Abstract
HIV-1 integrase is essential for viral replication and can be inhibited by antiviral nucleotides. Photoaffinity labeling with the 3′-azido-3′-deoxythymidine (AZT) analog 3′,5-diazido-2′,3′-dideoxyuridine 5′-monophosphate (5N3-AZTMP) and proteolytic mapping identified the amino acid 153–167 region of integrase as the site of photocrosslinking. Docking of 5N3-AZTMP revealed the possibility for strong hydrogen bonds between the inhibitor and lysines 156, 159, and 160 of the enzyme. Mutation of these residues reduced photocrosslinking selectively. This report elucidates the binding site of a nucleotide inhibitor of HIV-1 integrase, and possibly a component of the enzyme polynucleotide binding site.
The HIV type 1 (HIV-1) integrase (IN) is a 32-kDa protein encoded in the 3′ end of the pol gene of the virus. IN is responsible for the insertion of the viral DNA into host chromosomes and is essential for effective viral replication (1–5). During viral infection, IN catalyzes two consecutive reactions. Following reverse transcription, IN first processes the linear viral DNA ends by removing the nucleotides (generally two nucleotides) immediately 3′ to the conserved CA dinucleotide, leaving recessed 3′-OH termini (Fig. 1D). This reaction is referred to as 3′-processing and takes place in the preintegration complexes. After migration of the preintegration complexes into nuclei, integrase catalyzes the 3′-end-joining (strand transfer) reaction in which the IN-processed 3′ ends of the retroviral DNA are joined to the 5′-phosphate end of a break made by IN in the target chromosomal DNA. These two steps can be measured with in vitro assays employing purified recombinant HIV-1 IN and a 21-mer duplex oligonucleotide whose sequence corresponds to the U5 region of the HIV-1 long terminal repeat (LTR) (see Fig. 1D) (refs. 6–9; for review see refs. 1–4, 10, and 11).
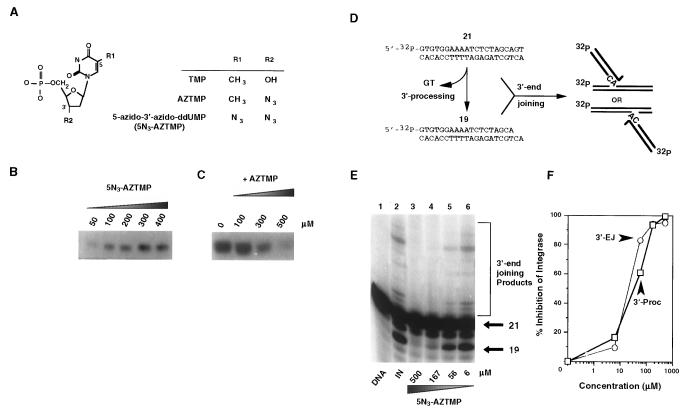
Figure 1.
Photoaffinity labeling and inhibition of HIV-1 IN with 5N3-AZTMP. (A) Structures of the nucleotides relevant to the present study. (B) Autoradiograph of HIV-1 IN photocrosslinked with the indicated amount (50–400 μM) of [32P]5N3-AZTMP. (C) Autoradiograph of HIV-1 IN (5 μg) photocrosslinked with 50 μM [32P]5N3-AZTMP and the indicated amount of competing AZTMP. (D) HIV-1 IN in vitro reactions: a 21-mer blunt-end oligonucleotide corresponding to the U5 end of the HIV-1 proviral DNA was 5′-32P-end-labeled and reacted with purified recombinant HIV-1 IN. The initial step (3′-processing) involves the nucleolytic cleavage of two bases from the 3′ end, resulting in a 19-mer oligonucleotide. The second step (3′-end-joining, also referred to as strand transfer) involves joining of these recessed 3′ ends to the 5′ end of an IN-induced break in another identical oligonucleotide that serves as the target DNA. (E) Concentration-dependent inhibition of HIV-1 IN by 5N3-AZTMP. (F) Quantitation of the results presented in E.
