Abstract
The three genomic and a single subgenomic RNA of brome mosaic virus (BMV), an RNA virus infecting plants, are packaged by a single-coat protein (CP) into three morphologically indistinguishable icosahedral virions with T = 3 quasi-symmetry. Genomic RNAs 1 and 2 are packaged individually into separate particles whereas genomic RNA3 and subgenomic RNA4 (coat protein mRNA) are copackaged into a single particle. We report here that packaging of dicistronic RNA3 requires a bipartite signal. A highly conserved 3′ tRNA-like structure postulated to function as a nucleating element (NE) for CP subunits (Y. G. Choi, T. W. Dreher, and A. L. N. Rao, Proc. Natl. Acad. Sci. USA 99:655-660, 2002) and a cis-acting, position-dependent packaging element (PE) of 187 nt present in the nonstructural movement protein gene are the integral components of the packaging core. Efficient incorporation into BMV virions of nonviral RNA chimeras containing NE and the PE provides confirmatory evidence that these two elements are sufficient to direct packaging. Analysis of virion RNA profiles obtained from barley protoplasts transfected with a RNA3 variant lacking the PE provides the first genetic evidence that de novo synthesized RNA4 is incompetent for autonomous assembly whereas prior packaging of RNA3 is a prerequisite for RNA4 to copackage.
Encapsidation of the genome by structural components (including the envelope) leading to the assembly of infectious progeny virions is an essential step in the life cycle of RNA viruses. Based on the configuration, the genome of a given RNA virus can be either nonsegmented or segmented. The genomes of nonsegmented polio and retroviruses are encapsidated into a single virion. Likewise, the genomes of bacteriophage φ6 and reoviridae are also encapsidated into a single virion despite being segmented among 3 and 10 to 12 double-stranded RNAs (dsRNAs), respectively (17, 18). By contrast to these viral systems, the segmented genomes of bromoviruses, a group of RNA viruses pathogenic to plants, are encapsidated into three separate virions of identical size and morphology. The fact that cellular RNAs are rarely found in the progeny virions of each of the above mentioned viral systems (with the exception of retroviruses) suggest that viral genomic RNAs are selectively recognized by viral coat protein during encapsidation. This recognition process is achieved through a specific interaction between the viral protein components and some sort of packaging signal within the nucleic acids (1).
In eukaryotic RNA viruses, the most thoroughly characterized packaging signal is that of tobacco mosaic virus, a helical plant RNA virus (30) in which the specific interaction between the CP and a 69-nucleotide (nt) region internal to the RNA genome leads to the specificity of virion assembly. Among viruses with icosahedral symmetry, RNA elements that act as specific packaging signals have been characterized for enveloped animal viruses such as coronaviruses (8) and alphaviruses (11, 28), human and murine retroviruses (1) and for a nonenveloped plant virus, turnip crinkle virus (TCV) (19, 25). Since the genomes of these RNA viruses are not segmented, their packaging into virions is considered to be straight forward. However, for segmented viruses there is the additional problem of ensuring that all genome segments are present in each particle or in a limited number of particles. Although specific packaging signals distributed on each of the 10 to 12 dsRNA segments of Reoviridae are not known, their packaging into a single particle is uniform and stoichiometric (1, 17). Similarly, the three dsRNAs of bacteriophage φ6 are copackaged precisely and sequentially into a single particle (15).
