtelomeric probe 305J7-T7 | Deleted | NC |
| RP11−59O6 | Deleted | Distal |
| RP11−910H2 | Partially Deleted | Distal |
| RP11−165F24 | Normal | Proximal |
| RP11−130C19 | NC | Proximal |
| RP11−125K10 | NC | Proximal |
| RP11−376O21 | NC | Proximal |
Subtelomeric Probes from Vysis Inc.; NC, not checked
Mapping of the 9p chromosomal breakpoint in a female patient with MR, anhidrotic ectodermal dysplasis and a de novo t(X;9)(q13;p24) translocation
We previously mapped the Xq13 chromosome breakpoint of patient CMS1485 within intron 1 of the anhidrotic ectodermal dyspalsia (EDA) gene (Xq13) and determined that the ectodermal defects in this female patient (CMS1485) were due to the disruption of the EDA gene [14, 15]. Thus, the MR is presumably associated with the 9p24 breakpoint. To refine the 9p24 chromosomal breakpoint, FISH analysis was performed, using three clones spaced across chromosomal band 9p24. All three clones initially used (RP11−130C19, RP11−125K10, and RP11−376O21) mapped proximal to the 9p breakpoint and gave signals on both the normal chromosome 9 and the derivative 9 (Table 1). FISH analysis with additional clones narrowed the location of the 9p breakpoint between clones RP11−174M15 and RP11−165F24. Clones RP11−143M1 and RP11−174M15 mapped distal to the breakpoint (Fig. 1a and Table 1). Both probes gave FISH signals on the normal chromosome 9, but also gave a 9p-specific signal on the derivative X chromosome. Clone RP11−165F24 mapped proximal to the breakpoint and gave signals on both the normal and derivative chromosome 9 (Table 1). Furthermore, clone RP11−59O6 also mapped distal to the breakpoint (Fig. 1a). Genomic clone RP11−910H2 partially overlaps both RP11−59O6 and RP11−165F24. FISH analysis with RP11−910H2 revealed signals on the normal chromosome 9 and the derivative X (Fig. 1e). These results suggested the chromosome 9 translocation breakpoint lies within the overlap of these two adjacent and partially overlapping clones (RP11−59O6 and RP11−165F24) (Fig. 1a), further narrowing the translocation breakpoint region (Fig. 1a) and also suggested the possibility of a microdeletion at the breakpoint.
Delineation of a putative critical MR region in subtelomeric 9p and identification of DOCK8 as a candidate MR gene
Sequence information corresponding to the critical subtelomeric deletion region (Fig. 1a) was obtained from the NCBI database. We further analyzed end sequences of BAC/PAC clones from the critical region and determined the physical map of the overlapping genomic clones. We deduced the minimal critical deletion region to approximately 230 kb in CMS6482.
In silico analysis of the gene content of the 9p24 deleted region and the flanking regions showed the presence of known and putative genes (Fig. 1a). A known gene, FOXD4 (GenBank accession no. AY344640), is located near the telomeric end of the deletion, and a gene CBWD1, encoding a COBW-like protein was also found in a homologous region on human chromosome 2p13 [12]. Two putative genes, LOC389701 and FLJ00026, mapped near the proximal end of the 9p subtelomeric deletion region (not shown). EST alignments of cDNAs and RT-PCR subsequently showed these two putative genes to be partial sequences of a singe gene, DOCK8 (Fig. 1a). We and others have characterized the DOCK8 gene in detail [13, 16]. The gene consists of 47 exons spanning approximately 190 kb of genomic DNA (Fig. 1a).
These results suggested that the subtelomeric deletion in patient CMS6482 disrupts the DOCK8 gene. Furthermore, the results demonstrated that the 9p24 translocation breakpoint in CMS 1485 lies within the DOCK8 gene (Fig. 1a). To further support the findings in CMS1485, pulsed-field gel electrophoresis (PFGE) analysis was performed. A DOCK8 cDNA probe detected a normal fragment of approximately 190 kb and a novel fragment of approximately 250 kb in patient's DNA digested with SalI (Fig. 1f) indicating a likely disruption of the DOCK8 gene in this patient.