HIV-1 IN is an important target for intervention by chemotherapeutics (10, 11) because IN mutants with impaired catalytic activity cannot replicate (1–5, 12–15). To date, several classes of HIV-1 IN inhibitors, including nucleotides, have been reported (for review see refs. 11 and 16). However, there is currently little information on the specific binding sites of these inhibitors to HIV-1 IN. In the absence of an x-ray crystal structure of an HIV-1 IN/inhibitor complex, several HIV-1 IN mutants have been studied to obtain further information on the mechanism of drug binding (16, 17). A previous study demonstrated that therapeutic concentrations of the 3′-azido-3′-deoxythymidine (AZT) metabolite AZT 5′-monophosphate (AZTMP) inhibited the enzymatic activities of HIV-1 IN by acting on the enzyme catalytic core (amino acid residues 50–212) and competing with the DNA substrate (18). A recent study comparing the HIV-1 IN inhibitory potential of other nucleotides further suggested the existence of a specific nucleotide binding site on HIV-1 IN (19, 20). This possibility was supported by the inhibition of HIV-1 IN by pyridoxal phosphate (19, 21), a compound that reacts with lysine residues and is known to inhibit DNA polymerases, and by inhibition of HIV-1 IN by coumermycin A1, an ATP binding site inhibitor of DNA gyrase (19).
In this study, a nucleotide photoaffinity probe was used to investigate the nucleotide binding site of HIV-1 IN (22). Identification of such specific residues in HIV-1 IN not only can provide information on the DNA binding sites of HIV-1 IN but also can be important in the design of inhibitors.‖
MATERIALS AND METHODS
Materials.
The photoaffinity analog 3′,5-diazido-2′,3′-dideoxyuridine 5′-mono[32P]phosphate ([32P]5N3-AZTMP) was synthesized by the enzymatic phosphorylation of the corresponding nucleoside derivative to its radiolabeled monophosphate by using [γ-32P]ATP and herpes simplex virus 1 thymidylate kinase (23, 24). Molecular weight markers and precast N-[tris(hydroxymethyl)methyl]glycine (Tricine)/SDS/10–20% polyacrylamide gels were purchased from Novex (San Diego). Endoproteinase Glu-C (V8 protease), endoproteinase AspN, and chymotrypsin were purchased from Sigma. All other reagents were purchased from Fisher. Purified recombinant wild-type HIV-1 IN deletion mutants HIV-1 IN50–212 and HIV-1 IN50–288 were prepared as previously described (25, 26) .
Oligonucleotide Substrates and IN Assays.
The HPLC-purified oligonucleotides AE117, 5′-ACTGCTAGAGATTTTCCACAC-3′, and AE118, 5′-GTGTGGAAAATCTCTAGCAGT-3′, were purchased from Midland Certified Reagent Company (Midland, TX). To analyze the extent of 3′-processing and strand transfer with 5′-end labeled substrates, AE118 was 5′-end labeled by using T4 polynucleotide kinase (GIBCO/BRL) and [γ-32P]ATP (DuPont/NEN). The kinase was heat-inactivated and AE117 was added to the same final concentration. The mixture was heated to 95°C, allowed to cool slowly to room temperature, and run on a G-25 Sephadex Quick Spin column (Boehringer Mannheim) to separate annealed double-stranded oligonucleotide from unincorporated label.
HIV-1 IN assays were performed essentially as previously described (27).
Photoaffinity Labeling and Enzymatic Digestion of HIV-1 IN.
For photoaffinity labeling reactions, 1–10 μg of HIV-1 IN in 20 mM Tris⋅HCl, pH 6.8/8 mM MnCl2 was incubated with 50 μM [32P]5N3-AZTMP for 10 s in 25 μl final volume on ice. The sample was then irradiated on ice for 90 s with a hand-held UV lamp (254 nm UVP-11; Ultraviolet Products, San Gabriel, CA) at a distance of 3 cm. The reaction was quenched by addition of an equal volume of 10% trichloroacetic acid. The protein was precipitated by centrifugation at 13,000 × g for 10 min, the supernatant was discarded, and the pellet was resuspended in 20 μl of protein solubilizing mix for separation in SDS/12% polyacrylamide gels (24). Saturation and inhibition of photoincorporation experiments were done as previously described (23, 24, 28). Dried gels were exposed for autoradiography for 1–3 days. The comparative intensities of the photoincorporated HIV-1 IN bands on the autoradiographs were determined by laser densitometry (Bio-Rad model GS-670 imaging densitometer).
When the photolabeled sample was to be prepared for proteolytic mapping, 30–50 μl of digestion buffer (50 mM Tris⋅HCl, pH 8.0/0.1% SDS) was substituted for the solubilizing mix. Proteases were also dissolved in this same buffer. Photolabeled HIV-1 IN was digested with chymotrypsin at a ratio of photolabeled HIV-1 IN to protease of 10:1 (wt/wt) at various times and at room temperature. For V8 protease and endoproteinase AspN, 7 units or 0.15 mg, respectively, was added and samples were digested at 37°C.