Thus, for understanding the mechanism of RNA packaging in segmented viruses, the multicomponent brome mosaic virus (BMV), a prototype of the plant virus family Bromoviridae (20) is an ideal model system since dissociated coat protein and RNA can be reassembled in vitro. Using this in vitro assembly approach, we have recently identified an additional role for a sequence of 201 nucleotides at the 3′end, which is essentially identical for all four virion RNAs and assumes a tRNA-like structure (TLS). When incubated with dissociated coat protein subunits, BMV genomic and subgenomic RNAs lacking the 3′ TLS failed to assemble into virions and this defective assembly was restored by the addition of 201-nt sequence encompassing the TLS to the assembly mixture (4). It was also observed that tRNAs of wheat germ and yeast supplied in trans were similarly active in promoting the assembly of truncated BMV RNAs into virions. Interestingly virions assembled from truncated BMV RNAs in the presence of tRNAs or TLS-containing short sequences did not incorporate the later molecules (4). Based on these observations, it was hypothesized that the highly conserved 3′ TLS, serves as a chaperone, in a transient association with virion CP, functioning as nucleating element (NE) to initiate the assembly of viral RNA into BMV virions (4). However, if viral TLS or tRNAs are the only elements that interact with BMV CP and promote packaging, how do we reconcile for the specific distribution of four BMV RNAs into three morphologically indistinguishable virions? The fact that BMV CP exhibits a high degree of specificity in vitro and in vivo to selectively encapsidate viral RNAs (3, 5) suggest that, in addition to viral TLS, the CP interacts with selective packaging signals located elsewhere on each packaged BMV RNA. In this study, using a well established in vitro assay for assembling infectious BMV virions, we identified that a 187-nt sequence from the nonstructural movement protein as being the distinct RNA element required for specific packaging of BMV RNA3. Consequently this report provides the first evidence for the existence of a bipartite packaging signal in an RNA virus with icosahedral symmetry.
MATERIALS AND METHODS
Plasmids.
All BMV RNA3 (B3) variants constructed in this study (Fig. 1 to 4) are derived from a plasmid pT7B3 which contains a cDNA clone of full-length B3 (6). For engineering a desired set of deletions spanning the entire B3 sequence, the cDNA clone was subjected to either PCR directed mutagenesis or sequential digestion with specified restriction enzymes (4) (Fig. 1). Mutant B3 transcripts derived from these clones were: B3/Δ5′ nontranslated region (B3/Δ5′NTR; bases 1 to 92 deleted); B3/ΔMP (bases 92 to 1007 deleted); B3/Δintercistronic region (B3/ΔICR; bases 1007 to 1253 deleted); B3/ΔCP (bases 1253 to 1819 deleted); B3/Δ3′ NTR (bases 1819 to 1917 deleted) and B3/ΔTLS (bases 1917 to 2117 deleted). Variant clone B3/Xe was constructed by precisely replacing the MP-coding region with a similarly sized fragment of Xenopus laevis elongation factor gene sequence amplified in a PCR from pTRI-xef (Ambion). The construction of four additional variant clones of B3/ΔMP as follows: (i) B3/ΔMP-BC was constructed by deleting 502 nt present between BamHI (located at position 92) and ClaI (located at position 601); (ii) B3/ΔMP-CBg was constructed by deleting 406 nt present between ClaI and BglII (located at position 1007); (iii) B3/ΔMP-ΔCBg.1 was constructed by deleting 291 nt located between ClaI and PflMI (located at position 817); (iv) B3/ΔMP-ΔCBg.2 was constructed by deleting 190 nt present between PflMI and BglII.
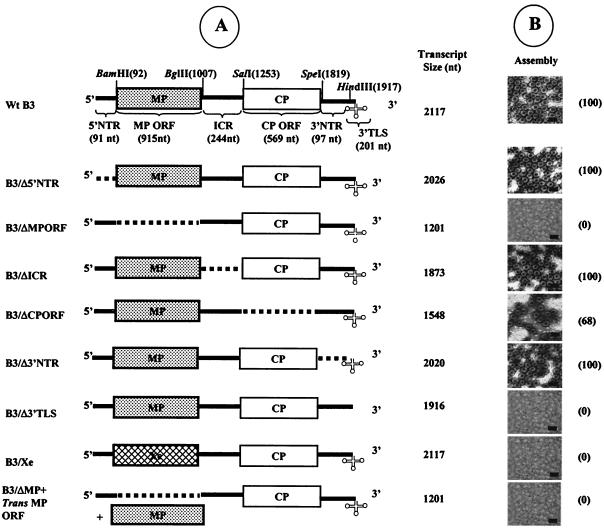
FIG. 1.
