Abstract
Simian immunodeficiency virus (SIV) of macaques isolate SIVmac239 is highly resistant to neutralization by polyclonal antisera or monoclonal antibodies, a property that it shares with most primary isolates of human immunodeficiency virus type 1 (HIV-1). This resistance is important for the ability of the virus to persist at high levels in vivo. To explore the physical features of the viral envelope complex that contribute to the neutralization-resistant phenotype, we examined a panel of SIVmac239 derivatives for sensitivity to neutralization by a large collection of monoclonal antibodies (MAbs). These MAbs recognize both linear and conformational epitopes throughout the viral envelope proteins. The variant viruses included three derivatives of SIVmac239 with substitutions in specific N-linked glycosylation sites of gp120 and a fourth variant that lacked the100 amino acids that encompass the V1 and V2 loops. Also included in this study was SIVmac316, a variant of SIVmac239 with distributed mutations in env that confer significantly increased replicative capacity in tissue macrophages. These viruses were chosen to represent a broad range of neutralization sensitivities based on susceptibility to pooled, SIV-positive plasma. All three of these very different kinds of mutations (amino acid substitutions, elimination of N-glycan attachment sites, and a 100-amino-acid deletion spanning variable loops V1 and V2) dramatically increased sensitivity to neutralization by MAbs from multiple competition groups. Thus, the mutations did not simply expose localized epitopes but rather conferred global increases in neutralization sensitivity. The removal of specific N-glycan attachment sites from V1 and V2 led to increased sensitivity to neutralization by antibodies recognizing epitopes from both within and outside of the V1-V2 sequence. Surprisingly, while most of the mutations that gave rise to increased sensitivity were located in the N-terminal half of gp120 (surface subunit [SU]), the greatest increases in sensitivity were to MAbs recognizing the C-terminal half of gp120 or the ectodomain of gp41 (transmembrane subunit [TM]). This reagent set and information should now be useful for defining the physical, structural, thermodynamic, and kinetic factors that influence relative sensitivity to antibody-mediated neutralization.
The acute stage of infection of humans with the human immunodeficiency virus (HIV) or monkeys with the simian immunodeficiency viruses (SIVs) is characterized by high levels of viral replication and plasma viremia (30, 52, 67). Within the first few weeks after acute infection, the levels of circulating virus drop dramatically, coincident with the appearance of adaptive immune responses, including cytotoxic T lymphocytes (CTLs) and antibodies. However, virus is not completely eradicated, and virus replication persists despite the presence of readily detectable adaptive immune responses. Although antigenic escape plays a proven role in allowing persistent lentiviral replication, it is increasingly clear that antigenic variation alone does not explain the capacity of these viruses to persist (32). Other factors that may contribute to chronic infection include destruction of activated CD4+ T lymphocytes, Nef-mediated downmodulation of major histocompatibility complexes (MHCs), and the accumulation of long-lived, cellular reservoirs of infection (5, 13, 15, 16, 30, 67).
HIV-infected patients and experimentally infected rhesus monkeys typically generate high levels of circulating antibodies directed to epitopes in the viral envelope proteins. Although such antibodies often react well with monomeric envelope proteins and envelope-derived peptides, they have little or no capacity to neutralize typical primary viral isolates in vitro (48). This has led to the conclusion that the fully assembled viral envelope complex is inherently resistant to recognition and binding by the very antibodies its constituent proteins evoke (55).
The viral envelope complex is a trimer of heterodimers, with each heterodimer consisting of one gp120 (surface [SU]) subunit and one gp41 (membrane-spanning, or transmembrane [TM]) subunit (11, 12, 41, 45, 70). The fully assembled envelope complex is expressed on the surface of infected cells and incorporated into budding virions. Physical features of the Env complex that contribute to its poor immunogenicity include the arrangement of masking variable loop sequences on the exposed surface of the complex, the occlusion of protein surfaces by trimer formation, the presence of extensive N- and O-linked glycosylation, and the use of a two-receptor entry mechanism (4, 10, 39). In particular, the CD4 binding site is relatively recessed in the Env structure, and the coreceptor site is probably transiently exposed following receptor engagement (56). However, these features alone are not sufficient to explain the resistance of primary isolate (PI) viruses to neutralization. T-cell line-adapted (TCLA) viral isolates retain the variable loops, heavy glycosylation, and two-receptor entry mechanism that typify PI viruses, yet TCLA isolates are generally much more sensitive to antibody-mediated neutralization than their PI counterparts (42, 48, 49). Therefore, it is probable that the difference in neutralization phenotype of PI versus TCLA viruses reflects conformational adjustments in their respective envelope complexes, differences in binding and entry kinetics, or a combination of these factors (49).
The first strains of SIV were isolated from captive rhesus macaques within 2 years of the reported identification of HIV-1 as the causative agent of AIDS (3, 17, 25). As a direct result, the SIV-infected macaque model for AIDS pathogenesis has been in continual use for almost 18 years. SIVs of macaques (SIVmac) are very closely related to the human pathogen HIV-2, and like HIV-1 and HIV-2, arose from cross-species transmission of SIV from a naturally infected, nonhuman primate species (26, 27). As with HIV-1 and HIV-2, the jump to a naive species resulted in the establishment and spread of a persistent, pathogenic infectious agent. Experimental infection of macaques with SIVmac causes a reproducible disease course identical in many aspects to HIV-1 infection in human patients (19). Similarities include an acute phase characterized by high levels of viremia, a reduction in viremia coincident with the onset of adaptive immune responses, a gradual decline in CD4+ T cells, and a prolonged asymptomatic phase culminating in increased viral load and the development of opportunistic infections. SIVmac239 is a molecular clone of a pathogenic SIV isolate (58). Our extensive experience in manipulating and analyzing SIVmac239 has led to the observation that SIVmac239 is highly resistant to antibody-mediated neutralization, a property it shares with the majority of clinically relevant, PIs of HIV-1 (32). Moreover, we have previously shown that mutations in the SIVmac239 envelope gene that impart sensitivity to antibody-mediated neutralization without affecting inherent replicative capacity result in long-term immunological control, suggesting that resistance is important for the virus' ability to persist at high levels in vivo (33). Thus, the union in SIVmac239 of two properties—a highly neutralization-resistant phenotype amenable to molecular genetic analyses and a robust animal model for AIDS pathogenesis—creates an ideal system for comprehensive studies of the role of the antibody response to infection and the corresponding viral mechanisms for evading this response.
To begin to understand the physical basis for neutralization resistance, we have assembled a panel of seven viruses that we show here display a broad range of susceptibility to antibody-mediated neutralization. These include the pathogenic isolate SIVmac239 and five engineered variants that differ from the parental SIVmac239 at defined positions in the envelope gene. This panel is particularly useful for such studies, because differential susceptibility to neutralization can be unambiguously attributed to specific alterations in the envelope amino acid sequence. In this report, we describe the effects of assorted mutations in the envelope gene sequence on the degree and breadth of antibody-mediated neutralization. In particular we show that SIVmac316 and SIVΔV1V2 are inherently sensitive to neutralization by antibodies with a broad range of specificities, whereas the SIVmac239 parental virus, which contains identical sequences for most if not all of the same epitopes, is not detectably neutralized by any of these same antibodies.
MATERIALS AND METHODS
Production of viral stocks.