Separation of Peptide Fragments by Tricine/SDS/PAGE.
Photolabeled HIV-1 IN peptides were separated on precast Tricine/SDS/10–20% polyacrylamide gels (Novex), stained with Coomassie blue, destained, and dried by using a cellulose drying kit from Promega (29). Dried gels were exposed for autoradiography for 1–3 days. The molecular masses of peptides were determined in reference to the low molecular weight markers (Mark 12, Novex) and mapped to the predicted molecular mass of proteolytic HIV-1 IN peptides as determined by using a Wisconsin Package program (Genetics Computer Group, Madison, Wisconsin).
Site-Directed Mutagenesis.
Site-directed mutagenesis was performed on the pINSD plasmid (a gift from R. Craigie, National Institutes of Health) containing the sequence coding for full-length HIV-1 IN, by using the Quikchange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. Codons for lysines 156 and 159 were mutated to arginines by using the following oligonucleotides. K156R: sense, GGA GTA ATA GAA TCT ATG AAT AGA GAA TTA AAG AAA ATT ATA GG; antisense, CC TAT AAT TTT CTT TAA TTC TCT ATT CAT AGA T TC TAT TAC TCC. K159R: sense, GAA TCT ATG AAT AAA GAA TTA AGG AAG ATT ATA GGA CAG G; antisense, C CTG TCC TAT AAT CTT CCT TAA TTC TTT ATT CAT AGA TTC. The codon for arginine-166 was mutated to threonine by using the following oligonucleotides: sense, GGA CAG GTA ACA GAT CAG GCT G; antisense, C AGC CTG ATC TGT TAC CTG TCC. The substituted nucleotide is shown in boldface and the mutated codon is underlined. After transformation, the DNA was isolated from single colonies arising from each mutagenesis reaction and was sequenced with an automated sequencer. Plasmid DNA that included the desired mutations was then introduced into BL21 (DE3) competent Escherichia coli by transformation (Novagen).
Molecular Modeling and Docking.
The coordinates of heavy atoms of HIV-1 IN50–212 were downloaded from the Protein Data Bank. Then hydrogen atoms that are not in the x-ray crystal structure were reconstructed and the resulting structure was fully minimized by using charmm. On the basis of the x-ray crystal structure of AZT (30), the 5N3-AZTMP structure was built by using the 3D editor in quanta 4.0. The partial charges of atoms of 5N3-AZTMP were calculated and assigned by Gaussian 94. 5N3-AZTMP was docked into the 153–167 region of the HIV-1 integrase catalytic core domain structure by using the gramm docking software. Molecular dynamics simulation was carried out with charmm 24, using the all-atom parameter set parm20 [parameter file for charmm version 20, Polygen (now Molecular Simulation), Waltham, MA]. The molecular dynamics simulation protocols are summarized as follows. The system (complex structure of HIV-1 IN bound to 5N3-AZTMP in the region encompassing residues 153–167) was first heated from 0 to 300K within 5 ps. The system was then equilibrated for 20 ps and a 150-ps simulation was performed. Snapshots were taken at 0.1-ps intervals during the simulation period and analyzed to extract information concerning the motion of ligand in the protein and the binding interactions between the two. The electrostatic forces were calculated by using the force switch method and a switching range of 8–12 Å. The van der Waals forces were treated with the shift method with a cutoff of 12 Å. Nonbonded lists were kept to 14 Å and updated heuristically. The dynamics simulation was propagated with a Verlet algorithm using a time step of 1 fs and was conducted in a layer of water 10 Å wide around the surface of HIV-1 IN catalytic core domain. Water was represented with a TIP3P model.
RESULTS AND DISCUSSION
Photoaffinity Labeling of HIV-1 IN.