Schematic representation of wt and deletion variants of B3 used for in vitro assembly assays. (A) The genome organization of wt B3 is shown, with NTR represented as solid lines and the stippled and open boxes represent the MP-coding region and the CP-coding regions, respectively. The clover leaf structure at the 3′ end represents the highly conserved tRNA-like structure (3′TLS). Restriction sites used for constructing variants of the full-length cDNA clone of B3 are shown above the map. The lengths of wt B3 and its variant transcripts are shown. Broken lines in each variant clone represent the extent of each deletion. (B) In vitro assembly assays. Electron micrographs display the negatively stained preparations of virions assembled in vitro from purified CP subunits and noncapped RNA transcripts of either wt B3 or its variant. The numbers shown in parentheses next to each micrograph represent the percentage of assembly efficiency for each variant RNA sequence with respect to wt control transcripts (scale bar = 50 nm).
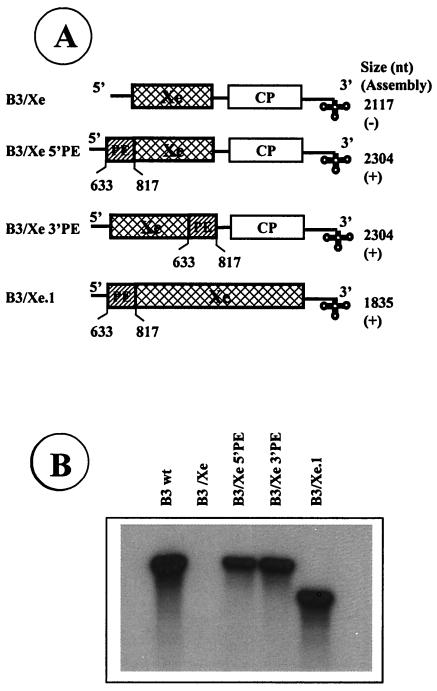
FIG. 4.
Assembly of heterologous RNA chimeras into BMV virions. Schematic representation of chimeras used to demonstrate that PE and 3′ TLS are sufficient to direct RNA packaging into BMV virions. (A) The structure of packaging incompetent B3/Xe is shown. In B3/Xe 5′PE and B3/Xe 3′PE, a 187-nt fragment encompassing the PE was, respectively, fused to either 5′ or 3′ end of the Xenopus sequence present in the assembly defective B3/Xe. As depicted, B3/Xe.1 was constructed such that the Xenopus sequence was flanked by regions encompassing PE and NE. The lengths of variant transcripts and results of in vitro assembly assays are shown. Symbols: +, detection of wt level virion reassembly; −, absence of virion assembly. (B) Analysis of RNA chimeras assembled into BMV virions. Autoradiograph of an agarose gel showing 32P labeled RNA chimera recovered from RNase resistant virions assembled in vitro.