Virus stocks were produced by DEAE-dextran transfection of appropriate target cells, either CEMx174 (SIVmac239, g46, g56, SIV-M5, and SIVmac316) or the herpesvirus saimiri-immortalized rhesus monkey T-cell line Rh221-89 (SIVΔV1V2). Full-length viral genomes were generated by SphI digestion and ligation of separate plasmids containing the 5′ and 3′ halves of the viral genomes, and the ligated DNA was used to transfect cells by the DEAE-dextran method. Twenty-four hours prior to harvesting viral stocks, the transfected cells were pelleted and resuspended in fresh media. Laboratory-adapted, neutralization-sensitive SIVmac251lab was generated previously by extensive passage of uncloned SIVmac251 in culture (46). Viral content of harvested stocks was determined by p27 antigen capture assay (Coulter), and stocks were aliquoted and stored at −80°C. The SIVmac316 isolate used in this study contains eight of the nine coding changes originally described for this virus (50), except that the stop codon at position 767 has been changed to code for glutamine in order to restore the full-length cytoplasmic tail.
Neutralization assays.
The neutralization sensitivity of each virus was tested by using the secreted alkaline phosphatase (SEAP) reporter cell assay as described previously (46). Aliquots of all virus stocks used in these experiments were subject to serial twofold dilutions, and their titers were determined on SEAP reporter cells. Virus equivalent to 2 ng of p27 capsid protein was chosen as the lowest level of viral input sufficient to give a clear SEAP signal within the linear range for all of the virus strains. SEAP activity was measured on the earliest days, when levels were sufficiently over background to give reliable measurements: usually ranging from 2.5 to 3 days for SIVmac239 and SIVmac316 to 7 days for SIVΔV1V2. The percent neutralization of SIVmac239, SIV-M5, and SIVΔV1V2 by pooled SIV-positive plasma did not vary significantly over multiple days postinfection.
To perform neutralization assays, 96-well plates were set up as follows. To the first three columns, 25 μl of medium (RPMI 1640-10% fetal calf serum [FCS]) was added. To each of the other columns (no. 4 through 12), 25-μl aliquots of successive twofold dilutions of test antibody or plasma in RPMI 1640-10% FCS were added. Virus equivalent to 2 ng of p27 in a total volume of 75 μl was then added to each well in columns 3 through 12. Virus-free medium was added to columns 1 and 2 (mock). The plate was incubated for 1 h at room temperature. After incubation, 40,000 target cells (pLNLTR-SEAP) in a volume of 100 μl were added to each well. The plate was then placed into a humidified chamber within a CO2 incubator at 37°C for 3 to 7 days. SEAP activity was measured according to the manufacturer's recommendations, with modifications as described previously (46). Neutralization activity for all antibodies and plasma samples was measured in duplicate and reported as the average of at least two measurements.
RhMAbs.
The production and characterization of rhesus macaque monoclonal antibodies (RhMAbs) recognizing SIV envelope glycoproteins have been described previously (14, 62). In the present study, the following RhMAbs were used: 3.8E, 3.10C, H31, 3.10A, 5.5B, 3.11G, 3.11H, 1.11A, 1.1C, 3.11E, E31, 1.10A, C26, 3H3, and 3B3. An additional five RhMAbs, designated 3.7C, 3.5B, 3.5F, 3.4E, and 1.9C, were similarly derived by rhesus Epstein-Barr virus (EBV) transformation of peripheral blood B cells from a rhesus monkey (Mm 387-96) infected with the quintuple-glycosylation site mutant SIV-M5 (60). This monkey developed neutralizing antibodies to fully glycosylated wild-type virus SIVmac239 (60). All RhMAbs were purified from supernatant culture fluids by protein A affinity chromatography as described previously (62). Based on their cross-competition binding patterns, the original panel of RhMAbs was placed into eight competition groups. Neutralizing activity against SIV isolate SIV/17e was found in three groups (V, VI, and VII) (14). Groups V and VII recognized independent sites, but group VI RhMAbs showed considerable competitive interactions with both groups V and VII and were further subdivided based on their binding to a 45-kDa protease digestion fragment of SIVgp110 and the loss of antibody binding to gp120 containing certain mutations in V4. The competition binding patterns of the five new RhMAbs were determined as previously described (14).
Rhesus Fab fragments.
Phage display libraries were prepared from bone marrow of two monkeys (Mm 346-95 and Mm 347-95) that exhibited neutralizing antibody titers against the fully glycosylated wild-type virus SIVmac239. These two monkeys were infected with SIVmac239 mutants lacking specific N-linked glycosylation sites designated g45 and g46, respectively (60). Phage display libraries were prepared as described by Glamann et al. (28). Selection of phage-display libraries was performed against SIVmac239 gp130 (obtained from the AIDS Research and Reference Reagent Program) (29). The expression constructs were converted to express soluble Fabs, after which macaque Fab fragments were expressed in Escherichia coli XL1 Blue and purified by affinity column chromatography. Phage library selections and manipulations were performed as described previously (20). Affinity chromatography was performed with a rabbit antibody raised against rhesus macaque Fab fragments. Affinity columns were prepared by capturing and cross-linking of the rabbit antibody to a protein A column as described previously (2). Purified antibody was concentrated, dialyzed against phosphate-buffered saline (PBS), and filter sterilized.
Murine MAbs.
The murine MAbs used in these experiments were obtained from the AIDS Reagent Repository. The production and characterization of these reagents have been described previously (35, 36). The limited amounts of each antibody provided as ascites fluid necessitated presentation of neutralization results as dilution titers.
ELISA.
Reactivity against whole-lysed virions was measured by enzyme-linked immunosorbent assay (ELISA) as described previously (18, 68). Briefly, flat-bottom 96-well plates were coated with detergent-disrupted virions. Antibody was added to the wells and allowed to react for 1 h at 37°C. The wells were then washed with PBS containing 0.05% Tween 20, and an appropriate dilution of alkaline peroxidase-conjugated secondary antibody was added. Next, each well received 200 ml of p-nitrophenyl phosphate substrate solution (Kirkegaard & Perry, Gaithersburg, Md.). After 30 min, 50 ml of 3 N NaOH was added to each well, and the A410 was read.
RESULTS
Comparative neutralization using SIV-positive monkey plasma.
The genetic differences between SIVmac316, SIVmac239, and the g46, g56, SIV-M5, and SIVΔV1V2 derivatives of SIVmac239 are confined to the env gene and are known precisely (Fig. 1). The replicative and pathogenic properties of these viruses in vivo have been described elsewhere (17, 32, 33, 37, 44, 50, 58-60). With the exception of laboratory-adapted SIVmac251 (SIVmac251lab), which is an uncloned, highly passaged stock, all the viruses used in the experiments described below were derived by transfection of immortalized T-cell lines with molecularly cloned, genetically defined proviral DNA. Viral stocks were harvested at or near the peak of virus production following DNA transfection and were not passaged further prior to use in neutralization assays.
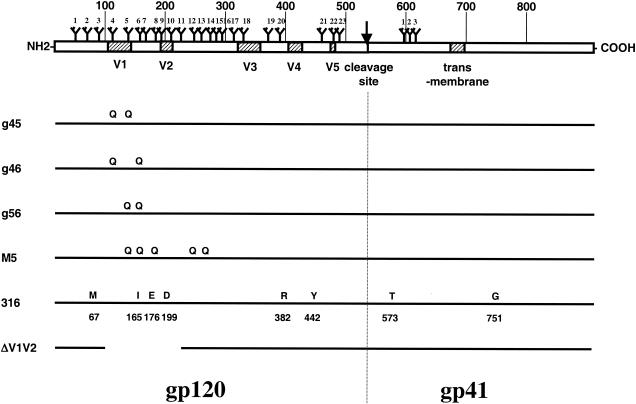
FIG. 1.