To determine the AZTMP binding site on the enzyme, a photoaffinity analog of AZTMP, 5N3-AZTMP (Fig. 1A) (24), was used to label purified recombinant wild-type HIV-1 IN. As shown in the autoradiograph in Fig. 1B, increasing concentrations of [32P]5-N3-AZTMP resulted in photoincorporation, with a half-maximal saturation concentration at 90 μM. These results suggested crosslinking at a specific site on the enzyme; if not, a linear instead of hyperbolic relationship between probe concentration and photoincorporation levels would have been observed. For further verification of site specificity, different nucleoside and nucleotide inhibitors were included in the photolabeling reactions with HIV-1 IN and [32P]5N3-AZTMP. The representative autoradiograph shown in Fig. 1C demonstrates that AZTMP inhibited photoincorporation of 50 μM [32P]5N3-AZTMP in a dose-dependent manner with half-maximal inhibition at 220 μM. For comparison, the concentrations that inhibited photoincorporation by 50% [ICP50, (24)] were 360 μM for AZT, 450 μM for thymidine, 410 μM for TMP, and 400 μM for dCMP (data not shown). These values are consistent with the HIV-1 IN inhibition data by these nucleotides (18, 19). Cumulatively, the photolabeling saturation and inhibition studies suggested that 5N3-AZTMP binds to an AZTMP-specific site.
Binding of 5N3-AZTMP to HIV-1 IN would be expected to inhibit enzyme activity, because AZTMP inhibits HIV-1 IN (18). Fig. 1 E and F shows that 5N3-AZTMP was an effective inhibitor of HIV-1 IN, with IC50 values for both 3′-processing and 3′-end-joining in the 50–70 μM range. These results are consistent with the inhibitory potencies of other antiviral nucleotide derivatives tested in these reactions (18–20) and further suggest that 5N3-AZTMP binds to a nucleotide site on HIV-1 IN.
Proteolytic Mapping of HIV-1 IN.
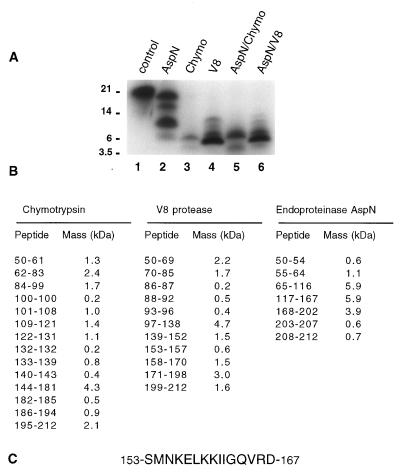
A proteolytic mapping procedure (24) was utilized to identify the photocrosslinked HIV-1 IN peptide(s) in the catalytic core domain of HIV-1 (IN50–212) (1–5). The parameters determined for optimal photolabeling with [32P]5N3-AZTMP were similar to those determined for the full-size enzyme, IN1–288 (half-maximal saturation 90–100 μM for IN50–212, data not shown). After photolabeling with 50 μM [32P]5N3-AZTMP, HIV-1 IN50–212 was digested with proteases and the resulting peptides were separated on Tricine/SDS/10–20% polyacrylamide gels. Peptide markers of known mass were used to generate a standard curve for size determination of photolabeled peptides (Fig. 2). The known primary amino acid sequence of HIV-1 IN was used to predict molecular masses of peptides after digestion with endoproteinase AspN (endo-AspN), chymotrypsin, or endoproteinase Glu-C (V8 protease) (Fig. 2B). Digestion with endo-AspN proved to be the most informative because of the small number of peptides generated (Fig. 2A). Photolabeled peptides of 6.0, 9.9, and 15.5 kDa were generated, with the 9.9-kDa peptide being the predominant labeled fragment. The logic for identifying the crosslinked endo-AspN peptide was as follows (24): The smallest photolabeled peptide, 6.0 kDa, was compared with the possible endo-AspN peptides shown in Fig. 2B. Two peptides comprising residues 65–116 and 117–167 were candidates. To distinguish between these two, adjacent peptides were added to derive possible peptide combinations. The only combination of peptides that could generate the observed pattern would be if the 117–167 peptide was photolabeled; addition of the 3.9-kDa 168–202 peptide would make a 9.8-kDa fragment (amino acids 117–202), and addition of the 65–116 peptide would yield a 15.7-kDa fragment (amino acids 65–202). If the 65–116 peptide had been the site of photocrosslinking, a predicted pattern of peptides of approximately 6, 7, 12, and 16 kDa would have been generated.
Figure 2.
Proteolytic mapping of 5N3-AZTMP binding to the catalytic core domain of HIV-1 IN. (A) Autoradiograph of HIV-1 IN50–212 photolabeled with [32P]5N3-AZTMP after protease digestions and Tricine/SDS/gel separation. HIV-1 IN50–212 (10 μg) was photolabeled with 50 μM [32P]5N3-AZTMP as described for Fig. 1B. Lane 1, undigested HIV-1 IN50–212; lane 2, plus endoproteinase AspN; lane 3, plus chymotrypsin; lane 4, plus V8 protease; lane 5, plus AspN for 30 min, then plus chymotrypsin (60 min total); lane 6, plus AspN for 30 min, then plus V8 protease (60 min total). (B) Predicted HIV-IN peptide masses after digestion with chymotrypsin, endoproteinase AspN, and V8 protease. (C) Deduced sequence of the HIV-1 IN peptide photocrosslinked with 5N3-AZTMP.