We used quantitative genomic PCR to further narrow the centromeric boundary of the 9p subtelomeric deletion in patient CMS6482 and the putative microdeletion at the 9p breakpoint in CMS1485. We analyzed five amplicons of the DOCK8 gene and identified a deletion of one copy of three amplicons representing intron 13, exon 18, and exon 23 in CMS6482 and determined the centromeric boundary of the subtelomeric deletion to lie within the gene DOCK8 (Fig. 1a, g) in CMS6482. The deletion in this patient extends through nearly half of the 5’ end of the DOCK8. In CMS1485, quantitative genomic PCR revealed a microdeletion of up to 37 kb located between exons 23 and 34 of DOCK8 (Fig. 1g) and mapped the microdeletion in this patient to a region overlapping with the centromeric end of the 230 kb subtelomeric deletion observed in CMS6482 (Fig. 1a).
Characterization of DOCK8 and its expression studies
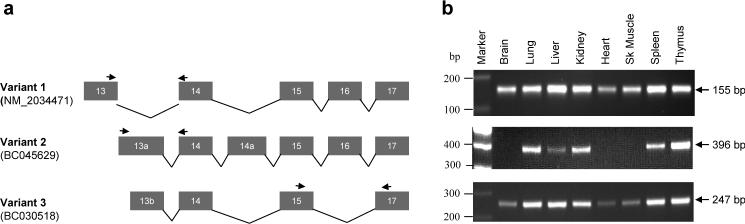
The longest DOCK8 transcript of approximate 8 kb encodes a protein of 2031aa that contains a DOCK domain in the C-terminal region (Fig. 1a). The NCBI database analysis revealed multiple splice variants of the DOCK8 gene. Two DOCK8 variants lacked residues 859−890 or residues 959−972. In addition, one variant had additional 68aa residues at its N-terminal [16]. Analysis of the available EST sequences further revealed two smaller variants of the gene (Fig. 2a). Northern analysis of fetal and adult human tissue blots using the DOCK8 cDNA as a hybridization probe detected a transcript of about 8.0 kb in all human tissues analyzed. To monitor DOCK8 gene expression in the human brain, we used Northern blots containing RNAs from different regions of the adult human brain. A variable level of expression was detected in all brain regions (Supplementary Fig. 2). Expression analysis in a panel of fetal cDNAs also revealed the presence of DOCK8 in most tissues including fetal brain (Fig. 2b).
Fig. 2.
Tissue expression profile of human DOCK8 variants. (A) Schematic diagram of primer locations used in variant specific RT-PCR. (B) Variant 1 and 3 are expressed in all fetal tissues tested. Variant 2 expression could only be detected in fetal lung, liver, kidney, spleen and thymus.
We used RT-PCR to investigate the tissue distribution of the three splice variants (Fig. 2a, b). All three variants of the DOCK8 were expressed at variable levels in all tissues analysed (Fig. 2b). No expression or a comparatively low level of expression was noted for variant 2 in brain, heart and skeletal muscle (Fig. 2b, middle panel).
Mutation screening in patients with MR of unknown cause
Using Incorporation PCR SSCP (IPS) or Denaturing High Performance Liquid Chromatography (DHPLC) (see Material and methods for details), we screened all 47 exons of the DOCK8 gene for alterations in a cohort of 145 patients with MR. Five single nucleotide variations were identified (Supplementary Table 1) including two previously known SNPs, p.P1876P and p.A1902V. Two intronic variations (IVS4+14A>G and IVS4+65G>A) were too frequent in the MR cohort and were considered likely to be polymorphisms rather than MR causing mutations. One missence alteration, p.L1262V, which appeared to be unique in the MR cohort (frequency 0.68%), was subsequently identified in controls (frequency 1.25%). Additionally, we also identified an alteration, p.S97L, in a random control individual (Supplementary Table 1).