SIVmac239 and the derivative strains used in this study. The diagram at the top depicts the SIVmac239 envelope open reading frame, with the approximate locations of the variable loops and membrane-spanning domains (shaded boxes), the gp120-gp41 cleavage site (arrow), and the attachment sites for N-linked glycosylation indicated. Large numbers indicate amino acid positions in intervals of 100, and small numbers identify individual N-glycan attachment sites. Mutations that define the derivative strains are depicted below. Amino acid substitutions relative to SIVmac239 are indicated by the single-letter amino acid code of the new amino acid. The letter Q indicates an N-to-Q substitution that specifically eliminates an N-glycan attachment site. The 100-amino-acid deletion in SIVΔV1V2 is indicated by a line break.
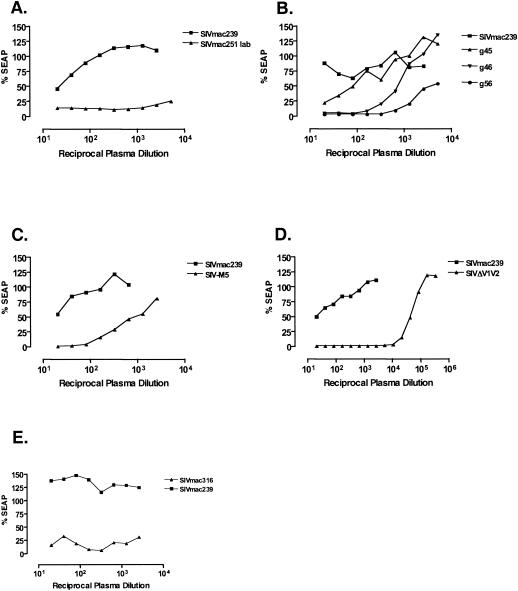
Despite the fact that the sequences of these viruses differed minimally from one another, they displayed a wide range of susceptibility to neutralization by SIV-positive monkey plasma. Typical results are depicted in Fig. 2. The parental virus SIVmac239 was routinely found to be resistant to antibody-mediated neutralization, with neutralization detectable only at the lowest dilutions (1:20 to 1:40) of pooled plasma from SIV-positive monkeys (Fig. 2). In contrast, SIVΔV1V2 was found to be at least as sensitive to SIV-positive plasma as SIVmac251lab. Fifty percent neutralization of SIVΔV1V2 was achieved at dilutions >1:10,000 (Fig. 2D). The sensitivity of SIVmac316 was similar to those of SIVΔV1V2 and SIVmac251lab (Fig. 2E). Fifty percent neutralization of SIVmac239 derivative viruses with substitutions in two N-glycan attachment sites (g45, g46, and g56) was achieved over a range of dilutions, g45 typically being the least sensitive of the three mutants (average 50% neutralization = 1:80), followed by g46 (1:430) and then g56 (>1:2,773). A comparison of the three mutants is also depicted in Fig. 2B. The sensitivity of the quintuple mutant SIV-M5, which lacks the 5th, 6th, 8th, 12th, and 13th glycosylation sites, was similar to that of g56 (Fig. 2C).
FIG. 2.
Comparative neutralization of SIVmac239 and derivatives by SIV-positive plasma. The assays depicted are representative of multiple comparisons performed with plasma samples from animals experimentally infected with SIV. Neutralization assays were performed as described in Materials and Methods. (A) SIVmac239 versus laboratory-adapted SIVmac251. (B) SIVmac239 versus the g45, g46, and g56 double-N-glycan attachment site mutants. (C) SIVmac239 and SIV-M5. (D) SIVmac239 and SIVΔV1V2. (E) SIVmac239 and SIVmac316-TMopen.
Neutralization by murine anti-gp120 MAbs.
We next measured neutralization of all seven viruses by a panel of 17 murine MAbs. The panel consisted of MAbs directed against various regions of gp120, including the N terminus; the C1 conserved domain; the V1, V2, and V3 loops; and conformational epitopes involving the V4 loop. Two MAbs recognizing epitopes in the ectodomain of gp41 were also tested (35, 36). None of the murine MAbs achieved 50% neutralization against SIVmac239, even at dilutions of ascites as low as 1:20 (Table 1). Twelve of the 15 anti-gp120 murine MAbs had measurable activity against at least one molecular variant of SIVmac239 (g46, g56, SIV-M5, SIV316, or SIVΔV1V2). Among the three MAbs that did not neutralize any variant, only one (VM18S) reacted strongly with whole-lysed SIVmac239 virions by ELISA (data not shown).
TABLE 1.
Neutralization by murine anti-SIV MAbs
| MAb | Epitope | Neutralization activity for virusa
|
||||||
|---|---|---|---|---|---|---|---|---|
| SIV239 | g46 | g56 | M5 | SIV251lab | SIV316 | ΔV1V2 | ||
| KK17 | NH2 | − | 30 | 10 | 10 | >2,560 | 2 × 106 | 2 × 105 |
| KK68 | C1 | − | − | − | − | − | − | − |
| KK65 | V1 loop | − | 10 | 24 | 10 | − | ND | − |
| KK8 | V1-V2 | − | − | 10 | − | 50 | − | − |
| KK42 | V3 loop | − | 20 | 15 | − | >2,560 | ND | 1.5 × 105 |
| KK45 | V3 loop | − | − | 10 | − | 160 | ND | 2.2 × 104 |
| KK46 | V3 loop | − | 20 | − | 10 | >2,560 | 6.5 × 104 | >1.3 × 106 |
| Senv7.1 | V4/conf.b | − | 60 | 20 | 25 | >2,560 | 6 × 106 | 15 |
| Senv101.1 | V4/conf. | − | 42 | 15 | − | >2,560 | ND | − |
| KK5 | V4/conf. | − | − | − | − | 12 | ND | 3 × 104 |
| KK9 | V4/conf. | − | − | − | − | − | ND | >1.3 × 106 |
| KK15 | gp41 | − | − | − | − | − | ND | − |
| KK41 | gp41 | − | 16 | 45 | 200 | >2,560 | >104 | 4.1 × 104 |
| VM18s | ?c | − | − | − | − | − | − | − |
| KK57 | ? | − | − | − | − | 640 | ND | 6 × 104 |
| Senv50.1 | ? | − | − | − | − | 50 | ND | − |
| Senv71.1 | ? | − | − | 10 | − | − | ND | − |
The numbers are the reciprocal of the dilution of ascites fluid required to reduce infectivity of the indicated virus by 50%. -, 50% neutralization was not achieved, even at the lowest dilution (highest concentration) tested; >, the highest dilution (lowest concentration) tested still gave greater than 50% neutralization, and sufficient dilutions were not performed to reach the 50% endpoint. ND, not determined.
conf., conformational.
?, the precise epitope has not been determined.
SIVΔV1V2 and SIVmac316 both displayed considerable neutralization sensitivity, in many cases equal to or exceeding that of SIVmac251lab, with 50% neutralizing dilutions on the order of 10−4 to 10−6 (Table 1). SIVΔV1V2 and SIVmac316 were detectably neutralized by 8 of 15 and 4 of 7 anti-gp120 murine MAbs tested, respectively. MAb KK8, which recognizes an epitope that maps to the V1-V2 region of gp120, did not react with the SIVΔV1V2 deletion mutant, confirming the specificity of this antibody for determinants in the V1-V2 region of envelope (Table 1) (35, 36). The g46, g56, and M5 glycosylation attachment site mutants were also neutralized by a majority of the murine MAbs, albeit at much lower dilutions (range, 1:10 to 1:200).