To confirm that the site of [32P]5N3-AZTMP photocrosslinking was within the 117–167 peptide, the complex was digested with chymotrypsin or V8 protease (Fig. 2A). For chymotrypsin, a major photolabeled peptide of 6.0 kDa was generated. Within the 117–167 region, amino acids 133–185 or 140–194 could account for a peptide of this size. V8 protease generated a major photolabeled peptide of 4.9 kDa, plus peptides of 6.5, 9.3, and 11.4 kDa. Consistent with the AspN and chymotrypsin patterns, a possible combination of V8 peptides would be residues 153–198 (5.1 kDa), 139–198 (6.6 kDa), 88–170 (9.2 kDa), and 97–198 (11.3 kDa). Double protease digestions were also done with endo-AspN for 30 min, followed by addition of either chymotrypsin or V8 protease for another 30 min (Fig. 2A). The AspN/chymotrypsin pattern was similar to that with chymotrypsin alone, except for two additional peptide bands at 3.0 and 7.0 kDa. These additional bands are consistent with internal cleavage of the major 10-kDa photolabeled AspN peptide (residues 117–202) to generate 2.9-kDa (residues 140–167) and 7.0-kDa (residues 140–202) hybrid AspN/chymotrypsin peptides. For AspN/V8, the V8 pattern predominated but an additional, faint, 3.2-kDa photolabeled peptide was apparent. This peptide would be consistent with a hybrid AspN/V8 peptide comprising amino acids 139–167 (3.2 kDa). For each protease digest presented, longer digestion times of up to 18 hr did not result in detectable peptides of lower molecular masses. On the basis of the reproducibility of gel separations, we estimate that the detection error is approximately ±0.5 mm migration, which translates to a ±0.2 kDa variability in the determined peptide masses for peptides less than 6 kDa. Photolabeled peptides below the molecular mass of 2.3 kDa migrate off the gel and are not detectable by the mapping procedure.
Cumulatively, the peptide digestion data indicate that AspN cleavage results in a [32P]5N3-AZTMP photolabeled peptide encompassing residues 117–167. Digestions with chymotrypsin and V8 protease narrow this region to residues 140–181 and 153–198, respectively. Combination digestions indicated residues 139–167 or 140–167. After consolidation of the overlapping fragments, these complementary digestion patterns thus indicate that the crosslinking site is located among amino acids 153–167 (Fig. 2C). Similar peptide mapping analyses were done with wild-type HIV-IN, HIV-1 IN1–212, and HIV-1 IN50–288 photolabeled with [32P]5N3-AZTMP (data not shown); the resulting peptide patterns all indicated the site of crosslinking to be within the same 153–167 peptide.
Docking of 5N3-AZTMP on the Peptide Residues 153–167 of the HIV-1 IN Core Domain.
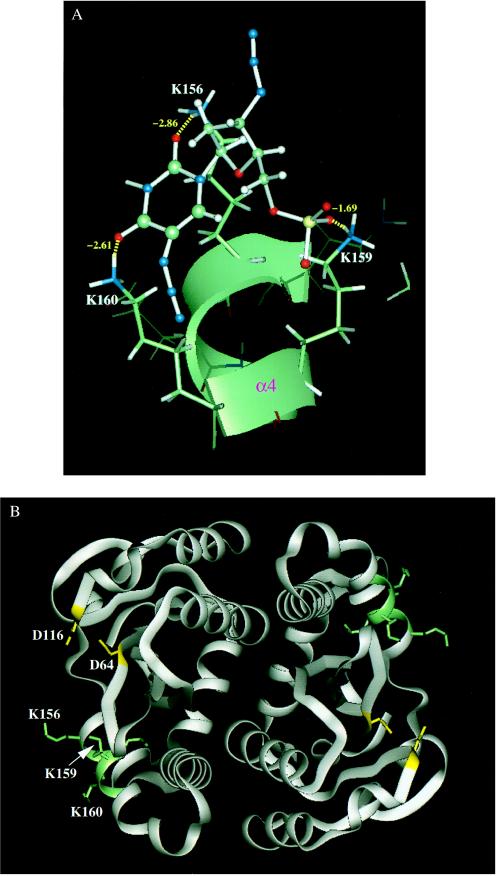
To identify the residues involved in the binding of 5N3-AZTMP to HIV-1 IN, molecular modeling and molecular dynamics simulations were carried out. The region of HIV-1 IN corresponding to residues 153–167 was selected for 5N3-AZTMP docking. The three-dimensional structure of HIV-1 IN was established from the coordinates obtained from the published crystal structures (31). After minimization, a 150-ps molecular dynamics simulation was conducted (Fig. 3). Residues K156, K159, and K160 appeared the most important for binding of 5N3-AZTMP to HIV-1 IN. The total hydrogen bond energy contributed by lysines 156, 159, and 160 was −7.16 kcal/mol, and the highest hydrogen bond energy was for lysine-156 (−2.86 kcal/mol). These results suggested that these three lysine residues (156, 159, and 160) are critical for nucleotide binding.