Variant clones ΔA, ΔB, ΔC, ΔAB, ΔAC, and ΔBC were constructed by precisely deleting desired sequences encompassing each stem-loop structure (Fig. 2B) using PCR. Thus, in ΔA, nt 601 to 632 deleted; in ΔB nt 634 to 734 deleted, ΔC nt 754 to 819 deleted, in ΔAB nt 601 to 734 deleted, in ΔAC, nt 601 to 632 and 754 to 819 deleted and in ΔBC, nt 634 to 820 deleted. Other variant clones of B3 such as B3/2MP, B3/2CP and B3/Rev (Fig. 3A) were constructed by amplifying the fragments of each coding region flanked by the desired restriction sites in a PCR, followed by subcloning into the desired location of pT7B3. A set of six B3 deletion variants spanning the region between the packaging element (PE) and the 3′ TLS was constructed by sequential digestion with specified restriction enzymes and mung bean nuclease. Variant B3 transcripts derived from these clones were: B3/ΔPS (bases 817 to 1253 deleted), B3/ΔPX (bases 817 to 1760 deleted), B3/ΔPH (bases 817 to 1917 deleted) and B3/ΔPK (bases 817 to 2075 deleted). Two variant clones of B3/Xe (Fig. 1A), B3/Xe5′PE and B3/Xe3′PE were constructed by fusing the packaging element (PE), respectively, to either 5′ or 3′ end of the Xenopus sequence present in B3/Xe (Fig. 1A). To construct B3/Xe.1, plasmid B3/ΔMP-BC was digested simultaneously with PflMI and HindIII and blunt-ended with mung bean nuclease to yield B3/ΔPH. A 1,327-nt cDNA fragment of Xenopus RNA obtained from pTRI-xef was blunt ended and ligated into B3/ΔPH. For the synthesis of riboprobes specific for B3, a plasmid pT73BMP was constructed by cloning a BamHI-EcoRI fragment encompassing the entire MP-coding region from pT7B3 into a pT7/T3-18U vector. For constructing a B3 variant lacking the PE, a ClaI-digested pT7B3 was treated with Bal31enzyme (23). After screening several clones, a plasmid referred to as B3/ΔPE characterized by lacking 597 nt (located between positions 421 and 1028) was selected. The nucleotide sequences of all variant clones were determined to verify the presence of expected mutations. Throughout these studies noncapped RNA transcripts (sometimes 32P labeled) were synthesized in vitro using T7 RNA polymerase (4, 6).
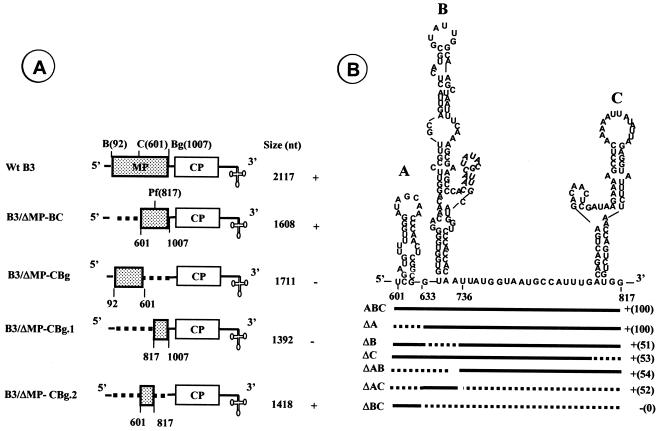
FIG. 2.
Delineation of packaging signal in B3. (A) B3 variants harboring deletions in the MP coding region. Broken lines in each variant clone represent the extent of deletions engineered using the restriction sites shown above the wt B3 map. The lengths of wt B3 and its variant transcripts are shown. The results of in vitro reassembly assays with wt CP subunits and noncapped RNA transcripts of each B3 variant are shown. (B) Predicted secondary structure of 220 nt of MP-coding region required for efficient packaging of B3 and delineation of packaging signal. The stem-loop structure was predicted by the RNA STRUCTURE program (13). Each stem-loop structure is identified as A through C. For delineating the signal, either one or more stem-loop structures were deleted. For example, in variant clone ΔA, nucleotides present between positions 601 to 632 are deleted whereas in ΔAB nucleotides present between positions 601 to 734 are deleted. The efficiency of virion assembly for each variant sequence was compared by visually counting the number of virions present in an area measuring 7.5 μm2 on each electron micrograph negative taken at ×50,000. Symbols: +, detection of virion assembly; −, absence of virion assembly. The numbers shown in parentheses represent the percentage of assembly efficiency for each variant RNA sequence with respect to wt control transcripts. Abbreviations: B, BamHI; C, ClaI; Bg, BglII; Pf, PflMI.