Neutralization by anti-gp120 RhMAbs.
Of particular relevance to understanding the neutralization-resistant phenotype of primary viral isolates are antibodies representing binding specificities generated during actual viral infection. Therefore, we extended our survey of MAb neutralization to include a panel of MAbs derived from experimentally infected, SIV-positive rhesus macaques. Twenty of the RhMabs used in this study have been described previously, including their organization into nine separate groups based on pairwise competition mapping (14). The experiments reported here included five additional anti-SIV RhMAbs, which we have characterized and assigned to the previously defined competition groups (summarized in Table 2).
TABLE 2.
Assignment of RhMAbs to competition groups
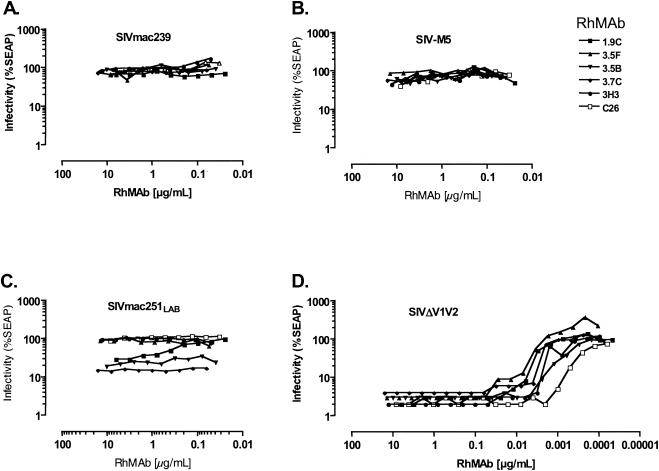
As with the murine MAbs, SIVmac239 was not effectively neutralized by any of the RhMAbs tested (Fig. 3). Fifteen of the 20 anti-gp120 RhMAbs had neutralizing activity against two or more of the SIVmac239 variants. Of these, 10 also neutralized SIVmac251lab (Table 3). Interestingly, the g46 virus, which lacks two glycosylation attachment sites in the V1 loop, was neutralized by a group VII antibody at high concentration (RhMab 1.10A [10 μg/ml]), and the closely related SIV-g56 mutant was also neutralized by 1.10A at high concentration (15 μg/ml), as well as by a group VI RhMab (3.11E [6.3 μg/ml]). Thus, substitution of N-glycan attachment sites in the N-terminal half of the gp120 amino acid sequence led to increased sensitivity to antibodies recognizing epitopes in the C-terminal half of the protein. Only 3 of the 20 MAbs showed no neutralization activity against any of the variants.
FIG. 3.
Comparative neutralization by rhesus anti-gp120 MAbs. The figure depicts a representative set of neutralization curves, comparing neutralization of the SIVmac239 parent (A), SIV-M5 (B), SIVmac251lab (C), and SIVΔV1V2 (D) by seven different RhMAbs.
TABLE 3.
Neutralizing activity of anti-gp120 RhMAbs by competition group
| MAb competition group (binding site) | Concn of MAb (μg/ml) reducing infectivity by 50%a
|
||||||
|---|---|---|---|---|---|---|---|
| SIV239 | g46 | g56 | M5 | SIV251lab | SIV316 | ΔV1V2 | |
| I (NH2) | |||||||
| H31 | − | − | − | − | 2.1 | 0.1 | 0.3 |
| 3.8E | − | − | − | − | − | − | − |
| 3.10C | − | − | − | − | − | − | − |
| II (V1 loop) | |||||||
| 3.10A | − | − | − | − | − | 0.1 | − |
| III (V2 loop) | |||||||
| 5.5B | − | − | − | − | − | 0.04 | − |
| IV (V3 loop) | |||||||
| 3.11G | − | − | − | − | 3.5 | 0.6 | 0.4 |
| 3.11H | − | − | − | − | 0.2 | 0.005 | 0.08 |
| V | |||||||
| 1.1C | − | − | − | >5.3 | 0.02 | 0.003 | 4.0 |
| 1.11A | − | − | − | − | 0.09 | 0.01 | 2.5 |
| 3.4E | − | − | − | − | − | 0.002 | 0.04 |
| VIa | |||||||
| 3.11E | − | − | 6.3 | 3.5 | 0.05 | 0.0009 | 0.001 |
| VI | |||||||
| 1.9C | − | − | − | 6.4 | 0.5 | 0.001 | 0.003 |
| 3.5B | − | − | − | − | 0.04 | 0.002 | 0.0005 |
| 3.5F | − | − | − | − | − | 0.0001 | 0.004 |
| 3.7C | − | − | − | − | 0.06 | 0.003 | 0.003 |
| E31 | − | − | − | − | − | 0.0002 | 0.05 |
| VII (V4) | |||||||
| 3H3 | − | − | − | 10.0 | − | 0.003 | 0.002 |
| C26 | − | − | − | 6.0 | − | 0.0008 | 0.0002 |
| 1.10A | − | 10.0 | >15.6 | 15.0 | 3.8 | 0.004 | 0.0003 |
| VIII (?) | |||||||
| 3B3 | − | − | − | − | − | − | − |
The numbers indicate the concentration of MAb required to reduce infectivity of the indicated virus by 50%. −, neutralization was not detected. >, neutralization was observed, but 50% neutralization was not achieved at the highest concentration tested (typically 10 μg/ml or higher).
All of the 14 RhMabs comprising groups IV, V, VI, and VII had significant neutralizing activity against SIV316 and SIVΔV1V2 (Table 3). Six of these also had detectable activity against SIV-M5, including at least one RhMAb from groups V, VI, and VII. However, the concentration of these antibodies required for effective neutralization of SIV-M5 was typically much higher than for SIVmac316 or SIVΔV1V2, requiring anywhere from 2- to 50,000-fold more RhMab to achieve similar reductions in infectivity (Table 3).
Collectively, competition groups IV, V, VI, and VII define binding determinants found in the C-terminal half of the gp120 linear sequence, including elements of the V3 and V4 loops (14, 62). SIV316 was neutralized by the group II and group III MAbs, which recognize epitopes in the V1 and V2 variable loops, respectively. It is interesting to note that the group II and III RhMAbs did not neutralize any of the other variants with intact V1 and V2 sequences (i.e., SIVmac239, g46, g56, and SIV-M5). Three viruses, SIVΔV1V2, SIVmac316, and SIVmac251lab, were neutralized by RhMAb H31, one of the three RhMAbs comprising competition group I; the other two group I RhMAbs did not detectably neutralize any of the viruses. Likewise, the sole MAb representing group VIII (3B3) did not have detectable neutralizing activity against any of the viral variants tested.
Neutralization by rhesus Fab fragments.
Fab fragments derived from rhesus monkeys experimentally infected with the g45 and g46 variants of SIVmac239 were screened for neutralizing activity against a panel of eight viruses. A total of 14 rhesus-derived Fab fragments were tested. This included Fab 346-4, which has been mapped to an epitope in the V3 loop, and 13 additional Fabs that compete with the KK9 MAb for binding to soluble gp120 and are therefore likely to recognize conformational epitopes involving the V3-V4 region of envelope (data not shown). Fab b6, which recognizes the CD4 binding site of HIV-1 gp120, was included as a negative control. These 14 Fab fragments were obtained independently of the rhesus MAbs described in the previous section.
