Abstract
Human papillomavirus type 16 (HPV-16), a DNA tumor virus, has a causal role in cervical cancer, and the viral oncoproteins E6 and E7 contribute to oncogenesis in multiple ways. E6 increases telomerase activity in keratinocytes through increased transcription of the telomerase catalytic subunit gene (TERT), but the factors involved in this have been elusive. We have found that mutation of the proximal E box in the TERT promoter has an activating effect in luciferase assays. This suggested that a repressive complex might be present at this site. HPV-16 E6 activated the TERT promoter predominantly through the proximal E box, and thus, might be acting on this repressive complex. This site is specific for the Myc/Mad/Max transcription factors as well as USF1 and USF2. Addition of exogenous USF1 or USF2 repressed activation of the TERT promoter by E6, dependent on the proximal E box. Using siRNA against USF1 or USF2 allowed for greater activation of the TERT promoter by E6. Conversely, loss of c-Myc function, through a dominant-negative Myc molecule, reduced activation by E6. Chromatin immunoprecipitations showed that in the presence of E6, there was a reduction in binding of USF1 and USF2 at the TERT promoter proximal E box, and a concomitant increase in c-Myc bound to this site. This shows that a repressive complex containing USF1 and USF2 is present in normal cells with little or no telomerase activity. In E6 keratinocytes, this repressive complex is replaced by c-Myc, which corresponds to higher levels of TERT transcription and consequently, telomerase activity.
One characteristic of tumor cells is their capacity for seemingly limitless division, or immortality. Normal cells, in culture, undergo the process of senescence and crisis following a given number of cell divisions that is dependent, in part, on the loss of telomeric DNA (reviewed in reference 24). Telomeres, the repeat sequences at the ends of eukaryotic chromosomes, are believed to signal senescence or crisis when they reach some critically short length, termed the Hayflick limit (26). Telomerase is an enzyme that maintains telomere length in cells, and thus protects cells from senescence (21). It has become clear that rather than maintaining long telomeres, telomerase protects cells from senescence by maintaining some critically short length, below which cells will cease to proliferate (reviewed in reference 7). Telomerase has been found to be up-regulated in 85 to 90% of tumor cells, the rest utilizing the alternative- lengthening-of-telomeres (ALT) pathway to protect their chromosomes (25). Thus, telomerase is an attractive target for antitumor therapy, and understanding its regulation would have relevance to many tumor models.
The human papillomavirus (HPV) is a DNA tumor virus, related to simian virus 40 and other polyomaviruses (reviewed in reference 40). Various HPV subtypes can infect the cutaneous or mucosal epithelium. The subtypes are further categorized based on their ability to cause serious disease. HPV type 6 (HPV-6) and HPV-11, for example, are associated with benign hyperplasia of the lower genital tract. In contrast, HPV-16 and -18 are associated with neoplasia, especially cervical intraepithelial neoplasia, a condition that progresses to cervical carcinoma if left untreated. HPV infection has a causal role in development of cervical cancer, and over 95% of cervical cancers contain HPV DNA (40).
E6 is an early HPV gene product that is involved in immortalization and transformation by the virus. E6 has several functions, including the ability to mediate degradation of the p53 tumor suppressor protein (57) through the E6-associated protein (E6-AP), an E3 ubiquitin ligase (28, 46, 47, 57), and the ability to bind to and inhibit transactivation by the coactivator proteins CBP and p300 (45, 61). In addition, E6 has been shown to interact with ERC-55 (E6-BP), a calcium binding protein (8), PDZ domain proteins, involved in cell to cell contacts in normal epithelium (17, 36), and the E6-TP1 protein, a Ran-like GTPase putatively involved in growth signal responsiveness (16, 49). Finally, E6 has been shown to increase telomerase activity when expressed in a variety of cell types, through transcriptional activation of the TERT gene, which encodes the catalytic subunit of the telomerase enzyme (18, 32, 33, 44, 51, 53).
The telomerase enzyme complex contains a protein core, TERT, which catalyzes reverse transcription (RT), and an RNA molecule, TERC, that serves as a template for synthesis (20, 21). An important regulatory point for telomerase is at the level of expression of the TERT subunit, which is the limiting factor in telomerase activity, as evidenced by the fact that ectopic expression of TERT alone is sufficient to induce high levels of telomerase activity in human foreskin fibroblasts and keratinocytes (15; reviewed in reference 12). The core TERT promoter, a region of ∼300 bp upstream of the translational start site, lacks a TATA box but contains two E boxes surrounding several Sp1 binding sites. E box sites bind several cellular proteins, including the Myc/Mad/Max family of transcription factors (9), as well as the upstream stimulatory factors (USF1 and USF2) (19). c-Myc is a potent oncogene, up-regulated in many human tumors. It functions as a heterodimer with partner molecule Max to activate transcription when bound to E box sites (reviewed in reference 9). Antagonizing c-Myc function are the Mad proteins, as well as several other family members, which can dimerize with Max and block c-Myc access to Max. Mad/Max dimers are active transcriptional repressors, recruiting histone deacetylases and other negative transcriptional regulators to chromatin to inhibit target gene transcription (reviewed in reference 3). Another family of proteins antagonizes c-Myc function in a less active way (4, 5). The USF1 and USF2 proteins can bind to E boxes, and occupy the site so that Myc/Max complexes cannot bind. Although USF1 and USF2 have lower affinity for E boxes than c-Myc complexes, they exist in such abundance relative to c-Myc that they can occupy E boxes under conditions where c-Myc expression is low, while increases in c-Myc levels can displace the USF complexes (50). Additionally, USF1 and USF2 have been shown to inhibit c-Myc-mediated transformation in vitro (37). Thus, these factors seem to be functionally antagonistic.
Several groups have recently shown that E6 can regulate transcription from the TERT promoter and correlated this with increased levels of telomerase activity in keratinocytes (18, 44, 53). These groups showed that E6-mediated transcriptional increases depended, at least in part, on the E boxes in the TERT promoter. These groups found no consistent increase in c-Myc levels in E6 keratinocytes, although in other cell types increased c-Myc has been observed in the presence of E6 (31, 56). No other factors that might be responsible for the effects of E6 on TERT transcription were described in these reports (18, 44, 53). In addition, a potentially confounding aspect of these studies was that the mutation in the proximal E box used also affects the binding site of the MT transcription factor, a newly described regulator of TERT transcription (52).
These studies left unanswered what specific factors were differentially regulated by E6, and whether the overlapping E box and MT sites played separate roles. Thus, we sought to identify the transcription factors involved in the regulation of TERT transcription by E6 and have found that the proximal E box in the TERT promoter, at position −34, is responsible for much of the ability of E6 to transactivate the promoter. We show here that a repressive complex containing USF1 and USF2 is present at this site in normal keratinocytes. In the presence of E6, these factors are replaced by c-Myc, which drives transcription of the TERT gene in vivo.
MATERIALS AND METHODS
Cell culture and infections.