Analysis of actin organization in CMS1485 fibroblast cell lines
In an attempt to examine a likely involvement of DOCK8 in organization of filamentous actin, we performed immunofluorescent microscopy with the actin stain phalloidin on the X;9 translocation cell line (CMS1485) and a control fibroblast cell line. No measurable difference in actin organization or cell size was observed between the control and patient cells (Fig. 3).
Fig. 3.

Actin structure of fibroblast cell lines CMS11195 (control) and CMS1485 (patient). Filamentous actin was detected by Alexa Fluor® 488 phalloidin green and the cells were examined by immunofluorescence microscopy. The nucleus is visualized using SYTOX® orange.
Discussion
An important cause of MR has been shown to be chromosomal abnormalities that result in segmental aneuploidy. MR/DD-associated subtelomeric deletions in patients provide critical genomic intervals for the identification of the causative genes that yield the various associated clinical features. Here we have defined the 9p subtelomeric region associated with MR/DD in two unrelated patients. The MR-associated critical region defined here mapped further telomeric to the previously defined 9p24.3 deletions associated with 46, XY gonadal dysgenesis and the ANKRD15 MR locus [17-19] and also appears to be unrelated to the more common 9p minus syndrome (OMIM # 158170).
There is some evidence that suggests some 9p subtelomeric deletions may be a normal variant. Techakittiroj and coworkers reported a case of a patient with multiple congenital anomalies and a 9p deletion [20]. Three asymptomatic individuals in this family were also found to have a 9p subtelomeric deletion. It is unknown if the 9p deletion in this family extends into DOCK8 gene region, or if the symptomatic individual's deletion has expanded as compared to the non-symptomatic family members. The finding of a genomic interval common to two patients with MR/DD in our study would suggest a likely role of a gene from the critical region that may influence cognitive function.
Several putative genes are located within the MR critical region. ESTs representing CBWD1 have been found in various tissues including fetal brain and especially in the hippocampus. Another candidate is FOXD4, a member of the forkhead family of transcription factors [21, 22]. Many members of the forkhead family are known to be key regulators of embryogenesis [23]. FOXD4 is widely expressed, most prominently in heart and skeletal muscle [21]. Mutations in FOX genes have been implicated in several human disorders including the involvement of the FOXP2 gene at 7q31 in a severe speech and language disorder [24]. C9orf66 (UniGene Hs.190877) was considered to be a less likely candidate since it has no similarity to known proteins, and ESTs representing this putative gene were isolated only from kidney, testis and prostate cDNA libraries.
One of two copies of DOCK8 was found to be physically disrupted by the 9p subtelomeric deletion in one patient and by the 9p24 translocation breakpoint in the second patient which suggests DOCK8 is a likely candidate for the MR features in both patients. DOCK8 is expressed in both fetal and adult brain and belongs to the dedicator of cytokinesis family of proteins. These proteins are potential guanine nucleotide exchange factors and activate some GTPases. DOCK8 has also been shown to interact with CDC42, a small GTPase of the Rho-subfamily [13]. Additionally, the authors [13] have shown through immunofluorescence staining that DOCK8 is present at the cell edges in areas undergoing lamellipodia formation. A potential role of DOCK8 in the organization of filamentous actin was suggested [13]. The lack of any measurable differences in the actin cytoskeleton in the patient CMS1485 with the t(X;9) and a control fibroblast cells in our study could be explained by the low level of expression of DOCK8 in the fibroblast cells. It is also likely that the haploinsufficiency of DOCK8 may not be sufficient to cause phenotypic changes visible at the microscopic level.
The physiological and biological roles of the DOCK8 variants remain unknown. Recently, a homozygous deletion including the DOCK8 locus at chromosome 9p24 has been found in a human lung cancer cell line [16]. Furthermore, localization of DOCK8 protein in lamellipodia suggests that it might have a role in cell migration, morphology, adhesion and growth. Further functional analysis of the DOCK8 isoforms is needed to elucidate the biological and any pathogenic consequences of DOCK8 mutations.