Figure 3.
Molecular modeling of 5N3-AZTMP binding to HIV-1 IN50–212. (A) Binding mode and hydrogen bond network between HIV-1 IN and 5N3-AZTMP. The modeling shows 5N3-AZTMP bound to the α4 helix of IN by forming hydrogen bonds with residues K156, K159, and K160. (B) Position of the 5N3-AZTMP binding site in the dimer structure of the HIV-1 IN catalytic core domain. HIV-1 IN50–212 is represented in ribbon model for the protein backbone. The D64 and D116 residues essential for catalysis are shown in yellow and stick model. The 5N3-AZTMP binding region and residues K156, K159, and K160 elucidated from our modeling studies are drawn in green and stick model.
The 153–167 peptide comprises primarily the fourth α-helical domain of HIV-1 IN (31, 32). Fig. 3B demonstrates that this domain is not part of the dimer interface, nor does it include the highly conserved catalytic core motif residues Asp-64, Asp-116, or Glu-152 that constitute the canonical DD(35)E motif of retroviral integrases (1–5). The clustered lysine residues 156, 159, and 160 near the active site of HIV-1 IN were predicted to be important for DNA binding (33, 34), and lysines 156 and 159 are essential for IN function (34). A study using systematic alanine substitutions for charged cluster groupings of HIV-1 IN also demonstrated the functional importance of the HIV-1 IN 153–167 region (35). The Arg-166 to alanine mutation alone or paired with Asp-167 resulted in replication-defective virus, whereas the 159/160 alanine mutations yielded viruses with greatly impaired integration frequency (35).
Role of Lysines 156, 159, and 160 of HIV-1 IN in Drug Binding.
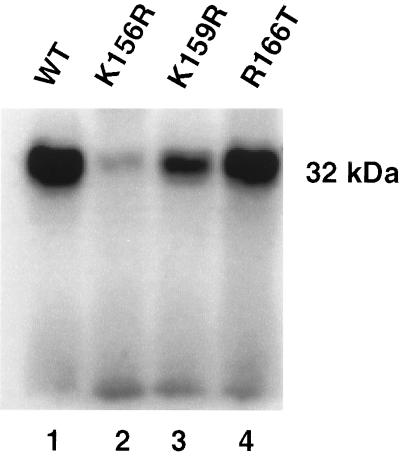
To provide direct evidence that lysines 156, 159, and 160 are involved in the binding of the AZT monophosphate analog, lysines 156 and 159 were conservatively replaced by arginines by using site-directed mutagenesis of the IN expression vector, and photocrosslinking was carried out with [32P]5N3-AZTMP. Fig. 4 demonstrates that substitution of arginine for lysine-156 practically abolished photocrosslinking of [32P]5N3-AZTMP, whereas substitution for lysine-159 reduced binding significantly. In contrast, substitution for arginine-166, which, according to our docking analysis, should not exhibit any interaction with [32P]5N3-AZTMP, did not affect HIV-1 IN binding. Thus, our mutational analysis suggests that although lysine-159, and possibly lysine-160, contributes to the binding of the AZT monophosphate analog, lysine-156 is critical for binding.
Figure 4.
Conservative mutations of critical residues of HIV-1 IN inhibit photocrosslinking with 5N3-AZTMP. Photocrosslinking and autoradiographies were performed as described for Fig. 1B. Lane 1, HIV-1 IN wild type; lanes 2, 3, and 4, crosslinked products of K156R, K159R, and R166T mutants with 5N3-AZTMP, respectively.