Primary human foreskin keratinocytes (HFK) were isolated from neonatal foreskins and cultured in EpiLife medium supplemented with human keratinocyte growth supplement (Cascade Biologics). To generate stable cells expressing HPV-16 E6, the pBabe retroviral system was employed (34). Using the ΦNX packaging cell line (American Type Culture Collection), vesicular stomatitis virus-pseudotyped amphotropic retrovirus was generated. Retrovirus encoding E6 or empty vector was used to infect second- passage HFK in 8 μg of Polybrene per ml of medium. Following selection for 4 days with 1.25 μg of puromycin per ml of medium, cell populations were expanded, and cells were harvested for RNA isolation, protein lysate preparation or chromatin preparation no later than two passages after selection (fourth passage in culture).
Plasmids.
The human TERT core promoter, from −799 to + 1 (gift of R. Dalla-Favera), was cloned into the pGL3-basic vector. Site directed mutagenesis was performed using the QuikChange kit (Stratagene) to alter the distal (−241 to −236) or proximal (−34 to −29) E box sites, or the MT box (−32 to −23), in the promoter, to sequences shown in Fig. 1A, identical to mutations described by Tzukerman et al. (52). HPV-16 E6 in pSG5 (gift of L. Laimins), USF1 and USF2 in pSG5 (gifts of M. Sawadogo), and pSG5 empty vector (Stratagene), the Gal4-luciferase construct in pGL2-basic (gift of T. Kouzarides), the p300 (1-743)-Gal4 fusion in pcDNA3 (gift of A. Giordano), the dominant-negative c-Myc construct, pBabe-Myc/Max-64 (gift of H. Land) and siRNA molecules targeting human USF1 and human USF2 generated in the pBS/U6 vector (gift of Y. Shi), were used in transient transfections as indicated. The sequences of the siRNA molecules were as follows: USF1 5′ GGA TTC TAT CCA AAG CTT GTG AAG CTT CAC AAG CTT TGG ATA GAA TCC CTT TT TG 3′, USF2 5′ GGA GAT ACT ACG GCT GTG TCC AAG CTT GGA CAC AGC CGT AGT ATC TCC TTT TTG 3′.
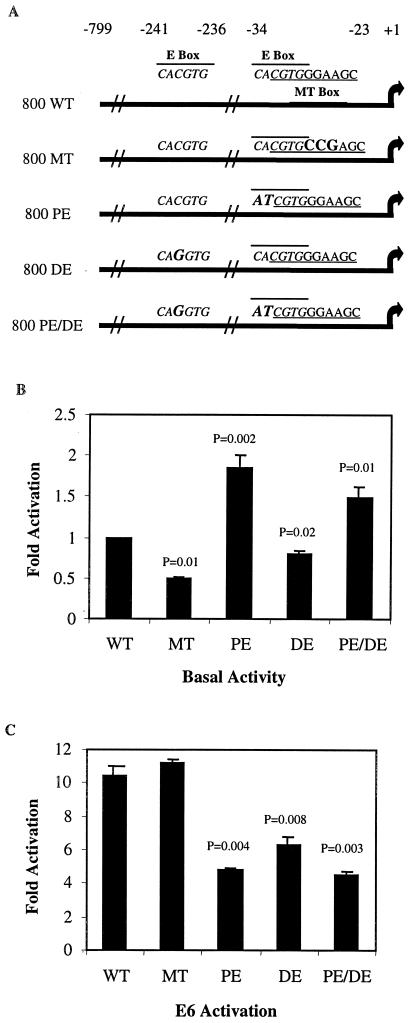
FIG. 1.
E6 activates the TERT promoter in keratinocytes. (A) Schematic of promoter constructs used: 800 WT, core TERT promoter region from −799 to + 1; 800 MT, mutation of the MT box, located at positions −32 to −23; 800 PE, mutation of the proximal Ebox located at positions −34 to −29; 800 DE, mutation of the distal E box located at positions −241 to −236; 800 PE/DE, mutation of both E boxes. Sequences of each binding site are shown, and mutations are indicated in larger, bold text for each construct. (B and C) Luciferase assays were carried out using mutants shown in panel A. One hundred nanograms of each promoter construct was cotransfected with 1.5 μg of vector (B) or E6 (C). Data shown are from one experiment, representative of three performed. Standard deviations (error bars) and P values are derived from three replicates within one experiment. Shown are the basal activity from each promoter, normalized to wild type (B), and activation by E6 relative to the basal activity of a given promoter construct (C).
Transfections and luciferase assays.
HFK were transfected using the Fugene 6 reagent (Roche Diagnostics Corporation) and DNA amounts as indicated. Cells were harvested 48 h posttransfection for luciferase assay extract preparation using reporter lysis buffer at a concentration of 2× (Promega) and lysates were mixed with luciferase assay reagent (Promega) and read in a luminometer (Packard Instruments).
siRNA production.
For examination of the functionality of the siRNA molecule, HFK were transiently transfected with the USF1 or USF2 siRNA construct, plus pEGFP-C1 (Invitrogen), a green fluorescent protein (GFP) expression vector, as a marker for transfection. Forty-eight hours posttransfection, cells were sorted using a FACSvantage cell sorter (BD Biosciences) and lysed for Western blots in whole-cell lysis buffer (250 mM NaCl, 50 mM Tris [pH 7.5], 0.5% NP-40, 20% glycerol, protease inhibitor cocktail). Lysates were run on sodium dodecyl sulfate (SDS)-12% polyacrylamide gels, transferred to nitrocellulose, and blotted for USF1 or USF2 using antibodies described below.
TRAP assay.
TRAP assays were done based on the protocol of Kim et al. (29) with modifications to allow for nonisotopic detection of PCR products. Cells were lysed in whole-cell lysis buffer (described above). TRAP PCRs contained PCR buffer (Fermentas), 1.5 mM MgCl2, 1 mM EGTA, 0.1 μg of TS primer with a 5′ covalently linked biotin molecule (5′-biotin-AATCCGTCGAGCAGAGTT-3′; Invitrogen), 5 μg of bovine serum albumin, 100 μM deoxynucleoside triphosphate (dNTP), 1 μg of T4 g32 single-strand binding protein (Roche Diagnostics Corporation), and 5 μg of cell extract. Reaction materials were incubated at room temperature for 30 min, and then 0.1 μg of CX primer [5′-(CCCTTA)3CCCTAA-3′] and 5 U of Taq DNA polymerase (Fermentas) were added. The reactions were subjected to 30 cycles of PCR (94°C for 40 s, 50°C for 40 s, and 72°C for 90 s), run on a 10% nondenaturing polyacrylamide gel, and transferred to Gene Screen Plus nylon membranes (NEN) in Tris-borate-EDTA. The membrane was washed with blocking buffer (5% SDS, 17 mM Na2HPO4, 8 mM NaH2PO4), and streptavidin-horseradish peroxidase (Chemicon) was applied in blocking buffer at a 1:1,000 dilution. The membrane was washed with wash buffer I (1:10 dilution of blocking buffer) followed by wash buffer II (100 mM Tris [pH 9.5], 100 mM NaCl, 10 mM MgCl2), developed with the ECL Plus reagent (NEN), and detected using the ChemiImager 5500 (Alpha Innotech).