Several members of the DOCK family of proteins play roles in neuronal development and function. A defect of the DOCK3 gene has been shown in a patient with an attention deficit hyperactivity disorder-like phenotype [25]. Furthermore, a Rac GTPase activator protein, DOCK7, has been shown to have a role in axon formation and neuronal polarity [26]. Several genes involved in Rho GTPase regulation and signaling have been linked to mental retardation [27, 28]. The interaction of DOCK8 with CDC42 suggests a possible mechanism whereby the disruption of DOCK8 in our patients may result in their impaired cognitive ability. Further analysis of the DOCK8 in patients with MR, as well as functional analysis in animal models, should clarify the possible involvement of DOCK8 in brain development and function.
Materials and methods
Case report
Case 1 (CMS6482) is a 55 year old white male with profound mental retardation (IQ 2). He was born at term to a 19 year old mother who had an appendectomy at 4 months gestation. He was reported to weigh 14 pounds at birth. He walked at age 11 months. At one year of age, he was found to have a seizure disorder, unusual posturing and no speech. As an adult, he developed cataract with presbyopia and a cholesteatoma of the right ear. Physical examination at age 47 revealed his height to be 166.5 cm, weight 61 kg and head circumference 55 cm (25th centile). He had no speech. He had significant balding, prominent supraorbital ridges, recessed eyes, midface hypoplasia, and a square chin. His ears measured 7.7 cm bilaterally with the left having a more simple architecture. His finger tips were somewhat squared at the tip with prominent pads. The distal phalanx of the left third finger was missing from an accidental amputation. He walked into the exam room with a stooped over posture. Normal laboratory studies included routine chromosome analysis (450−650 band level), normal FMR1 gene expansion analysis, urine creatine, urine organic acids and plasma amino acids. FISH analysis with a subtelomeric probe set (Vysis) revealed the 9p subtelomeric deletion (ish 9ptel (305J7×1). No other chromosomal abnormalities were detected elsewhere.
Case 2 (CMS1485, alias “ANLY”) has been previously described [14, 15, 29, 30]. This female patient was diagnosed with mental retardation and anhidrotic ectodermal dysplasia, and had a de novo balanced translocation t(X;9)(q13.1;p24). The patient was reported to have delayed speech and psychomotor development. At age 7, she reportedly had education in a special school and psychological assessment showed her functioning at the lower end of the moderate MR range [29].
Cell culture
Peripheral blood lymphocytes of case 1 (CMS6482) were immortalized by Epstein-Barr virus transformation to establish a lymphoblastoid cell line. Fibroblast cell lines corresponding to patient CMS1485 (GM00705A) and a control sample CMS11195 (GM03652F) were obtained from NIGMS human genetic mutant cell repository, Coriell Institute for Medical Research (Camden, NJ). These cell lines were used for FISH experiments, RNA preparations, and genomic DNA preparations using standard protocols.
Molecular cytogenetics
Genomic contigs, clones, markers, mapping and sequence information were obtained from the NCBI (http://www.ncbi.nlm.nih.gov/genome/guide/human) databases.
BAC and PAC clones were obtained from commercial resources. DNA was isolated using Qiagen Mini-Prep columns and was labeled by incorporation of digoxigenin-11-dUTP or biotin-16-dUTP (Boehringer Mannheim) by nick translation using DNA polymerase (Life Technologies). Labeled products were combined with a 50× human Cot-1 DNA (Gibco-BRL) and the chromosome-specific labeled centromeric alphoid probe to a final concentration of 20 ng/μl in Hybrisol VII. Probes were denatured at 76°C for 10 minutes and preassociated at 37 °C for 15−20 minutes before hybridization.