In summary, the cumulative data from the enzymatic, photoaffinity mapping, structural, and mutation studies identify the HIV-1 IN segments encompassing residues 153–167 as forming part of a specific nucleotide binding site. We also found that lysines 156 and 159 are critical for this mononucleotide binding. During the course of our study, two independent groups investigated the DNA binding site of the HIV-1 IN (34, 36). Using 10 DNA substrates containing azidophenacyl or 5-iodocytosine substitutions, Heuer and Brown (36) identified six different IN peptides that were involved in DNA binding. Two of these peptides are in the core HIV-1 IN domain (residues 49–69 and 139–152). However, they do not overlap with the peptide identified in our present study (residues 153–167) or the peptide identified by Jenkins et al. (residues 158–198) (34). The apparent discrepancy could be because of differences in DNA substrates, photoaffinity probes, and experimental conditions. For example, Heuer and Brown (36) used a branched oligonucleotide disintegration substrate. Their azidophenacyl-derivatized photoprobes had a large diameter and could potentially bind at various sites. Also, the contribution of a racemic mixture of phosphorothioate stereoisomers used to attach the photoprobes might contribute to crosslinking at different sites (36). The report of Jenkins et al. (34) used a 21-mer duplex oligonucleotide with 5-iododeoxyuridine substituted for the deoxyadenosine within the conserved CA dinucleotide adjacent to the scissile bond and suggested Lys-159 as the most likely site of crosslinking (34). Our model agrees with the results of Jenkins et al. that Lys-156 and Lys-159 are part of the nucleotide binding site. The data of Jenkins et al. show no role for Lys-160 in substrate binding, whereas we find that this residue is important for the binding of 5N3-AZTMP to the enzyme. Thus, the binding of 5N3-AZTMP to Lys-156, Lys-159, and Lys-160 places 5N3-AZTMP in the DNA binding site, where it can act as a competitor. Together, these observations indicate that, in addition to the catalytically important DD(35)E motif, the HIV-1 catalytic core domain appears to contain regions that contact the DNA substrate directly. The structure presented here might offer opportunities for designing novel HIV-1 IN inhibitors for the treatment of AIDS. As has been successfully done with HIV protease inhibitors, using the data obtained with 5N3-AZTMP should permit the optimization of interactions of potential inhibitors with HIV-1 IN to increase drug potency and selectivity.
ABBREVIATIONS
- IN
integrase
- AZT
3′-azido-3′-deoxythymidine
- AZTMP
AZT 5′-monophosphate
- 5N3-AZTMP
3′,5-diazido-2′,3′-dideoxyuridine 5′-monophosphate
- Tricine
N-[tris(hydroxymethyl)methyl]glycine
Footnotes
Portions of this work were presented at the Tenth International Society for Antiviral Research Meeting, April 6–11, 1997, Atlanta, GA.
References
- 1.Varmus H E, Brown P O. In: Mobile DNA. Berg D, Howe M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 53–108. [Google Scholar]
- 2.Coffin J M. In: Retroviridae and Their Replication. Fields B N, Knipe D M, editors. New York: Raven; 1990. pp. 1437–1500. [Google Scholar]
- 3.Goff S P. Annu Rev Genet. 1992;26:527–544. doi: 10.1146/annurev.ge.26.120192.002523. [DOI] [PubMed] [Google Scholar]
- 4.Katz R A, Skalka A M. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 5.Rice P, Craigie R, Davies D R. Curr Opin Struct Biol. 1996;6:76–83. doi: 10.1016/s0959-440x(96)80098-4. [DOI] [PubMed] [Google Scholar]
- 6.Craigie R, Mizuuchi K, Bushman F D, Engelman A. Nucleic Acids Res. 1991;19:2729–2734. doi: 10.1093/nar/19.10.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelman A, Mizuuchi K, Craigie R. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 8.Katzman M, Katz R A, Skalka A M, Leis J. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vink C, van Gent D C, Elgersma Y, Plasterk R H A. J Virol. 1991;65:4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bushman F. Science. 1995;267:1443–1444. doi: 10.1126/science.7878462. [DOI] [PubMed] [Google Scholar]