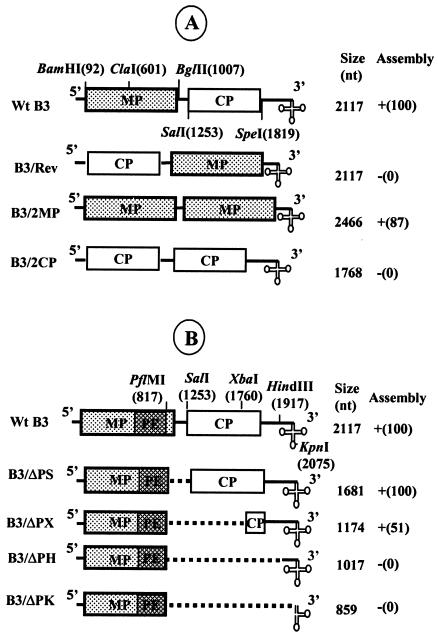
FIG. 3.
Effect of relocation of the MP coding region on B3 packaging. (A) Schematic representation of B3 variant clones used for testing the effect of relocation of the MP coding region on B3 packaging. In B3/Rev, the position of MP and CP has been reversed. Variant clones B3/2MP and B3/2CP are characterized by having duplication of either MP ORF or CP ORF, respectively. The restriction sites used to construct these variant clones are above the wt B3 clone. The size of each variant transcript and the results of in vitro assembly assays between wt CP subunits and noncapped RNA transcripts of each B3 variant are shown. Symbols: +, detection of wt level virion assembly; −, absence of virion assembly. (B) Determination of minimal distance between the PE (packaging element) and the 3′ TLS required for efficient B3 packaging. Characteristics of B3 variants used for testing the required spatial constraints between the PE and the 3′ TLS. In each B3 variant, the distance between the PE and the 3′ TLS was progressively condensed by deleting a specific region located between two unique restriction sites indicated above the wt B3 map. The size of each RNA transcript and the results of assembly assays are shown. The numbers shown in parentheses represent the percentage of assembly efficiency for each variant RNA sequence with respect to wt control transcripts.
In vitro transcription, transfection of protoplasts, and progeny RNA analysis.
Full-length cDNA clones corresponding to the three genomic RNAs of BMV, pT7B1, pT7B2, and pT7B3(-Tth), from which infectious RNAs can be transcribed in vitro, have been described previously (6). All wild-type (wt) and variant clones were linearized with BamHI prior to transcription. Capped full-length transcripts were synthesized in vitro using a MEGAscript T7 kit (Ambion Inc., Austin, Tex.). Unless specified otherwise, RNA3 variants were always coinoculated with wt RNAs 1 and 2. Control inoculations contained in vitro transcripts of all three wt BMV RNAs. Isolation and transfection of barley (Hordeum vulgare cv. Dickson) protoplasts and procedures used to extract progeny RNA and their analysis by Northern hybridization using riboprobes corresponding to the 3′ conserved region were performed as described previously (22), except prehybridization and hybridizations were carried out at 55°C in a hybridization oven. Northern blots were quantitated by densitometry and the correction for differences in inoculation efficiency and protoplast variability was made by normalizing the yield of progeny RNA3 against the value obtained for RNAs 1 and 2 (3, 22).
Coat protein preparation, in vitro virion assembly assays and electron microscopy.
BMV virions were purified from symptomatic barley leaves as described (3). Preparation of CP subunits and in vitro assembly assays, performed in a buffer containing 50 mM Tris-HCl (pH 7.2), 50 mM NaCl, 10 mM KCl, 5 mM MgCl2, and 1 mM DTT, were as performed as described previously (3, 29). Each assembly reaction (100 μl) contained 14 pmol of genomic RNA and 3 μg of purified BMV CP subunits. For trans complementation experiments, 14 pmol of the viral TLS or nonviral RNA were also present, unless otherwise stated. Each in vitro assembled virion preparation was negatively stained with 1% uranyl acetate and examined with a Hitachi transmission electron microscope.