For in situ hybridization, the metaphase chromosome spreads were obtained from lympoblastoid cells (CMS6482) or from fibroblast cells (CMS1485) using conventional methods. Slides were incubated in 2× SSC at 37°C for 30 minutes, serially dehydrated in 70%, 80%, and 95% ethanol at room temperature, denatured in 70% formamide/2 × SSC at 72°C for two minutes, then serially dehydrated in 70%, 80%, 95% ethanol. In situ hybridization and washing procedures were performed as previously described [31, 32]. Replication banding was performed simultaneously with FISH. Commercially available chromosome-specific labeled centromeric alphoid probes were used to identify specific chromosomes. The labeled probes were visualized with rhodamine-labeled anti-digoxigenin, and chromosomes were counterstained with DAPI. Images were examined at 100× magnification under a Zeiss Axiphot fluorescent microscope equipped with FITC, DAPI and triple band pass filter sets. Digital images were captured by computer using Applied Imaging Probevision software (Pittsburgh), and photographs were printed on a Kodak XL 7700 color image printer.
Pulsed-field gel electrophoresis (PFGE)
High molecular weight DNA was isolated from patient fibroblasts in agarose plugs by standard techniques. The plugs were digested with 80U of SalI for 16 hours in appropriate endonuclease buffer. Digested fragments were separated by electrophoresis on a 1% Seakem® Gold agarose gel (Cambrex) at 6 V/cm for 22 hours, at 12°C in 0.5X TBE running buffer, using a 45 sec pulse time on a CHEF-DR® III apparatus (BioRad). After electrophoresis, samples were transferred to a Hybond-N+ membrane (GE Healthcare) using an alkaline downward transfer procedure. The filters were pre-hybridized at 65°C for 2−3 hours in aqueous hybridization solution before adding the denatured probe.
A 938 bp cDNA amplicon from DOCK8 exon 41 to a portion of exon 47 was amplified using primers D8ex41.47 Forward – 5’- TCACATAGACTAGAGGCATTTTA -3’, Reverse – 5’- GATCAGGGAGTGTACAGTCAGTC -3’. The cDNA hybridization probe was labeled with [32P]-dCTP using the RadPrime DNA Labeling System (Invitrogen).
Quantitative Real-Time PCR
Genomic Quantitative PCR was performed using iQ™ SYBR® Green Supermix (BioRad) on an iCycler iQ™ Real-Time PCR detection System (BioRad). The experimental primer pairs sequences were: IVS13/F26IVS7 Forward – 5’- TTTTTGTCTTTGCCCCTCTGTA -3’, Reverse – 5’-TGTCTGGCCACCTCGCTGTAA -3’(333 bp product); Exon18/D8ex18 Forward – 5’-TTGCTTGTTAGTAATCAGAAAAG -3’, Reverse – 5’-GTCCCCACAAAGAACTAAAG -3’(294 bp product); IVS23/D8IVS23a Forward – 5’-TCTCGCAGTGACATCCTC -3’, Reverse – 5’-ATTTGTGCTTTCTTCTCAGTG -3’(151 bp product); Exon29/D8ex29 Forward – 5’-TGGGGAAATGTCATGTTTGA -3’, Reverse – 5’-CAGACACATGTGCACCAACA -3’(214 bp product); Exon34/D8ex34 Forward – 5’-CTGAGCTAATTGTCAGGAAC -3’, Reverse – 5’-GAAGCTAGATGAGAATCTTACTC -3’(272 bp product). For the reference gene, a primer pair amplifying exon 16 of the NEPH2 gene was used. The primer sequences were: Forward – 5’-GGGCCATCAGAGCTAAAGACC -3’, Reverse – 5-CTTGGGGGAAGTGGAGGTTTA -3’ (210 bp product). The cycling conditions used an initial 5 minute denaturation step at 95°C followed by the two-step amplification cycles at 95°C for 10 seconds and 58°C for 30 seconds repeated 40 times. Data was analyzed using the iCycler iQ™ software to generate a standard curve for each gene and calculate the fluorescence generated from the experimental primer pairs relative to NEPH2 using the comparative Ct method.
Expression Studies
Northern blots (Multiple Tissue Blots, BD Biosciences) containing poly A+ RNAs from different human brain sections (from individuals of ages 16−75 years) were hybridized with a 457 bp DOCK8 cDNA probe spanning exons 22−25. The probe was labeled with [32P]-dCTP using the RadPrime DNA Labeling System (Invitrogen) and hybridization was performed following the ExpressHyb (BD Biosciences) protocol. Filters were washed at high stringency according to the manufacturer's recommendations.