The pattern of neutralization by the rhesus anti-SIV Fab panel was similar to that observed with the murine and rhesus MAbs. None of the Fab fragments achieved 50% neutralization against SIVmac239. The g45 double-glycosylation mutant was also not detectably neutralized by any of the Fabs. The g46 double-glycosylation mutant was neutralized by two Fabs, the g56 virus by four, and the M5 quintuple-glycosylation mutant by eight. SIVΔV1V2 was neutralized by seven Fabs, at concentrations 1.5- to 20-fold less than those required for neutralization of g46 or g56 (Table 4). Eight Fabs were tested against SIVmac316. The sensitivity of SIVmac316 was similar to that of SIVΔV1V2, with 50% neutralizing concentrations ranging from 0.0007 to 0.4 μg/ml.
TABLE 4.
Neutralizing activity of the rhesus anti-SIV Fab panol
| Fab | Concn of Fab fragment (μg/ml) reducing infectivity by 50%a
|
||||||
|---|---|---|---|---|---|---|---|
| SIV239 | g45 | g46 | g56 | M5 | SIV316 | ΔV1V2 | |
| 346-4 | − | − | − | − | ND | ND | 0.06 |
| 346-16h | − | − | 1.5 | 0.5 | 0.2 | 0.003 | 0.05 |
| 346-19h | − | − | − | 0.6 | 1.3 | 0.02 | 0.4 |
| 346-23h | − | − | − | − | − | ND | − |
| 346-24h | − | − | − | − | 3.4 | 0.4 | − |
| 346-25h | − | − | − | 12.5 | ND | ND | 0.7 |
| 347-1h | − | − | − | − | 6.9 | 0.01 | 0.7 |
| 347-2h | − | − | 10.4 | − | ND | ND | ND |
| 347-5h | − | − | − | − | − | 0.04 | − |
| 347-6h | − | − | − | − | 0.9 | ND | − |
| 347-10h | − | − | − | − | 12.5 | 0.01 | − |
| 347-19h | − | − | − | − | 33.6 | ND | 4.7 |
| 347-23h | − | − | − | 2.0 | 0.7 | 0.0007 | 0.1 |
| B6 | − | − | − | − | − | − | − |
The numbers indicate the concentration of Fab fragment required to reduce infectivity of the indicated virus by 50%. −, 50% neutralization was not achieved even at the highest concentration tested. ND, not determined.
Neutralization by an anti-gp41 MAb.
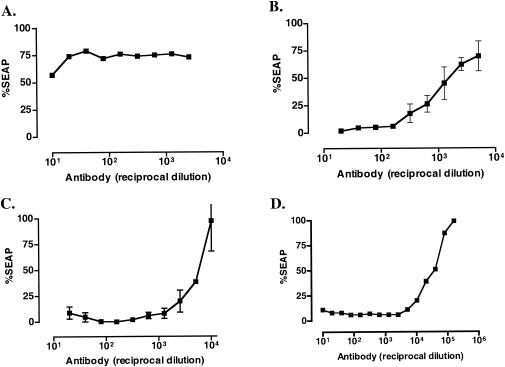
The murine MAbs KK15 and KK41 have been previously shown to react with peptides derived from the N-terminal helical region of the SIV TM protein gp41, and both antibodies recognize gp41 in Western blots (36). Neither antibody neutralized SIVmac239 (Table 1), consistent with previous reports (36). However, KK41 had significant neutralizing activity against all five SIVmac239 variants tested in this report (Table 1 and Fig. 4). SIV-M5, SIVmac316, and SIVΔV1V2 were particularly sensitive to KK41-mediated neutralization, with 50% neutralization achieved at dilutions on the order of 10−3 to 10−4 (Fig. 4). MAb KK15 binds to an epitope that closely overlaps the epitope recognized by KK41 (36). However, KK15 did not neutralize any of the variants tested. KK15 also reacted weakly with whole-lysed virions by ELISA and did not react at all with SIVmac239 gp41 in Western blots (data not shown).
FIG. 4.
Neutralization of SIVmac239 (A) and derivatives SIV-M5 (B), SIVmac316 (C), and SIVmac239ΔV1V2 (D) by the murine anti-gp41 MAb KK41.
RhMAbs 1.7H, 4.9C, and 3.8F are thought to recognize determinants in gp41 based on competition mapping, but do not react in Western blots (J. Robinson, unpublished observations) and fail to bind whole-lysed virions of SIVmac239 or recombinant envelope protein when measured by ELISA (data not shown). None of these three antibodies had detectable neutralizing activity against SIVmac239 or any of the viral variants tested.
DISCUSSION
A collection of genetically defined derivatives was used to investigate the neutralization-resistant phenotype of SIVmac239, a pathogenic molecular clone of SIV with the properties of a primary isolate. The viral variants included in the panel differed from one another and the parental virus only at specific sites in the envelope gene, allowing alterations in neutralization profile to be attributed unambiguously to specific changes in amino acid sequence. The sensitivity of each variant to neutralization by SIV-positive animal plasma and a panel of 51 MAbs was determined by a quantitative neutralization assay. Although the specificities of the 51 MAbs were not all precisely defined, collectively they represented at a minimum 10 distinct epitope competition groups throughout the SIV envelope proteins (14, 35, 36, 62). These included antibodies with specificity for the ectodomain of gp41; the N terminus, V1, V2, and V3 loops of gp120; and multiple conformational epitopes involving the V3-V4 region of gp120.
The cumulative results of pairwise neutralization assays indicated that three very different mutational patterns in the SIV envelope gene independently gave rise to similar increases in breadth of neutralization sensitivity. SIVmac316, with eight amino acid replacements throughout env; SIV-M5, which lacks five N-glycan attachment sites; and SIVΔV1V2, from which the entire V1-V2 region has been removed, were all sensitive to neutralization by antibodies from multiple competition groups. SIVmac316 and SIVΔV1V2 in particular displayed significant increases in both breadth and degree of sensitivity compared to the parental SIVmac239 strain from which they were derived. Similar increases in sensitivity have been observed for HIV-1 strains engineered to contain deletions of the V1 and V2 loops (9, 65, 69).
Strikingly, not one of the 37 MAbs or 14 rhesus Fabs tested had significant neutralizing activity against SIVmac239. However, in most cases, neutralization was detected against at least two or more viruses derived from SIVmac239. Since these neutralization-sensitive variants only differ from SIVmac239 (and from one another) at a few defined sites in the envelope gene, there is a very high probability that the primary amino acid sequences that form the antibody binding sites are also present in the parental SIVmac239 env gene. Therefore, for most of these antibodies, it is unlikely that the lack of detectable neutralizing activity is due to sequence divergence between SIVmac239 and the immunogen or immunogens against which the antibodies were initially raised.
Why then didn't these antibodies neutralize the parental SIVmac239 virus? One reasonable hypothesis is that the relevant epitopes are simply not accessible to antibody in the context of the envelope complex on SIVmac239 virions, whereas the mutations that define the derivative strains induced alterations in the envelope complex that resulted in increased epitope exposure (49). A body of evidence now exists to suggest that the envelope complex of primary HIV-1 isolates assumes a closed conformation that is not readily accessible to antibodies (42, 48, 56, 70) and that even small perturbations of this conformation can shift the complex to favor a more open, neutralization-sensitive form (40). The g45, g46, g56, and SIVmac316 envelope proteins have previously been shown to have both decreased dependence on CD4 and increased sensitivity to antibody-mediated neutralization in cell-cell fusion assays (57). This observation and the results we describe here are consistent with the hypothesis that mutations that confer neutralization sensitivity result in conformational alterations in the viral envelope proteins.