Western blot analysis.
Whole-cell lysates were prepared as described above. Nuclear lysates were prepared as described previously (14). Briefly, cells were resuspended in hypotonic buffer (10 mM Tris [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol), swelled on ice for 10 min and Dounce homogenized. Nuclei were resuspended in buffer B (20 mM Tris [pH 7.9], 20% glycerol, 1.5 mM MgCl2, 300 mM KCl, 1% NP-40, protease inhibitor cocktail [P8340; Sigma]), rotated at 4°C and spun at high speed to clear lysates. Whole-cell extract (100 μg) or nuclear extract (25 μg) was run on SDS-10% polyacrylamide gels, transferred to nitrocellulose and probed with the following antibodies: c-Myc (9E10) (Roche Diagnostics Corporation), USF1 (C-20), USF2 (C-20), Max (C-17) (Santa Cruz Biotechnology). Blots were imaged on the ChemiImager 5500 (Alpha Innotech), and quantitation was done using these images and the FluorImager software package (version 2.1), using a local background method of calculation.
RT-PCR.
RNA was prepared using the RNeasy kit (Qiagen). RT reactions contained 5 μg RNA, 8 mM dithiothreitol, 500 μM dNTP, 16.7 μM random hexamer primer, RNase inhibitor, avian myeloblastosis virus RT, and buffer (Promega) and were incubated at 42°C for 60 min. PCRs were run with Promega buffer, 1.5 mM MgCl2, 200 μM dNTP, 10 μM primers, and 2.5 U of Taq polymerase (Promega). Primers for HPV-16 E6 were as follows: forward, 5′-GAG AGG ATC CAT GTT TCA GGA CCC ACA GG-3′; reverse, 5′-CAT GAA TCC TTA CAG CTG GGT TTC TCT AC-3′. Primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were as follows: forward, 5′-CCC CTC TGC TGA TGC CCC CAT GTT-3′; reverse, 5′-GAG CTT CCC GTC TAG CTC AGG GAT-3′. PCRs were run for 30 cycles of 94°C for 15 s, 54°C for 15 s, and 68°C for 45 s.
Chromatin immunoprecipitation.
Chromatin immunoprecipitations were carried out following a protocol from Upstate Biotechnology. Cells were treated with formaldehyde at a final concentration of 1% for 20 min to cross-link chromatin and the reaction was stopped with glycine at a final concentration of 125 mM. Cells were washed twice with PBS and then scraped and collected by centrifugation. Fixed cells were incubated in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris [pH 8.0]) on ice for 10 min. Lysates were sonicated to shear DNA to lengths of ∼1,000 bp, and the sonicated lysates were diluted 1:10 in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris [pH 8.0], 167 mM NaCl) and precleared on protein G beads blocked with sonicated salmon sperm DNA and bovine serum albumin. Immunoprecipitations (IP) were performed overnight at 4°C, using antibodies as described above, with the addition of GST (B-14; Santa Cruz Biotechnology), as a control. Immunoprecipitation reactions were washed with low-salt wash buffer (0.1% SDS, 1% Triton X-100, 20 mM Tris [pH 8.0], 2 mM EDTA [pH 8.0], 150 mM NaCl), high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris [pH 8.0], 500 mM NaCl), lithium chloride wash buffer (10 mM Tris [pH 8.0], 250 mM LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA), and finally with TE (10 mM Tris [pH 8.0], 1 mM EDTA). Samples were eluted in elution buffer (1% SDS, 0.1 M sodium bicarbonate) and incubated at 65°C overnight to reverse cross-link the chromatin. Samples were digested with 20 mg of proteinase K, phenol-chloroform extracted and DNA was precipitated with ethanol, dried and resuspended. Samples were analyzed via PCR with the following primers: TERT 3891 Forward, 5′ CCT CCC CTT CCT TTC CGC GG 3′; TERT 3891 Reverse, 5′ GGA AAG CCG CCG GGT CCC 3′; involucrin (INV) AP-1/5 Forward, 5′ TCT CCC ATA TAC GTG AAT GCC 3′; INV AP-1/5 Reverse, 5′ TTG TGC TCT GCT GCT GAC TT 3′. PCRs were run for TERT (31 cycles of 94°C for 45 s, 60°C for 45 s, and 72°C for 90 s) and for INV (31 cycles of 94°C for 45 s, 54°C for 45 s, and 68°C for 90 s).
RESULTS
E6 activation of the TERT promoter is E box dependent.
In this study, the contribution of several known transcription factor binding sites to E6 transactivation of the TERT promoter was assessed. To do this, TERT luciferase constructs containing an 800-bp region of the TERT promoter (−799 to + 1), with specific mutations in the distal E box (DE) at positions −241 to −236, or the proximal E box (PE) at positions −34 to −29, as well as a construct with both sites mutated (PE/DE), were generated. In addition, there was a report of the identification of a new transcription factor, the MT factor, that seemed to impact activation of TERT transcription (52), whose binding site, at −32 to −23, partially overlaps the proximal E box. Thus, mutations were engineered that were unique to each site in the PE and an MT construct, to clarify the relative contributions of the proximal E box and the MT box (Fig. 1A). The mutations chosen were identical to those used by Tzukerman et al. (52), who demonstrated that these mutations separate binding to the proximal E box or the MT box but are different mutant constructs from those used previously for study of E6 effects on TERT transcription (18, 44, 53).
These constructs were first examined for basal activity by transient transfections in HFK (Fig. 1B). In agreement with Tzukerman et al. (52), mutation of the MT box in this assay reduced the basal activity of the promoter by ∼50% (P = 0.01). Mutation of the distal E box reduced the basal promoter activity by 20% (P = 0.02). Loss of the proximal E box allowed for 80% greater basal activity from the promoter (P = 0.002), suggesting the presence of a repressive complex at this site. Mutation of both E boxes simultaneously yielded 50% higher activity than the wild-type promoter (P = 0.01), suggesting that the putative repressive complex on the proximal E box played a dominant role.
When E6 was cotransfected with these promoter mutants, the wild-type TERT promoter was activated by approximately 10-fold (Fig. 1C). Compared to its basal promoter activity, E6 was able to activate the MT mutant promoter to a comparable level, showing that this site was not integral for E6-mediated activation of transcription. In contrast, the PE, DE and PE/DE constructs each showed a 40 to 60% reduction in activation by E6 (P < 0.01 for each). This suggested that E6 is acting predominantly through the E boxes in the core TERT promoter, consistent with published results (18, 44, 53). Oh et al. have shown that the residual activity in the presence of mutated E boxes came from the five Sp1 sites between −236 and −34, since mutation of all five sites together with both E boxes was necessary to completely abolish the effects of E6 on TERT transcription (44).
Since the proximal E box had never been mutated on its own, and the potential repressive function seen here has not previously been reported, we chose to focus on the effects of transcription factors at this site, and to further examine what the putative repressive function we had observed might be. Recent papers suggested that the USF proteins might play a role in regulation of TERT transcription (35, 60). Thus, it seemed plausible that USF transcription factors could play a role in E6 regulation of the TERT promoter, occupying the proximal E box and passively or actively repressing transcription in a way that was reduced in the presence of HPV-16 E6.