Expression of DOCK8 in fetal tissues was also measured by RT-PCR using a human fetal multiple tissue cDNA panel (BD Bioscience) and the D8ex41.47 primer pair. Isoform specific primers were used to examine expression of the DOCK8 gene splice variants. Variant 1, Dock8ex13−14 Forward 5’- AACGGCTCCTGTGTGTTCTT -3’, Reverse 5’-TTGTAATGTTCCGGGCTGAT -3’ (155 bp product; annealing temperature 60°C); Variant 2, Dock8ex13a-14 Forward 5’- GGGAAGACCATCTGCACCT -3’, Reverse 5’-TCAGCCTCTGTGGGTAGACA -3’ (396 bp product; annealing temperature 60°C); Variant 3, Dock8ex15−17 Forward 5’- CAGCGGGCCTGAATTTCT -3’, Reverse 5’-TGGGAGACAGTAGGATCCAGTT -3’ (247 bp product; annealing temperature 60°C);
Mutation Screening
A patient cohort consisting of 145 females patients with MR was screened for mutations in DOCK8. These patients were previously tested negative for the MR-causing FMR1 expansion. Four random normal individuals were also included as controls. Primers flanking all DOCK8 exons were designed. Amplicons corresponding to each exon were analyzed using IPS or DHPLC as previously described. [33, 34]. Primer pair details are available on request.
Both strands of PCR products from patients with abnormally migrating fragments and a normal control were amplified and sequenced using DYEnamic ET Dye Terminator Cycle sequencing Kit (GE Healthcare) and an automated MegaBACE 1000 DNA Analysis system (GE Healthcare). Sequences were analyzed using the DNASTAR program (Madison WI).
Flourescent microscopy
The fibroblast cell lines CMS1485 and CMS11195 were seeded onto Nunc Lab-Tek II CC2 chambered slides (Nalge Nunc International) at a density of 15,000 cells per well. Cells were grown overnight and then stained with Alexa Fluor® 488 phalloidin green and SYTOX® orange (Invitrogen). Briefly, cells were washed two times with PBS and fixed with 4% para-formaldehyde for 5 minutes. Cells were then permeablized with 0.1% Triton X-100 in PBS for 5 minutes followed by washing twice with PBS. The cells were then incubated with a quenching solution (1.02 g sodium acetate in 50 ml PBS) twice for 8 minutes each. Cells were then incubated for 30 minutes in a blocking solution (2% horse sera, 0.4% BSA in PBS) with 200μg/ml RNaseA. Alexa Fluor® 488 phalloidin green diluted 1:200 in the blocking solution was applied to the cells for 1 hour at room temperature. Cells were rinsed 3 times 5 minutes each with Block followed by a final rinse in TS (10mM Tris-HCl pH 7.5, 15 mM NaCl). SYTOX® orange was diluted to 2 μM in TS and added to cells for 7 minutes at room temperature. The cells were rinsed 4 times with PBS. Imaging was performed on an Axiovert 200 Laser Scanning Microscope (Zeiss).