Quantification of in vitro assembly efficiencies.
Following assembly, virions were examined by EM and visually quantitated by counting within an area of 7.5 mm2 on each EM negative taken at a magnification of 50,000. The average number of virions (from at least 8 individual grid areas) assembled with purified CP and either virion RNA or wt transcripts was considered as the reference for 100% assembly efficiency. Virions assembled from purified CP subunits and BMV RNA variant sequences were subjected to a similar quantification procedure.
RESULTS
The sequence encoding MP Gene, not the CP Gene, is required for Packaging B3.
In BMV, B3 and B4 are predicted to copackage into a single virion and therefore a commonly shared packaging signal is envisioned. To identify this commonly shared packaging signal in B3, we divided the entire 2,117-nt sequence of B3 into six distinct regions (Fig. 1A). These are 5′ nontranslated regions (NTR) (91 nt), MP open reading frame (ORF) (915 nt), ICR (244 nt), CP ORF (569 nt), 3′ NTR (97 nt) and 3′ tRNA-like structure (3′ TLS; 201 nt). A set of six variant clones of wt B3 from which RNA transcripts can be synthesized in vitro (Fig. 1A) was constructed. These six variant clones fall into two categories. The first category contains a set of four variant clones and is characterized by having a precise deletion of each NTR (Fig. 1A). These are B3/Δ5′NTR, B3/ΔICR, B3/Δ3′NTR, and B3/Δ3′TLS. The second category contains two variant clones, each lacking either the MP ORF (B3/ΔMP) or the CP ORF (B3/ΔCP). Transcripts of each B3 variant clone were incubated under in vitro assembly conditions to form virions with wt CP subunits using a physiological environment that dictates in vivo packaging specificity (4). Final products of each assembly reaction were subjected to electron microscopic examination and the results are summarized in Fig. 1B.
Assembly with an efficiency similar to that of the wt B3 control was observed for three out of four B3 NTR variant transcripts (i.e., B3/Δ5′NTR, B3/ΔICR, and B3/Δ3′NTR; Fig. 1B). As reported previously (4), no assembly was observed with B3 transcripts lacking the 3′ TLS (i.e., B3/Δ3′TLS; Fig. 1B). Since the CP-coding region is commonly shared between B3 and B4, signals for copackaging are predicted to reside in this region. However, contrary to the conjecture, virion assembly with an efficiency approaching 70% was observed with B3 transcripts lacking the CP coding region (i.e., B3/ΔCP). Interestingly, deletion of MP coding region (i.e., B3/ΔMP) completely abolished assembly (Fig. 1B).
In TCV and flock house nodavirus, isometric viruses of plants and insects, respectively, the size of the RNA has been shown to profoundly influence virion assembly (19, 24). To examine whether the lack of virion assembly observed with transcripts of B3/ΔMP is due to its reduced RNA size, a heterologous sequence identical in length to that of the MP ORF (i.e., 911 nt) was amplified from a cDNA containing Xenopus elongation factor 1α gene sequence (Ambion) and substituted for the MP ORF sequence to yield a B3/Xe construct (Fig. 1A). The incompetence of B3/Xe transcripts to assemble into BMV virions (Fig. 1B) suggest that the defective assembly observed with transcripts of B3/ΔMP is not due to RNA size. These observations were consistently reproduced in at least five independent in vitro assembly assays performed with different transcript preparations and dissociated CP subunit preparations. Collectively, the data suggest that for efficient packaging into virions, B3 must retain the MP-coding region and the 3′TLS.
The MP-coding region contains a cis-acting RNA element required for packaging B3.