USF1 and USF2 antagonize E6 activation of TERT transcription.
To assess whether USF proteins might affect E6 regulation of the TERT promoter, we took two related approaches. First, we cotransfected exogenous USF1 or USF2 with the wild-type TERT promoter in the presence of E6, to examine whether either USF family member could alter E6 mediated activation of TERT transcription. Second, we utilized a siRNA approach to ask whether a reduction in endogenous USF1 or USF2 levels could also affect basal and E6 induced TERT transactivation.
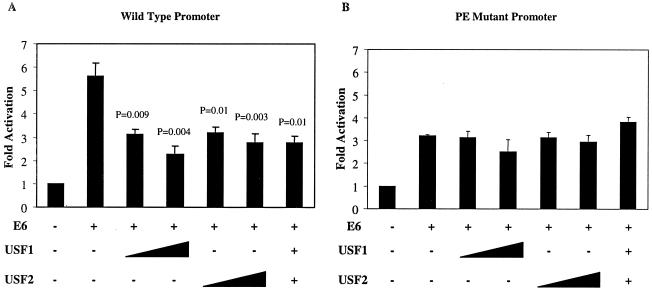
Luciferase assays were carried out in HFK using the wild-type TERT 800 promoter or the PE mutant construct, E6 and increasing amounts of USF1 or USF2 or both USF genes together. Addition of exogenous USF1 or USF2 in the presence of E6 was able to reduce the wild-type promoter activation in a titratable manner (Fig. 2A). Addition of both USF1 and USF2 together had a similar effect. This effect was not observed when the proximal E box was mutated, where the lower level of E6 activation was not reduced by addition of exogenous USF1 or USF2 (Fig. 2B). Thus, USF1 and USF2 could specifically antagonize the effects of E6 on TERT transcription, dependent on the proximal E box.
FIG. 2.
Exogenous USF1 or USF2 can repress E6-mediated activation of TERT promoter through the proximal E box. (A and B) One hundred nanograms of the wild-type TERT 800 promoter construct (A) or the PE mutant promoter (B) and 1 μg of vector or E6 were cotransfected into HFK with 333 or 500 ng of USF1 or USF2 or 333 ng of each together. Data shown are from one experiment, representative of three performed. Standard deviations (error bars) and P values are derived from three replicates within one experiment.
To examine this in another way, siRNA molecules were designed against USF1 and USF2 and expressed in the pBS/U6 vector. To examine the functionality of these siRNA molecules, each plasmid was transfected into HFK with a GFP expression vector, GFP-positive and negative cells were collected and Western blots were performed to analyze levels of USF1 or USF2 in these cells. As shown in Fig. 3A, endogenous USF1 levels were reduced ∼60% in the presence of the siRNA USF1 plasmid and the USF2 siRNA reduced levels of its target by ∼75%. As a loading control, Western blots for the Max protein are shown.
FIG. 3.
Reduction in USF1 or USF2 expression potentiates E6 activation of the TERT promoter. (A) Expression of siRNA USF1 or siRNA USF2 reduces levels of these proteins in keratinocytes. Western blots were done of keratinocytes cotransfected with an enhanced GFP expression construct and either the siRNA USF1 or siRNA USF2 molecule and were sorted to isolate transfected cells from nontransfected cells. Numbers below blots represent relative protein amounts. Max is shown as a loading control. (B to E) Reduction in USF1 or USF2 levels allows for greater TERT promoter activation by E6, dependent on the proximal E box. Luciferase assays were performed on the wild-type TERT promoter (B and D) or the PE mutant promoter (C and E) with pSG5 empty vector or E6, in the presence of a control siRNA expression vector, or the anti-USF1 (B and C) or anti-USF2 (D and E) siRNA molecules. Data shown are from one experiment, representative of three performed. Standard deviations (error bars) and P values are derived from three replicates within one experiment.
These siRNA molecules were then used to determine what effect decreased expression of USF1 and/or USF2 would have on TERT transcription. Plasmids encoding either USF1 or USF2 siRNA were transfected into HFK with the wild-type TERT 800 luciferase vector or the PE mutant of this construct, in the absence or presence of E6. Both siRNA molecules had modest but reproducible effects, ∼1.5-fold activation, on the basal promoter activity from the wild-type promoter (Fig. 3B and D), showing that reduction in USF1 or USF2 levels alone is not sufficient to increase TERT promoter activity to the levels induced by E6. The siRNA constructs had no effect on the PE mutant promoter (Fig. 3C and E), demonstrating that USF complexes are acting at the proximal E box site.
In the presence of E6, the wild-type promoter was activated five to eightfold over the basal promoter (Fig. 3B and D). Addition of either siRNA increased promoter activity 1.5- to 2-fold over that seen with E6 alone (P < 0.05 in each case), demonstrating that reduction in USF levels potentiates E6 transactivation of the TERT promoter. When the PE mutant construct was used, the ability of E6 to activate was reduced, as previously noted, and addition of the USF1 or USF2 siRNA molecules did not have a significant effect on the ability of E6 to activate the TERT promoter in the absence of the proximal E box (Fig. 3C and E). These results suggest that E6 antagonizes USF1 and USF2 function at the proximal E box of the TERT promoter.
Myc levels are higher in E6 keratinocytes.
We considered several possible mechanisms for the effects of E6 on USF function. First, since E6 is known to induce the ubiquitin mediated degradation of many proteins, we examined whether USF1 and USF2 might also be targets. By Western blotting, we found that there was no significant difference in endogenous USF1 or USF2 levels in E6 keratinocytes, compared to vector control cells (Fig. 4A). Another possibility was that E6 might be impeding binding of the USF complex to the promoter through an associated protein such as p300, or inhibiting the transactivation capability of such a USF/p300 complex. Mutational analysis suggests that this is not the case, as there are mutants of E6 that have wild-type levels of telomerase activity but bind less than 20% of wild-type amounts of p300 and CBP (data not shown).
FIG. 4.
Myc levels are increased in E6 keratinocytes. Extracts of E6 keratinocytes, or matched vector controls, were used for a TRAP assay and Western blots. (A) Whole-cell lysates were used for TRAP assays and Western blots for c-Myc, USF1 and USF2. Numbers below c-Myc blots indicate relative amounts of protein, as quantitated by the ChemiImager 5500. Max is shown as a loading control. RT-PCR of E6 is shown to indicate that E6 was expressed as expected. Spliced forms of E6 are readily detected by these primers and are indicated in the figure. GAPDH is shown as a control for the RT-PCRs. (B) Nuclear extracts were used for Western blots of c-Myc, USF1, USF2, and Max (as a loading control). Numbers below Myc blots indicate relative amounts of protein, quantitated as above.