Acknowledgments
We are grateful to the patients and their family members for participation in this study. We thank Debbie Bealer for her assistance in the initial and ongoing evaluation of the patient and Cindy Skinner for assistance in the collection of patient material. We also thank Dr. Karl Franek and Joy Norris for assistance and advice on the cell culture work. This study has been approved by the Institutional Review Board of Self Regional Healthcare, the IRB of record for the GGC and was supported, in part, by a grant from NICHD (R01 HD39331) to AKS and a grant from the South Carolina Department of Disabilities and Special Needs (SCDDSN).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
References
- 1.Ropers HH. X-linked mental retardation: many genes for a complex disorder. Curr. Opin. Genet. Dev. 2006;16:260–269. doi: 10.1016/j.gde.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 2.Stevenson RE. Advances in X-linked mental retardation. Curr. Opin. Pediatr. 2005;17:720–724. doi: 10.1097/01.mop.0000184290.57525.fb. [DOI] [PubMed] [Google Scholar]
- 3.Brewer C, Holloway S, Zawalnyski P, Schinzel A, FitzPatrick D. A chromosomal deletion map of human malformations. Am. J. Hum. Genet. 1998;63:1153–1159. doi: 10.1086/302041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colleaux L, et al. A novel automated strategy for screening cryptic telomeric rearrangements in children with idiopathic mental retardation. Eur. J. Hum. Genet. 2001;9:319–327. doi: 10.1038/sj.ejhg.5200591. [DOI] [PubMed] [Google Scholar]
- 5.Anderlid BM, et al. Subtelomeric rearrangements detected in patients with idiopathic mental retardation. Am. J. Med. Genet. 2002;107:275–284. doi: 10.1002/ajmg.10029. [DOI] [PubMed] [Google Scholar]
- 6.Xu J, Chen Z. Advances In molecular cytogenetics for the evaluation of mental retardation. Am. J. Med. Genet. 2003;117C:15–24. doi: 10.1002/ajmg.c.10016. [DOI] [PubMed] [Google Scholar]
- 7.Knight SJ, et al. Subtle chromosomal rearrangements in children with unexplained mental retardation. Lancet. 1999;354:1676–1660. doi: 10.1016/S0140-6736(99)03070-6. [DOI] [PubMed] [Google Scholar]
- 8.Rossi E, et al. Cryptic telomeric rearrangements in subjects with mental retardation associated with dysmorphism and congenital malformations. J. Med. Genet. 2001;38:417–420. doi: 10.1136/jmg.38.6.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vries BB, Winter R, Schinzel A, van Ravenswaaij-Arts C. Telomeres: a diagnosis at the end of the chromosomes. J. Med. Genet. 2003;40:385–398. doi: 10.1136/jmg.40.6.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleefstra T, et al. Disruption of the gene Euchromatin Histone Methyl Transferase1 (Eu-HMTase1) is associated with the 9q34 subtelomeric deletion syndrome. J. Med. Genet. 2005;42:299–306. doi: 10.1136/jmg.2004.028464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleefstra T, et al. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am. J. Hum. Genet. 2006;79:370–377. doi: 10.1086/505693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin CL, Wong A, Gross A, Chung J, Fantes JA, Ledbetter DH. The evolutionary origin of human subtelomeric homologies-or where the ends begin. Am. J. Hum. Genet. 2002;70:972–984. doi: 10.1086/339768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruusala A, Aspenström P. Isolation and characterization of DOCK8, a member of the DOCK-180 related regulators of cell morphology. FEBS Letters. 2004;572:159–166. doi: 10.1016/j.febslet.2004.06.095. [DOI] [PubMed] [Google Scholar]
- 14.Kere J, et al. X-linked anhidrotic (hypohydrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nat. Genet. 1996;13:409–416. doi: 10.1038/ng0895-409. [DOI] [PubMed] [Google Scholar]
- 15.Srivastava AK, et al. Fine mapping of the EDA gene: a translocation breakpoint is associated with a CpG island that is transcribed. Am. J. Hum. Genet. 1996;58:126–132. [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi K, et al. Homozygous deletion and reduced expression of the DOCK8 gene in human lung cancer. Int. J. Oncol. 2006;28:321–328. [PubMed] [Google Scholar]