Although the hypothesis that there are global conformational changes in the mutant envelope spikes is an attractive explanation for the broad increases in neutralization sensitivity seen here, it is also possible that the increased sensitivity of the mutants is due, at least in part, to a decrease in the number of functional envelope spikes on the surface of virions. It has been proposed that neutralization results when antibody binding reduces the number of functional envelope spikes below the minimum required for infectivity (38, 54). If this is the case, then mutations in env that cause a decrease in the average number of functional spikes per virion could contribute to an increase in neutralization sensitivity. Such decreases in the number of functional spikes could take the form of a physical decrease in the overall number of incorporated spikes or could result from an effective decrease in the number of functional spikes due to an increase in the ratio of nonfunctional to functional spikes.
The ease with which a virus gains entry into a target cell may also be expected to influence its relative sensitivity to neutralization by antibodies. This could in turn be influenced by affinity of the virus for receptor and coreceptor, or by the kinetics of the fusion and entry process (21). For example, it has been reported that CD4-independent strains of SIV such as SIVmac316 are easier to neutralize in the absence of CD4 than when both CD4 and a coreceptor are available (57). One interpretation of this result is that interaction with CD4 effectively improves the probability that the virus will engage a coreceptor and begin the entry process before being bound by a neutralizing antibody (57). It has also been shown that lower concentrations of several antibodies were required to neutralize SIV when cells expressing a suboptimal coreceptor for SIV were used as target cells (23, 24). Subsequent to receptor and coreceptor engagement, the virus and cell membranes must fuse to permit release of the viral nucleocapsid into the cytoplasm of the target cell. As a result of structural rearrangements that take place during the fusion process, epitopes that are normally occluded within the envelope oligomer may be transiently exposed to antibodies (22, 70). If so, one could imagine that viruses that are more efficient at completing this process may be proportionally less sensitive to neutralization (21, 23, 57). It will be critical to differentiate the contributions of entry efficiency, spike density, and epitope accessibility before the underlying mechanisms of neutralization resistance of the parental virus can be fully appreciated.
The mutations that define the g45, g46, g56, SIV-M5, and SIVΔV1V2 variants used in this study are located in the N-terminal half of the gp120 linear sequence and are concentrated in the V1-V2 region (Fig. 1). Moreover, four of the eight substitutions in SIVmac316 are also located in the N-terminal half of gp120. It is interesting therefore that the recognition sites of a majority of the antibodies with strong neutralizing activity against these strains were mapped to the C-terminal half of gp120, particularly to determinants in the V3-V4 region (Table 3). This raises the possibility that the two distinct regions of gp120, the V1-V2 domain in the N-terminal half and the region encompassing V3 throughV4 in the C-terminal half, may be in close proximity and that elements of these two regions could potentially interact with one another. In this scenario, the substitutions in V1-V2 or the deletion of V1-V2 either results in unmasking of epitopes in V3-V4 or otherwise disrupt interactions between distinct regions of gp120 leading to exposure of epitopes that lie in the interface between the two domains. These interactions could be intramolecular, involving elements of the N-terminal and C-terminal halves within the same gp120 protomer, but it is important to note that the results are also consistent with a model in which the V1V2 region of one gp120 molecule is juxtaposed with the V3-V4 region of an adjacent gp120 molecule within the context of a fully assembled envelope trimer (41). This model is also consistent with the observation that RhMAbs directed against V1-V2 and RhMAbs directed against V3-V4 don't compete with one another for binding to monomeric gp120 (which indicates that the V1-V2 loops don't mask V3-V4 epitopes in the context of the monomer) (14, 62).
It is interesting that all of the variants of SIVmac239 described here, most of which have mutations confined to the gp120 subunit of Env, were also sensitive to neutralization by the MAb KK41 (Table 1 and Fig. 4). Most epitopes of HIV-1 gp41 that have been mapped can be sorted into two groups, referred to as cluster I and cluster II, depending on the region of gp41 in which they are found. KK41 is specific for an epitope that has been mapped to the “leucine-zipper” region in the N-terminal portion of the gp41 ectodomain (36) and is therefore analogous to an HIV cluster I antibody. Cluster II epitopes are located in the C-terminal, heptad repeat region of the HIV-1 gp41 ectodomain. Although anti-gp41 antibodies are abundant in HIV-infected patients, these antibodies typically have little or no neutralizing activity. In fact, it is thought that much of the gp41 ectodomain is occluded by interactions with gp120 or adjacent gp41 molecules (63). The only anti-gp41 antibodies with significant, broad neutralizing activity against HIV-1 bind to a region of gp41 adjacent to the membrane-spanning domain (51, 71). These include the well-characterized anti-gp41 MAb 2F5 (51) and at least two additional antibodies with similar specificity (71).
We previously reported that changes in gp41 acquired during passage of SIVΔV1V2 in culture were able to compensate for decreased infectivity engendered by the original, 100-amino-acid deletion of V1-V2 (34). This observation, along with our results showing that a variety of mutations in gp120 (including removal of N-glycans or deletion of V1-V2) gave rise to increased sensitivity to KK41, raises the possibility that the N-terminal half of SIV gp120 interacts with the ectodomain of gp41. These observations do not indicate whether this interaction exists in the native envelope spike or occurs transiently at some point during the entry process. For example, these mutations may weaken the association between subunits of the envelope complex, leading to increased accessibility to epitopes that are otherwise occluded in the parental envelope spike. Alternatively, these mutations could increase the half-life of an intermediate in the entry process, during which elements of the fusion apparatus are more exposed to binding by anti-gp41 antibodies. Recently published results of a comparison of matched, neutralization-resistant and neutralization-sensitive variants of the HIV-1 MN strain were similarly suggestive of a role for the N-terminal half of HIV-1 gp120 in interacting with the ectodomain of gp41 (43). Altogether, these observations are indicative of a direct or indirect influence of gp41 sequences on the conformation and function of the gp120 subunits.
Levels of resistance to MAbs similar to those we describe here have also been observed in cell-cell fusion assays and neutralization assays using virions pseudotyped with the SIVmac239 envelope complex (23, 57). The results of neutralization assays with MAbs, Fab fragments and SIV-positive plasma described here indicate that the unaltered SIVmac239 virion is inherently resistant to neutralization by antienvelope antibodies. Unlike the situation with HIV-1, individual MAbs with potent neutralizing activity against SIVmac239 have not yet been identified. Nonetheless, a number of observations suggest that SIVmac239 and primary isolates of HIV-1 utilize similar mechanisms to achieve the neutralization-resistant state. Antigenic escape from neutralizing antibody responses has been well documented for SIVmac239 (6, 7), and patterns of sequence variation associated with escape map to variable regions in envelope analogous to variable regions in the HIV-1 envelope (8). As with primary isolates of HIV-1, SIVmac239 can be readily neutralized by soluble CD4 (sCD4) (47, 64), and pretreatment of SIVmac239 with subneutralizing concentrations of sCD4 results in exposure of the coreceptor binding site and increased sensitivity to neutralization by MAbs (57). Concentrations of sCD4 that neutralize infectivity of SIVmac239 by 50%, approximately 280 ng/ml (47), are similar to or even less than the concentrations needed to neutralize primary isolates of HIV-1 (1, 66). We have also frequently observed neutralization of SIVmac239 by individual or pooled autologous serum samples at low dilutions (1:20 to 1:100). Among primary isolates of HIV-1, similar levels of resistance to neutralization by sera have also been observed (48, 53, 61), indicating that the degree of resistance displayed by SIVmac239 is not unique or aberrant. Finally, the arrangement of conserved and variable regions, positioning of N-glycans and disulfide bonds, use of CD4 and CCR5 as receptors, and overall organization of the HIV and SIV envelope trimers are very similar. Based on these observations, it seems likely that the underlying mechanisms of neutralization resistance for SIVmac239 and primary isolates of HIV-1 are similar.