A third possible model that we examined was that E6 might increase the expression of c-Myc, which would bind to the TERT promoter E boxes with higher affinity (50), replacing the USF complex, and this stoichiometric shift might account for the loss of USF1 and USF2 from the TERT promoter. Western blot analysis of whole-cell extract from keratinocytes stably expressing E6, or a vector infected control (pBabe) for each, showed that in E6 cells there was about a twofold increase in c-Myc levels (Fig. 4A). As a loading control, Max protein levels are shown. RT-PCR analysis demonstrated that the appropriate cells expressed full-length E6, as well as two spliced forms of the RNA, as previously observed (48), and a TRAP assay, to assess telomerase activity in these cells, is shown to demonstrate that each E6 line had higher levels of telomerase activity than its matched control. Since c-Myc functions in the nucleus, we also analyzed levels of nuclear c-Myc, USF1 and USF2 in E6 and matched control keratinocytes. As shown in Fig. 4B, USF1 and USF2 levels in the nucleus are similar in vector and E6 cells, while nuclear c-Myc levels are increased in E6 keratinocytes 4.5- to 7-fold. Max is shown as a loading control. Thus, in the presence of E6, c-Myc levels are increased modestly on the whole-cell and nuclear levels.
Loss of Myc function reduces E6 activation of the TERT promoter.
To assess whether E6 activation of TERT transcription required c-Myc activity, we utilized a dominant-negative chimeric protein (DN Myc), a Myc-Max fusion containing the DNA binding domain of Max fused to a transactivation deficient c-Myc. When overexpressed, this molecule can bind to E boxes with high affinity but cannot induce transcription and has been shown to antagonize c-Myc function in a dominant-negative way in transcriptional and transformation assays (2).
The DN Myc was cotransfected into HFK with the wild-type TERT promoter luciferase construct and E6, and luciferase assays were carried out to examine the ability of E6 to activate transcription in the presence of this molecule. As shown in Fig. 5A, activation of the TERT promoter by E6 was reduced up to 60% with increasing amounts of the dominant-negative Myc. This result indicates that c-Myc function is required for E6 to activate the TERT promoter, and suggests that increases in c-Myc by E6 can account for activation of the TERT promoter.
FIG. 5.
Loss of c-Myc function inhibits E6 activation of TERT promoter. (A) Luciferase assays were performed with 100 ng of the wild-type TERT promoter cotransfected with 1.3 μg of vector or E6 and 12.5, 50, or 100 ng of a dominant-negative Myc construct in HFK. Data shown are from one experiment, representative of three performed. Standard deviations (error bars) and P values are derived from three replicates within one experiment. (B) As a control, an irrelevant promoter, the Gal4-luciferase, was used to show that normal cellular function was not disrupted by the dominant-negative Myc molecule. One hundred nanograms of Gal4-luc was cotransfected with 100 ng of vector or p300 (1-743)-Gal4 fusion protein and 100 ng of DN Myc.
To show that the DN Myc did not induce a general transcriptional defect in cells, or simply kill transfected cells, a control transfection was done. This used a Gal4 reporter and the p300 (1-743)-Gal4 fusion, which activates the promoter to very high levels (45) and does not interact with c-Myc (39), with the maximum amount of the DN Myc used above. Activation of transcription by the Gal-4/p300 fusion was not inhibited by the DN Myc molecule, showing that c-Myc independent transcription was unaffected in these cells (Fig. 5B).
Increased Myc binding to the TERT promoter in E6 keratinocytes.
The results so far suggested that changes in transcription factor binding at the proximal E box of the TERT promoter were important for E6 mediated increases in TERT transcription. Mutation of the proximal E box led to a higher level of basal TERT promoter activity, and this mutant could not be activated as well by E6 (Fig. 1B and C). Transient transfection suggested that this might happen through a shift in the transcription factors bound to the proximal E box of the TERT promoter in the presence of E6, from repressive USF1 and/or USF2 complexes to c-Myc containing complexes (Fig. 2, 3, and 5). This model was tested directly by examining what transcription factors were bound to the endogenous TERT promoter in vivo, using the chromatin immunoprecipitation assay. This assay allowed examination of specific transcription factors bound to the proximal E box site within the TERT promoter in keratinocytes stably expressing E6, compared to vector-infected controls. Cross-linked cells were lysed, chromatin was purified and immunoprecipitations were done with antibodies specific for USF1, USF2, c-Myc, or GST as a control. PCR primers flanking the proximal E box region of the TERT promoter were used to examine binding to this site.
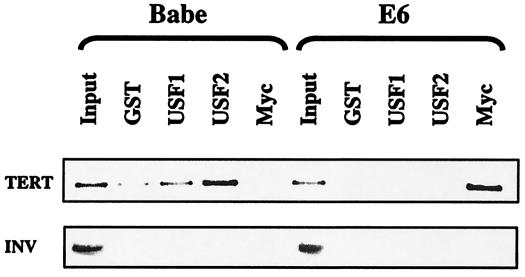
When E6- or vector-infected keratinocytes were used for the chromatin immunoprecipitation assay, reduced amounts of both USF1 and USF2 were found at the proximal E box of the TERT promoter in E6-expressing keratinocytes, compared to control cells (Fig. 6, upper panel). In addition, an increase in the amount of c-Myc bound to the promoter in E6-expressing keratinocytes was observed. As a control, primers for the INV promoter, which contains no E box sequences and is not regulated by E6, were used to show specificity of immunoprecipitations. These data confirm that c-Myc is replacing USF complexes on the proximal E box of the TERT promoter in the presence of E6. Taken together, these results show that E6 regulation of TERT transcription does in fact depend, at least in part, on c-Myc replacement of repressive USF1 and/or USF2 complexes on the TERT promoter.
FIG. 6.
Chromatin immunoprecipitation of the TERT promoter. Chromatin was prepared from Babe or E6 keratinocytes and immunoprecipitated with antibodies against USF1, USF2, c-Myc, or GST as a control. Coprecipitated DNA was used for PCRs with primers flanking the proximal E box of the TERT promoter (upper panel), or the INV promoter, as a control (lower panel). Data from one experiment, of four performed, are shown.
DISCUSSION
The results presented demonstrate that E6 activates transcription of the TERT gene, the catalytic subunit of the telomerase enzyme, at least in part through the c-Myc transcription factor. Utilizing different mutations in the promoter than those used previously for analysis of E6 regulation, we were able to show that the proximal E box of the TERT promoter bound a repressive complex. Chromatin immunoprecipitation results and additional reporter assays show that this complex contains USF1 and/or USF2. In the presence of HPV-16 E6, c-Myc levels are increased, and this increase leads to the replacement of the inhibitory USF complex by an activating, c-Myc containing complex at the proximal E box and induction of high level TERT transcription. Our results build on published analysis of the interplay between TERT transcription and HPV-16 E6 (18, 44, 53, 56), which suggested that E6 regulation of the TERT promoter was dependent, at least in part, on the E boxes in the promoter. Specific mutations that inhibit binding to the E box independently of the MT box were introduced, to allow for analysis of the role of each site individually. By doing this, we determined that mutation of the proximal E box revealed a twofold increase in basal promoter activity, a novel finding (Fig. 1B). Additionally, these mutations showed that E6 activation of the TERT promoter depended on an intact proximal E box, with a two- to threefold reduction in activation by E6 upon mutation of this site (Fig. 1C), whereas mutation of the MT box had no effect on the ability of E6 to activate TERT transcription.