- 17.Ottolenghi C, et al. The region on 9p associated with 46,XY sex reversal contains several transcripts expressed in the urogenital system and a novel doublesex-related domain. Genomics. 2000;64:170–178. doi: 10.1006/geno.2000.6121. [DOI] [PubMed] [Google Scholar]
- 18.Shan Z, et al. FISH mapping of the sex-reversal region on human chromosome 9p in two XY females and in primates. Eur. J. Hum. Genet. 2000;8:167–173. doi: 10.1038/sj.ejhg.5200431. [DOI] [PubMed] [Google Scholar]
- 19.Lerer I, et al. Deletion of the ANKRD15 gene at 9p24.3 causes parent-of-origin dependant inheritance of familial cerebral palsy. Hum. Mol. Genet. 2005;14:3911–3920. doi: 10.1093/hmg/ddi415. [DOI] [PubMed] [Google Scholar]
- 20.Techakittiroj C, Kim KC, Andersson H, Li MM. 9p subtelomere deletion: pathogenic mutation or normal variant? Beijing Da Xue Xue Bao. 2006;38:92–93. [PubMed] [Google Scholar]
- 21.Pierrou S, Hellqvist M, Samuelsson L, Enerback S, Carlsson P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J. 1994;13:5002–5012. doi: 10.1002/j.1460-2075.1994.tb06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsson C, Hellqvist M, Pierrou S, White I, Enerback S, Carlsson P. Chromosomal localization of six human forkhead genes, freac-1 (FKHL5), -3 (FKHL7), -4 (FKHL8), -5 (FKHL9), -6 (FKHL10), and -8 (FKHL12) Genomics. 1995;30:464–469. doi: 10.1006/geno.1995.1266. [DOI] [PubMed] [Google Scholar]
- 23.Kaufmann E, Knochel W. Five years on the wing of fork head. Mech. Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- 24.Lai CSL, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a sever speech and language disorder. Science. 2002;413:519–522. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 25.de Silva MG, et al. Disruption of a novel member of a sodium/hydrogen exchanger family and DOCK3 is associated with an attention deficit hyperactivity disorder-like phenotype. J. Med. Genet. 2003;40:733–740. doi: 10.1136/jmg.40.10.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watabe-Uchida M, John KA, Janas JA, Newey SE, Van Aelst L. The Rac activator DOCK7 regulates neuronal polarity through local phosphorylation of stathmin/Op18. Neuron. 2006;51:727–739. doi: 10.1016/j.neuron.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 27.Govek E, Newey S, Van Aelst L. The role of Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- 28.Benarroch EE. Rho GTPase: Role in dendritic and axonal growth, mental retardation, and axonal regeneration. Neurology. 2007;68:1315–1318. doi: 10.1212/01.wnl.0000259588.97409.8f. [DOI] [PubMed] [Google Scholar]
- 29.MacDermot KD, Hulten M. Female with hypohidrotic ectodermal dysplasia and de novo (X;9) translocation. Clinical documentation of the AnLy cell line case. Hum. Genet. 1990;84:577–579. doi: 10.1007/BF00210814. [DOI] [PubMed] [Google Scholar]
- 30.Thomas NS, et al. Characterisation of molecular DNA rearrangements within the Xq12-q13.1 region, in three patients with X-linked hypohidrotic ectodermal dysplasia (EDA) Hum. Mol. Genet. 1993;2:1679–1685. doi: 10.1093/hmg/2.10.1679. [DOI] [PubMed] [Google Scholar]
- 31.Ross ME, et al. Linkage and physical mapping of X-linked Lissencephaly/SBH (XLIS): A novel gene causing neuronal migration defects in human brain. Hum. Mol. Genet. 1997;6:555–562. doi: 10.1093/hmg/6.4.555. [DOI] [PubMed] [Google Scholar]
- 32.Vervoort VS, et al. AGTR2 mutations in X-linked mental retardation. Science. 2002;296:2401–2403. doi: 10.1126/science.1072191. [DOI] [PubMed] [Google Scholar]
- 33.Sossey-Alaoui K, et al. Molecular cloning and characterization of TRPC5 (HTRP5), the human homologue of a mouse brain receptor-activated capacitative Ca2+ entry channel. Genomics. 1999;60:330–340. doi: 10.1006/geno.1999.5924. [DOI] [PubMed] [Google Scholar]
- 34.Vervoort VS, et al. Sorting nexin 3 (SNX3) is disrupted in a patient with a translocation t(6;13)(q21;q12) and microcephaly, microphthalmia, ectrodactyly, prognathism (MMEP) phenotype. J. Med. Genet. 2002;39:893–899. doi: 10.1136/jmg.39.12.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.