To better understand the contribution of envelope structure to the neutralization-resistant phenotype, it will be important to precisely define the features of PI envelope complexes that give rise to the resistant state. Because of its highly neutralization-resistant phenotype, SIVmac239 is an ideal reagent for molecular and biochemical exploration of the factors that confer neutralization-resistance. This work and published studies on both HIV-1 and SIV are beginning to reveal what some of these factors may be. Better understanding of the neutralization-resistant phenotype will hopefully instruct efforts to design Env-containing vaccines with the ability to elicit antibodies with broad and potent neutralizing activity.
Acknowledgments
We thank Melissa Laird and Sue Czajak for assistance with neutralization assays, Dawn Slifka for assistance with Fab fragment isolation, and Eloisa Yuste for helpful discussions.
W.E.J. was supported by an Elizabeth Glaser Pediatric AIDS Foundation Scholar Award. This work was also supported by an NIH grant to the New England Primate Research Center (RR00168) and NIH grants AI35365 and AI50421 (R.C.D.), AI28243 (J.E.R.), AI44293 (P.W.H.I.P.), and AI33292 (D.R.B.).
REFERENCES
- 1.Arthos, J., C. Cicala, T. D. Steenbeke, T. W. Chun, C. Dela Cruz, D. B. Hanback, P. Khazanie, D. Nam, P. Schuck, S. M. Selig, D. Van Ryk, M. A. Chaikin, and A. S. Fauci. 2002. Biochemical and biological characterization of a dodecameric CD4-Ig fusion protein: implications for therapeutic and vaccine strategies. J. Biol. Chem. 277:11456-11464. [DOI] [PubMed] [Google Scholar]
- 2.Barbas, C. F., D. R. Burton, J. K. Scott, and G. J. Silverman. 2001. Phage display: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Barre-Sinoussi, F., J. C. Chermann, F. Rey, M. T. Nugeyre, S. Chamaret, J. Gruest, C. Dauguet, C. Axler-Blin, F. Vezinet-Brun, C. Rouzioux, W. Rozenbaum, and L. Montagnier. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868-871. [DOI] [PubMed] [Google Scholar]
- 4.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 5.Blankson, J. N., D. Persaud, and R. F. Siliciano. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 53:557-593. [DOI] [PubMed] [Google Scholar]
- 6.Burns, D. P. W., C. Collignon, and R. C. Desrosiers. 1993. Simian immunodeficiency virus mutants resistant to serum neutralization arise during persistent infection of rhesus monkeys. J. Virol. 67:4104-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns, D. P. W., and R. C. Desrosiers. 1994. Envelope sequence variation, neutralizing antibodies, and primate lentivirus persistence. Curr. Top. Microbiol. Immunol. 188:185-219. [DOI] [PubMed] [Google Scholar]
- 8.Burns, D. P. W., and R. C. Desrosiers. 1991. Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J. Virol. 65:1843-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao, J., N. Sullivan, E. Desjardin, C. Parolin, J. Robinson, R. Wyatt, and J. Sodroski. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71:9808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chackerian, B., L. M. Rudensey, and J. Overbaugh. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 71:7719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 12.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, G. B., R. T. Gandhi, D. M. Davis, O. Mandelboim, B. K. Chen, J. L. Strominger, and D. Baltimore. 1999. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10:661-671. [DOI] [PubMed] [Google Scholar]
- 14.Cole, K. S., M. Alvarez, D. H. Elliott, H. Lam, E. Martin, T. Chau, K. Micken, J. L. Rowles, J. E. Clements, M. Murphey-Corb, R. C. Montelaro, and J. E. Robinson. 2001. Characterization of neutralization epitopes of simian immunodeficiency virus (SIV) recognized by rhesus monoclonal antibodies derived from monkeys infected with an attenuated SIV strain. Virology 290:59-73. [DOI] [PubMed] [Google Scholar]
- 15.Collins, K. L., and D. Baltimore. 1999. HIV's evasion of the cellular immune response. Immunol. Rev. 168:65-74. [DOI] [PubMed] [Google Scholar]
- 16.Collins, K. L., B. K. Chen, S. A. Kalams, B. D. Walker, and D. Baltimore. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397-401. [DOI] [PubMed] [Google Scholar]
- 17.Daniel, M. D., N. L. Letvin, N. W. King, M. Kannagi, P. K. Sehgal, R. D. Hunt, P. J. Kanki, M. Essex, and R. C. Desrosiers. 1985. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science 228:1201-1204. [DOI] [PubMed] [Google Scholar]
- 18.Daniel, M. D., N. L. Letvin, P. K. Sehgal, G. Hunsmann, D. K. Schmidt, N. W. King, and R. C. Desrosiers. 1987. Long-term persistent infection of macaque monkeys with the simian immunodeficiency virus. J. Gen. Virol. 68:3183-3189. [DOI] [PubMed] [Google Scholar]
- 19.Desrosiers, R. C. 1990. The simian immunodeficiency viruses. Annu. Rev. Immunol. 8:557-578. [DOI] [PubMed] [Google Scholar]
- 20.Ditzel, H. J., P. W. Parren, J. M. Binley, J. Sodroski, J. P. Moore, C. F. Barbas III, and D. R. Burton. 1997. Mapping the protein surface of human immunodeficiency virus type 1 gp120 using human monoclonal antibodies from phage display libraries. J. Mol. Biol. 267:684-695. [DOI] [PubMed] [Google Scholar]
- 21.Doms, R. W. 2000. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology 276:229-237. [DOI] [PubMed] [Google Scholar]
- 22.Doms, R. W., and J. P. Moore. 2000. HIV-1 membrane fusion: targets of opportunity. J. Cell Biol. 151:F9-F14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edinger, A. L., M. Ahuja, T. Sung, K. C. Baxter, B. Haggarty, R. W. Doms, and J. A. Hoxie. 2000. Characterization and epitope mapping of neutralizing monoclonal antibodies produced by immunization with oligomeric simian immunodeficiency virus envelope protein. J. Virol. 74:7922-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edinger, A. L., T. L. Hoffman, M. Sharron, B. Lee, B. O'Dowd, and R. W. Doms. 1998. Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology 249:367-378. [DOI] [PubMed] [Google Scholar]
- 25.Gallo, R. C., S. Z. Salahuddin, M. Popovic, G. M. Shearer, M. Kaplan, B. F. Haynes, T. J. Palker, R. Redfield, J. Oleske, B. Safai et al. 1984. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 224:500-503. [DOI] [PubMed] [Google Scholar]
- 26.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 27.Gao, F., L. Yue, A. T. White, P. G. Pappas, J. Barchue, A. P. Hanson, B. M. Greene, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 1992. Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature 358:495-499. [DOI] [PubMed] [Google Scholar]
- 28.Glamann, J., D. R. Burton, P. W. H. I. Parren, H. J. Ditzel, K. A. Kent, C. Arnold, D. Montefiori, and V. M. Hirsch. 1998. Simian immunodeficiency virus (SIV) envelope-specific Fabs with high-level homologous neutralizing activity: recovery from a long-term-nonprogressor SIV-infected macaque. J. Virol. 72:585-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill, C. M., H. Deng, D. Unutmaz, V. N. Kewalramani, L. Bastiani, M. K. Gorny, S. Zolla-Pazner, and D. R. Littman. 1997. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J. Virol. 71:6296-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 31.Javaherian, K., A. J. Langlois, D. C. Montefiori, K. A. Kent, K. A. Ryan, P. D. Wyman, J. Stott, D. P. Bolognesi, M. Murphey-Corb, and G. J. Larosa. 1994. Studies of the conformation-dependent neutralizing epitopes of simian immunodeficiency virus envelope protein. J. Virol. 68:2624-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, W. E., and R. C. Desrosiers. 2002. Viral persistence: HIV's strategies of immune system evasion. Annu. Rev. Med. 53:499-518. [DOI] [PubMed] [Google Scholar]