Other work has identified repressive proteins that may bind to the proximal E box of the TERT promoter, including Mad1 (10, 23, 43, 59), a member of the c-Myc/Mad/Max family of transcriptional regulators, and the USF proteins (35, 60). Mad1 binding to the TERT promoter E boxes is associated with active repression and recruitment of histone deacetylase complexes, and is seen in systems where cells are induced to differentiate (23, 59) or upon overexpression of Mad1 (10, 43). The work presented here was carried out exclusively in cycling cells, and in this system, inhibition of deacetylase activity has no effect on TERT transcription, and no Mad1 is bound to the TERT promoter proximal E box in chromatin immunoprecipitations (data not shown). Thus, we chose to pursue the observation that USF family members could bind to this site, by analyzing the effects of these proteins on E6 mediated transactivation of the promoter.
To dissect the mechanism by which E6 affected USF complexes, and to understand why USF loss was not functionally equivalent to expression of E6, we examined several possibilities. First, since E6 is known to induce the degradation of many cellular proteins, we asked whether USF1 or USF2 levels were altered in E6 keratinocytes. This was not the case, as shown in Fig. 4A. Another possibility was that USF was mislocalized in E6 keratinocytes, compared to vector controls, but again, this was not the case. Nuclear levels of USF1 and USF2 are similar in Babe- and E6-expressing keratinocytes (Fig. 4B). Taken together, these results suggest that E6 is not directly affecting USF complexes. A third possibility was that E6 might be impeding binding of the USF complex to the promoter through an associated protein such as p300, or inhibiting the transactivation capability of such a USF/p300 complex. Although no direct interaction between either USF protein and p300 has been demonstrated, there is a report that adenovirus E1A can inhibit USF1 activation of transcription from a positively regulated promoter, and that this inhibition can be bypassed by addition of exogenous p300 (6). Several lines of evidence suggest that this is not the underlying mechanism of E6 alteration of USF function on the TERT promoter. First, the role of USF proteins on the TERT promoter seems to be repressive, and antagonistic to the activation by E6, while Breen et al. examined a promoter that is normally activated by USF1 and was repressed in the presence of E1A (6). Second, mutational analysis of E6 suggests that the p300 binding function and the up-regulation of telomerase are not related. If E6 were binding to the USF complex through its interaction with p300 and sequestering the p300 complex away from the promoter, E6 mutants that do not bind p300 should induce less TERT transcription and hence less telomerase activity. However, mutational analysis of E6 indicates that p300 binding and telomerase activity do not correlate. Specifically, there are mutants of E6 that have wild-type levels of telomerase activity but bind less than 20% of wild-type amounts of p300 and CBP (data not shown).
Several results suggested another possible model to explain our observations. In E6 keratinocytes, c-Myc levels were increased, compared to controls (Fig. 4A), and importantly, this observation held for nuclear levels of c-Myc protein as well (Fig. 4B). It is unclear how E6 is increasing c-Myc levels in the cell, although unpublished results from our laboratory suggest that this is at a transcriptional level (data not shown), and this observation is supported in the literature (31). Since c-Myc is a known positive regulator of TERT transcription (11, 27, 42, 56, 58), with a higher affinity for E box sites than USF complexes (50), we hypothesized that the increased c-Myc levels seen in the presence of E6 might be sufficient to account for increased TERT transcription. This was supported by the results that a dominant-negative Myc blocked TERT transactivation by E6 (Fig. 5) and that more c-Myc was bound to the proximal E box of the TERT promoter in the presence of E6 than in control cells (Fig. 6).
This model does not preclude the possibility that E6 is altering c-Myc function in additional ways. In fact, several results suggest that this is the case. The fact that there is a greater increase in nuclear c-Myc in the presence of E6 than that at the whole-cell level (Fig. 4A and B) suggests that, in the presence of E6, c-Myc is more concentrated in the nucleus. Similarly, disparity between the change in nuclear c-Myc levels in E6 cells and the difference in c-Myc associated with the TERT promoter in these cells (Fig. 4B and 5) points to additional levels of c-Myc regulation by E6. This suggests that, besides bulk increases in c-Myc levels, E6 alters the promoter affinity of c-Myc.
One possible mechanism for this altered affinity might be through E6 induced changes in cofactor recruitment by c-Myc. As discussed above, E6 activation of the TERT promoter does not strictly correlate with E6 binding to p300/CBP, but there may be other important interactions that are made or disrupted in the presence of E6 that affect c-Myc binding affinity, such as with the important regulator of c-Myc transactivation, the TRRAP protein (39). Alternatively, much has become known recently about ubiquitination as a regulator of transcriptional activity, specifically with respect to c-Myc (30, 41, 55). E6 has been shown to induce c-Myc ubiquitination, through the E6-E6AP interaction (22), and there is some evidence that this relates to transcriptional activation of the TERT promoter, as Gewin and Galloway have shown by mutational analysis that E6 transactivation of the TERT promoter, in transient assays, is dependent on E6-AP binding but not p53 degradation (18). These results suggest that E6 may be regulating c-Myc transcriptional activation through ubiquitination.
The work presented here extends the results of several groups, who demonstrated that HPV-16 E6 could mediate activation of the TERT promoter, dependent on the presence of E boxes in the promoter (18, 44, 53). These groups found no consistent increase in c-Myc protein in E6 keratinocytes (18, 44, 53) and thus concluded that E6 activated transcription of TERT in an E-box-dependent but c-Myc-independent, manner. However, there have been other reports of higher c-Myc levels in the presence of E6 (31, 56), and c-Myc has been previously shown to induce TERT transcription (10, 27, 43, 56, 58). These discrepancies may simply be due to differences in cell culture methods, perhaps in the passage and population doubling level at which cells were infected and harvested. Veldman et al. specify that they use late-passage, telomerase negative cells for their experiments (53). Gewin and Galloway reported variable levels of c-Myc protein in various oncoprotein-expressing cells, dependent on the rate at which they were proliferating (18). For the experiments presented here, only second passage cells were retrovirally infected and cells were harvested no later than fourth passage in culture, at which time points cell counting shows that the cultures are proliferating at similar rates (data not shown). These differences in experimental protocols may account for the differences in our observations. Because, in addition to increased levels of c-Myc protein at the whole-cell and nuclear levels, we have shown that TERT transcription depends on c-Myc function, by using a dominant-negative Myc, and in the presence of E6 more c-Myc is bound to the TERT promoter proximal E box, we believe that the increases in c-Myc observed are real and significant.