- 11.Neamati N, Sunder S, Pommier Y. Drug Discovery Today. 1997;11:487–498. [Google Scholar]
- 12.Stevenson M, Stanwick T L, Dempsey M P, Lamonica C A. EMBO J. 1990;9:1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LaFemina R L, Schneider C L, Robbins H L, Callahan P L, LeGrow K, Roth E, Schleif W A, Emini E A. J Virol. 1992;66:7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin C-C, Taddeo W A, Haseltine W A, Farnet C M. J Virol. 1994;68:1633–1642. doi: 10.1128/jvi.68.3.1633-1642.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelman A, Englund G, Orenstein J M, Martin M A, Craigie R. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pommier Y, Pilon A A, Bajaj K, Mazumder A, Neamati N. Antiviral Chem Chemother. 1997;8:483–503. [Google Scholar]
- 17.Chow S A. Methods. 1997;12:306–317. doi: 10.1006/meth.1997.0484. [DOI] [PubMed] [Google Scholar]
- 18.Mazumder A, Cooney D, Agbaria R, Gupta M, Pommier Y. Proc Natl Acad Sci USA. 1994;91:5771–5775. doi: 10.1073/pnas.91.13.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazumder A, Neamati N, Sommadossi J-P, Gosselin G, Schinazi R, Imbach J-L, Pommier Y. Mol Pharmacol. 1996;49:621–628. [PubMed] [Google Scholar]
- 20.Mazumder A, Uchida H, Neamati N, Sunder S, Jaworska-Maslanka M, Wickstrom E, Zeng F, Jones R A, Mandes R F, Chenault H K, Pommier Y. Mol Pharmacol. 1997;51:567–575. doi: 10.1124/mol.51.4.567. [DOI] [PubMed] [Google Scholar]
- 21.Lipford J R, Worland S T, Farnet C M. J AIDS. 1994;7:1215–1223. [PubMed] [Google Scholar]
- 22.Drake R, Neamati N, Sunthankar P, Mazumder A, Pommier Y. Antiviral Res. 1997;34:A42. [Google Scholar]
- 23.Mao F, Rechtin T M, Jones R, Cantu A, Anderson S, Radominiska A, Moyer M P, Drake R R. J Biol Chem. 1995;270:13660–13664. doi: 10.1074/jbc.270.23.13660. [DOI] [PubMed] [Google Scholar]
- 24.Rechtin T M, Black M E, Mao F, Lewis M, Drake R R. J Biol Chem. 1995;270:7055–7060. doi: 10.1074/jbc.270.13.7055. [DOI] [PubMed] [Google Scholar]
- 25.Bushman F D, Engelman A, Palmer I, Wingfield P, Craigie R. Proc Natl Acad Sci USA. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engelman A, Hickman A B, Craigie R. J Virol. 1994;68:5911–5917. doi: 10.1128/jvi.68.9.5911-5917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazumder A, Neamati N, Sunder S, Owen J, Pommier Y. In: Retroviral Integrase: A Novel Target in Antiviral Development; Basic In Vitro Assays with the Purified Enzyme. Kinchington D, Schinazi R, editors. Totowa, NJ: Humana; 1998. , in press. [DOI] [PubMed] [Google Scholar]
- 28.Radominska A, Drake R R. Methods Enzymol. 1994;230:330–339. doi: 10.1016/0076-6879(94)30022-4. [DOI] [PubMed] [Google Scholar]
- 29.Rechtin T M, Black M E, Drake R R. Anal Biochem. 1996;237:135–140. doi: 10.1006/abio.1996.0211. [DOI] [PubMed] [Google Scholar]
- 30.Camerman A, Mastropaolo D, Camerman N. Proc Natl Acad Sci USA. 1987;84:8239–8245. doi: 10.1073/pnas.84.23.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 32.Bujacz G, Alexandratos J, Qing Z L, Clement-Mella C, Wlodawer A. FEBS Lett. 1996;398:175–178. doi: 10.1016/s0014-5793(96)01236-7. [DOI] [PubMed] [Google Scholar]
- 33.Mazumder A, Neamati N, Pilon A, Sunder S, Pommier Y. J Biol Chem. 1996;271:27330–27338. doi: 10.1074/jbc.271.44.27330. [DOI] [PubMed] [Google Scholar]
- 34.Jenkins T M, Esposito D, Engelman A, Craigie R. EMBO J. 1997;16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiskerchen M, Muesing M A. J Virol. 1995;69:376–386. doi: 10.1128/jvi.69.1.376-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heuer T S, Brown P O. Biochemistry. 1997;36:10655–10665. doi: 10.1021/bi970782h. [DOI] [PubMed] [Google Scholar]