- 33.Johnson, W. E., J. D. Lifson, S. M. Lang, R. P. Johnson, and R. C. Desrosiers. 2003. Importance of B-cell responses for immunological control of variant strains of simian immunodeficiency virus. J. Virol. 77:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson, W. E., J. Morgan, J. Reitter, B. A. Puffer, S. Czajak, R. W. Doms, and R. C. Desrosiers. 2002. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J. Virol. 76:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kent, K. A., L. Gritz, G. Stallard, M. P. Cranage, C. Collignon, C. Thiriart, T. Corcoran, P. Silvera, and E. J. Stott. 1991. Production of monoclonal antibodies to simian immunodeficiency virus envelope glycoproteins. AIDS 5:829-836. [DOI] [PubMed] [Google Scholar]
- 36.Kent, K. A., E. Rud, T. Corcoran, C. Powell, C. Thiriart, C. Collignon, and E. J. Stott. 1992. Identification of two neutralizing and 8 non-neutralizing epitopes on simian immunodeficiency virus envelope using monoclonal antibodies. AIDS Res. Hum. Retrovir. 8:1147-1151. [DOI] [PubMed] [Google Scholar]
- 37.Kestler, H. W., III, Y. N. Naidu, T. Kodama, N. W. King, M. D. Daniel, Y. Li, and R. C. Desrosiers. 1989. Use of infectious molecular clones of simian immunodeficiency virus for pathogenesis studies. J. Med. Primatol. 18:305-309. [PubMed] [Google Scholar]
- 38.Klasse, P. J., and J. P. Moore. 1996. Quantitative model of antibody- and soluble CD4-mediated neutralization of primary isolates and T-cell line-adapted strains of human immunodeficiency virus type 1. J. Virol. 70:3668-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolchinsky, P., E. Kiprilov, and J. Sodroski. 2001. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J. Virol. 75:2041-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 41.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Labrijn, A. F., and P. W. H. I. Parren. 1999. Neutralizing epitopes of HIV-1, p. 13-34. In B. Korber, C. Brander, B. F. Haynes, J. P. Moore, R. Koup, B. Walker, and D. I. Watkins (ed.), HIV Molecular Immunology database. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 43.Leavitt, M., E. J. Park, I. A. Sidorov, D. S. Dimitrov, and G. V. Quinnan, Jr. 2003. Concordant modulation of neutralization resistance and high infectivity of the primary human immunodeficiency virus type 1 MN strain and definition of a potential gp41 binding site in gp120. J. Virol. 77:560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Letvin, N. L., M. D. Daniel, P. K. Sehgal, R. C. Desrosiers, R. D. Hunt, L. M. Waldron, J. J. MacKey, D. K. Schmidt, L. V. Chalifoux, and N. W. King. 1985. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science 230:71-73. [DOI] [PubMed] [Google Scholar]
- 45.Malashkevich, V. N., D. C. Chan, C. T. Chutkowski, and P. S. Kim. 1998. Crystal structure of the simian immunodeficiency virus (SIV) gp41 core: conserved helical interactions underlie the broad inhibitory activity of gp41 peptides. Proc. Natl. Acad. Sci. USA 95:9134-9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Means, R. E., T. Greenough, and R. C. Desrosiers. 1997. Neutralization sensitivity of cell culture-passaged simian immunodeficiency virus. J. Virol. 71:7895-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Means, R. E., T. Matthews, J. A. Hoxie, M. H. Malim, T. Kodama, and R. C. Desrosiers. 2001. Ability of the V3 loop of simian immunodeficiency virus to serve as a target for antibody-mediated neutralization: correlation of neutralization sensitivity, growth in macrophages, and decreased dependence on CD4. J. Virol. 75:3903-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas III, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore, J. P., and D. D. Ho. 1995. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS 9:S117-S136. [PubMed] [Google Scholar]
- 50.Mori, K., D. J. Ringler, T. Kodama, and R. C. Desrosiers. 1992. Complex determinants of macrophage tropism in env of simian immunodeficiency virus. J. Virol. 66:2067-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pantaleo, G., C. Graziosi, J. F. Demarest, L. Butini, M. Montroni, C. H. Fox, J. M. Orenstein, D. P. Kotler, and A. S. Fauci. 1993. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature 362:355-358. [DOI] [PubMed] [Google Scholar]
- 53.Parren, P. W. H. I., M. Wang, A. Trkola, J. M. Binley, M. Purtscher, H. Katinger, J. P. Moore, and D. R. Burton. 1998. Antibody neutralization-resistant primary isolates of human immunodeficiency virus type 1. J. Virol. 72:10270-10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parren, P. W. H. I., and D. R. Burton. 2001. The antiviral activity of antibodies in vitro and in vivo. Adv. Immunol. 77:195-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parren, P. W. H. I., D. R. Burton, and Q. J. Sattentau. 1997. HIV-1 antibody—debris or virion? Nat. Med. 3:366-367. [DOI] [PubMed] [Google Scholar]
- 56.Poignard, P., E. O. Saphire, P. W. Parren, and D. R. Burton. 2001. gp120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 57.Puffer, B. A., S. Pöhlmann, A. L. Edinger, D. Carlin, M. D. Sanchez, J. Reitter, D. D. Watry, H. S. Fox, R. C. Desrosiers, and R. W. Doms. 2002. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J. Virol. 76:2595-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Regier, D. A., and R. C. Desrosiers. 1990. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res. Hum. Retrovir. 6:1221-1231. [DOI] [PubMed] [Google Scholar]
- 59.Reitter, J. N., and R. C. Desrosiers. 1998. Identification of replication-competent strains of simian immunodeficiency virus lacking multiple attachment sites for N-linked carbohydrates in variable regions 1 and 2 of the surface envelope protein. J. Virol. 72:5399-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 61.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson, J. E., K. S. Cole, D. H. Elliott, H. Lam, A. M. Amedee, R. Means, R. C. Desrosiers, J. Clements, R. C. Montelaro, and M. Murphey-Corb. 1998. Production and characterization of SIV envelope-specific rhesus monoclonal antibodies from a macaque asymptomatically infected with a live SIV vaccine. AIDS Res. Hum. Retrovir. 14:1253-1262. [DOI] [PubMed] [Google Scholar]
- 63.Sattentau, Q. J., S. Zolla-Pazner, and P. Poignard. 1995. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology 206:713-717. [DOI] [PubMed] [Google Scholar]
- 64.Schenten, D., L. Marcon, G. B. Karlsson, C. Parolin, T. Kodama, N. Gerard, and J. Sodroski. 1999. Effects of soluble CD4 on simian immunodeficiency virus infection of CD4-positive and CD4-negative cells. J. Virol. 73:5373-5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stamatatos, L., and C. Cheng-Mayer. 1998. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J. Virol. 72:7840-7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 67.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 68.Wyand, M. S., K. H. Manson, M. Garcia-Moll, D. Montefiori, and R. C. Desrosiers. 1996. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J. Virol. 70:3724-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 71.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. H. I. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]