The increase in c-Myc levels by E6, and the functional consequences of this, requires further study. Increased c-Myc levels affect various cellular processes, including arrest in response to a variety of signals, immortalization, transformation and apoptosis (9), and may play a role in E6 effects on these processes. For instance, the increased levels of c-Myc seen in E6 cells may explain some observations concerning the ability of E6 to bypass certain types of arrest signals and induce transcription of other known c-Myc target genes. Malanchi et al. showed that E6 could bypass arrest of NIH 3T3 cells by serum starvation, or expression of p16INK4a or p27KIP1, and that the cyclin E and cyclin A promoters were activated by E6 expression, although none of these effects was as potent as seen with E7 (38). They did not identify a mechanism for these effects, but overexpression of c-Myc is known to bypass a cell cycle checkpoint under these various conditions (1, 54), and cyclin E and A are reported to be c-Myc target genes (13). More work will be needed to ascertain what role, if any, increases in c-Myc have in E6 biology.
In summary, we have demonstrated a novel mechanism for E6 regulation of TERT transcription, replacement of repressive USF complexes with activating c-Myc containing complexes. We have shown that USF antagonizes the ability of E6 to transactivate the TERT promoter, and that loss of c-Myc function blocks the ability of E6 to activate TERT transcription. The same transcription factor switch suggested by transient transfection analysis is shown to take place in cells that stably express E6, where c-Myc is bound to the TERT promoter, replacing USF1 and USF2.
Acknowledgments
We thank colleagues for their generous gifts of reagents; Branda Hu and Izabela Debkiewicz for technical assistance; and Laurel Baglia, Craig Menges, Don Nguyen, and Barry Thrash for critical reading of the manuscript.
This work was supported by grant NIDR DE13526. H.R.M. was supported by the Rochester Training Program in Oral Infectious Diseases, PHS/NIDCR T32 DE07165.
REFERENCES
- 1.Alevizopoulos, K., J. Vlach, S. Hennecke, and B. Amati. 1997. Cyclin E and c-Myc promote cell proliferation in the presence of p16INK4a and of hypophosphorylated retinoblastoma family proteins. EMBO J. 16:5322-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amati, B., M. W. Brooks, N. Levy, T. D. Littlewood, G. I. Evan, and H. Land. 1993. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell 72:233-245. [DOI] [PubMed] [Google Scholar]
- 3.Baudino, T. A., and J. L. Cleveland. 2001. The Max network gone mad. Mol. Cell. Biol. 21:691-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, K. E., and P. J. Farnham. 1999. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol. Cell. Biol. 19:8393-8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, K. E., and P. J. Farnham. 1997. Myc versus USF: discrimination at the cad gene is determined by core promoter elements. Mol. Cell. Biol. 17:2529-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breen, G. A., and E. M. Jordan. 2000. Upstream stimulatory factor 2 stimulates transcription through an initiator element in the mouse cytochrome c oxidase subunit Vb promoter. Biochim. Biophys. Acta 1517:119-127. [DOI] [PubMed] [Google Scholar]
- 7.Chan, S. W., and E. H. Blackburn. 2002. New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene 21:553-563. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J. J., C. E. Reid, V. Band, and E. J. Androphy. 1995. Interaction of papillomavirus E6 oncoproteins with a putative calcium-binding protein. Science 269:529-531. [DOI] [PubMed] [Google Scholar]
- 9.Cole, M. D., and S. B. McMahon. 1999. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene 18:2916-2924. [DOI] [PubMed] [Google Scholar]
- 10.Cong, Y. S., and S. Bacchetti. 2000. Histone deacetylation is involved in the transcriptional repression of hTERT in normal human cells. J. Biol. Chem. 275:35665-35668. [DOI] [PubMed] [Google Scholar]
- 11.Cong, Y. S., J. Wen, and S. Bacchetti. 1999. The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum. Mol. Genet. 8:137-142. [DOI] [PubMed] [Google Scholar]
- 12.Cong, Y. S., W. E. Wright, and J. W. Shay. 2002. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 66:407-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang, C. V. 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng, J., W. D. Funk, S. S. Wang, S. L. Weinrich, A. A. Avilion, C. P. Chiu, R. R. Adams, E. Chang, R. C. Allsopp, J. Yu, et al. 1995. The RNA component of human telomerase. Science 269:1236-1241. [DOI] [PubMed] [Google Scholar]
- 16.Gao, Q., L. Singh, A. Kumar, S. Srinivasan, D. E. Wazer, and V. Band. 2001. Human papillomavirus type 16 E6-induced degradation of E6TP1 correlates with its ability to immortalize human mammary epithelial cells. J. Virol. 75:4459-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardiol, D., C. Kuhne, B. Glaunsinger, S. S. Lee, R. Javier, and L. Banks. 1999. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene 18:5487-5496. [DOI] [PubMed] [Google Scholar]
- 18.Gewin, L., and D. A. Galloway. 2001. E box-dependent activation of telomerase by human papillomavirus type 16 E6 does not require induction of c-myc. J. Virol. 75:7198-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregor, P. D., M. Sawadogo, and R. G. Roeder. 1990. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 4:1730-1740. [DOI] [PubMed] [Google Scholar]
- 20.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405-413. [DOI] [PubMed] [Google Scholar]
- 21.Greider, C. W., and E. H. Blackburn. 1987. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 51:887-898. [DOI] [PubMed] [Google Scholar]
- 22.Gross-Mesilaty, S., E. Reinstein, B. Bercovich, K. E. Tobias, A. L. Schwartz, C. Kahana, and A. Ciechanover. 1998. Basal and human papillomavirus E6 oncoprotein-induced degradation of Myc proteins by the ubiquitin pathway. Proc. Natl. Acad. Sci. USA 95:8058-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunes, C., S. Lichtsteiner, A. P. Vasserot, and C. Englert. 2000. Expression of the hTERT gene is regulated at the level of transcriptional initiation and repressed by Mad1. Cancer Res. 60:2116-2121. [PubMed] [Google Scholar]
- 24.Hahn, W. C., and R. A. Weinberg. 2002. Modelling the molecular circuitry of cancer. Nat. Rev. Cancer 2:331-341. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 26.Hayflick, L. 2000. The illusion of cell immortality. Br. J. Cancer 83:841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horikawa, I., P. L. Cable, C. Afshari, and J. C. Barrett. 1999. Cloning and characterization of the promoter region of human telomerase reverse transcriptase gene. Cancer Res. 59:826-830. [PubMed] [Google Scholar]
- 28.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1991. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 10:4129-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, N. W., M. A. Piatyszek, K. R. Prowse, C. B. Harley, M. D. West, P. L. Ho, G. M. Coviello, W. E. Wright, S. L. Weinrich, and J. W. Shay. 1994. Specific association of human telomerase activity with immortal cells and cancer. Science 266:2011-2015. [DOI] [PubMed] [Google Scholar]
- 30.Kim, S. Y., A. Herbst, K. A. Tworkowski, S. E. Salghetti, and W. P. Tansey. 2003. Skp2 regulates myc protein stability and activity. Mol. Cell 11:1177-1188. [DOI] [PubMed] [Google Scholar]
- 31.Kinoshita, T., H. Shirasawa, Y. Shino, H. Moriya, L. Desbarats, M. Eilers, and B. Simizu. 1997. Transactivation of prothymosin alpha and c-myc promoters by human papillomavirus type 16 E6 protein. Virology 232:53-61. [DOI] [PubMed] [Google Scholar]
- 32.Kiyono, T., S. A. Foster, J. I. Koop, J. K. McDougall, D. A. Galloway, and A. J. Klingelhutz. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396:84-88. [DOI] [PubMed] [Google Scholar]
- 33.Klingelhutz, A. J., S. A. Foster, and J. K. McDougall. 1996. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature 380:79-82. [DOI] [PubMed] [Google Scholar]
- 34.Land, H., L. F. Parada, and R. A. Weinberg. 1983. Cellular oncogenes and multistep carcinogenesis. Science 222:771-778. [DOI] [PubMed] [Google Scholar]
- 35.Lee, D., H. Z. Kim, K. W. Jeong, Y. S. Shim, I. Horikawa, J. C. Barrett, and J. Choe. 2002. Human papillomavirus E2 down-regulates the human telomerase reverse transcriptase promoter. J. Biol. Chem. 277:27748-27756. [DOI] [PubMed] [Google Scholar]
- 36.Lee, S. S., B. Glaunsinger, F. Mantovani, L. Banks, and R. T. Javier. 2000. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 74:9680-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo, X., and M. Sawadogo. 1996. Antiproliferative properties of the USF family of helix-loop-helix transcription factors. Proc. Natl. Acad. Sci. USA 93:1308-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malanchi, I., S. Caldeira, M. Krutzfeldt, M. Giarre, M. Alunni-Fabbroni, and M. Tommasino. 2002. Identification of a novel activity of human papillomavirus type 16 E6 protein in deregulating the G1/S transition. Oncogene 21:5665-5672. [DOI] [PubMed] [Google Scholar]
- 39.McMahon, S. B., H. A. Van Buskirk, K. A. Dugan, T. D. Copeland, and M. D. Cole. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94:363-374. [DOI] [PubMed] [Google Scholar]
- 40.McMurray, H. R., D. Nguyen, T. F. Westbrook, and D. J. McCance. 2001. Biology of human papillomaviruses. Int. J. Exp. Pathol. 82:15-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muratani, M., and W. P. Tansey. 2003. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4:192-201. [DOI] [PubMed] [Google Scholar]
- 42.Oh, S., Y. H. Song, U. J. Kim, J. Yim, and T. K. Kim. 1999. In vivo and in vitro analyses of Myc for differential promoter activities of the human telomerase (hTERT) gene in normal and tumor cells. Biochem. Biophys. Res. Commun. 263:361-365. [DOI] [PubMed] [Google Scholar]
- 43.Oh, S., Y. H. Song, J. Yim, and T. K. Kim. 2000. Identification of Mad as a repressor of the human telomerase (hTERT) gene. Oncogene 19:1485-1490. [DOI] [PubMed] [Google Scholar]
- 44.Oh, S. T., S. Kyo, and L. A. Laimins. 2001. Telomerase activation by human papillomavirus type 16 E6 protein: induction of human telomerase reverse transcriptase expression through Myc and GC-rich Sp1 binding sites. J. Virol. 75:5559-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel, D., S. M. Huang, L. A. Baglia, and D. J. McCance. 1999. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 18:5061-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 47.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 48.Shirasawa, H., M. H. Jin, K. Shimizu, N. Akutsu, Y. Shino, and B. Simizu. 1994. Transcription-modulatory activity of full-length E6 and E6*I proteins of human papillomavirus type 16. Virology 203:36-42. [DOI] [PubMed] [Google Scholar]
- 49.Singh, L., Q. Gao, A. Kumar, T. Gotoh, D. E. Wazer, H. Band, L. A. Feig, and V. Band. 2003. The high-risk human papillomavirus type 16 E6 counters the GAP function of E6TP1 toward small Rap G proteins. J. Virol. 77:1614-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sommer, A., H. Burkhardt, S. M. Keyse, and B. Luscher. 2000. Synergistic activation of the mkp-1 gene by protein kinase A signaling and USF, but not c-Myc. FEBS Lett. 474:146-150. [DOI] [PubMed] [Google Scholar]
- 51.Stoppler, H., D. P. Hartmann, L. Sherman, and R. Schlegel. 1997. The human papillomavirus type 16 E6 and E7 oncoproteins dissociate cellular telomerase activity from the maintenance of telomere length. J. Biol. Chem. 272:13332-13337. [DOI] [PubMed] [Google Scholar]
- 52.Tzukerman, M., C. Shachaf, Y. Ravel, I. Braunstein, O. Cohen-Barak, M. Yalon-Hacohen, and K. L. Skorecki. 2000. Identification of a novel transcription factor binding element involved in the regulation by differentiation of the human telomerase (hTERT) promoter. Mol. Biol. Cell. 11:4381-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veldman, T., I. Horikawa, J. C. Barrett, and R. Schlegel. 2001. Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein. J. Virol. 75:4467-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vlach, J., S. Hennecke, K. Alevizopoulos, D. Conti, and B. Amati. 1996. Growth arrest by the cyclin-dependent kinase inhibitor p27Kip1 is abrogated by c-Myc. EMBO J. 15:6595-6604. [PMC free article] [PubMed] [Google Scholar]
- 55.von der Lehr, N., S. Johansson, S. Wu, F. Bahram, A. Castell, C. Cetinkaya, P. Hydbring, I. Weidung, K. Nakayama, K. I. Nakayama, S. A. d. O, T. K. Kerppola, and L. G. Larsson. 2003. The F-Box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol. Cell 11:1189-1200. [DOI] [PubMed] [Google Scholar]
- 56.Wang, J., L. Y. Xie, S. Allan, D. Beach, and G. J. Hannon. 1998. Myc activates telomerase. Genes Dev. 12:1769-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Werness, B. A., A. J. Levine, and P. M. Howley. 1990. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248:76-79. [DOI] [PubMed] [Google Scholar]
- 58.Wu, K. J., C. Grandori, M. Amacker, N. Simon-Vermot, A. Polack, J. Lingner, and R. Dalla-Favera. 1999. Direct activation of TERT transcription by c-Myc. Nat. Genet. 21:220-224. [DOI] [PubMed] [Google Scholar]
- 59.Xu, D., N. Popov, M. Hou, Q. Wang, M. Bjorkholm, A. Gruber, A. R. Menkel, and M. Henriksson. 2001. Switch from Myc/Max to Mad1/Max binding and decrease in histone acetylation at the telomerase reverse transcriptase promoter during differentiation of HL60 cells. Proc. Natl. Acad. Sci. USA 98:3826-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yago, M., R. Ohki, S. Hatakeyama, T. Fujita, and F. Ishikawa. 2002. Variant forms of upstream stimulatory factors (USFs) control the promoter activity of hTERT, the human gene encoding the catalytic subunit of telomerase. FEBS Lett. 520:40-46. [DOI] [PubMed] [Google Scholar]
- 61.Zimmermann, H., R. Degenkolbe, H. U. Bernard, and M. J. O'Connor. 1999. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J. Virol. 73:6209-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]