The above described set of experiments demonstrated that in addition to the 3′ TLS, the MP-coding region is essential to direct packaging of B3 into virions. In order to examine whether defective packaging exhibited by the transcripts of B3/ΔMP can be rescued by supplying sequences of the MP-coding region in trans, in a fashion similar to that of the TLS (4), a 915-nt RNA transcript encompassing the entire MP-coding region was added to an assembly mix containing wt CP subunits and transcripts of B3/ΔMP (Fig. 1A). EM examination of the reaction products of in vitro assembly revealed no evidence for virion formation (Fig. 1B). Therefore it was concluded that the packaging signal present in the MP-coding region is a cis-acting RNA element.
Delineation of minimal cis-acting sequences required for B3 packaging.
As a first step toward further characterizing the size and location of the essential cis-acting sequence contained within the MP gene required for directing B3 packaging, we engineered two independent deletions (Fig. 2A). These are B3/ΔMP-BC in which 509 nt deleted (bases 92 to 601) and B3/ΔMP-CBg in which 406 nt deleted (bases 601 to 1007). RNA transcripts synthesized from each of these two deletion variant clones were subjected to in vitro assembly reactions followed by electron microscopic examination. The data presented in Fig. 2A suggested that signals required for packaging B3 are localized within the 406-nt sequence present between ClaI and BglII sites of the MP coding region.
To further delineate the sequences located between ClaI and BglII sites required for packaging, two additional variant clones of B3/ΔMP-CBg were constructed (Fig. 2A). These are B3/ΔMP-CBg.1 in which a 220-nt region present between ClaI and PflMI was deleted (bases 601 to 817) and B3/ΔMP-CBg.2 in which a 190-nt region present between PflMI and BglII was deleted (bases 817 to 1007). As shown in Fig. 2A, virion assembly was observed only with transcripts of B3/ΔMP-CBg.2 but not with those of B3/ΔMP-CBg.1, suggesting that signals required for packaging B3 are localized within the 220-nt sequence present between the ClaI and PflMI sites.
The RNA structure program (13) was used to predict the RNA secondary structure for the 220-nt sequence of the MP coding region required for packaging B3 (Fig. 2B). The 220-nt sequence contained three potential stem-loop structures (A through C; Fig. 2B) and these stem-loop structures appeared in all of the structures generated by analyzing either only the MP coding region or the entire B3 sequence (data not shown). Based on this secondary structure, we evaluated which of the three stem-loop structures is needed for assembly by engineering six additional deletions. These are ΔA, ΔB, ΔC, ΔAB, ΔAC, and ΔBC (Fig. 2B). When only stem-loop A was deleted, the efficiency of virion assembly was indistinguishable from that of wt control (Fig. 2B). Whereas independent deletion of either stem-loop B or stem-loop C independently resulted in approximately 50% reduction in packaging efficiency (Fig. 2B). Like wise simultaneous deletion of stem-lops A and B (ΔAB) or A and C (ΔAC) also reduced the efficiency of packaging by 50% (Fig. 2B), By contrast, simultaneous deletion of stem-loops B and C (ΔBC) completely abolished virion assembly (Fig. 2B). Taken together these data indicate that the 187-nt region encompassing stem-loops B and C (Fig. 2B) is the required cis-acting sequence and this region will be referred to as the PE.
Relocation of PE affects B3 packaging.
In HIV type 1, the hairpins required for encapsidation function efficiently only when present in their correct context (14). Thus, to verify whether the PE of B3 functions in position dependent manner we carried out assembly experiments with three B3 variant clones (Fig. 3A). In variant clone B3/Rev, the PE was relocated near the 3′ end by reversing the positions of the two ORFs. In variant clone B3/2MP, the CP ORF was replaced with that of MP so that one each of the PE is located at the 5′ and 3′ end of B3. As a control, variant clone B3/2CP was constructed to have two CP ORFs, one each at the 5′ and 3′ ends of B3. In vitro assembly assays similar to those described above were performed with transcripts of each of these variant clones. The incompetence of B3/2CP to assemble into virions confirmed that packaging B3 requires MP ORF (Fig. 3A). Interestingly, the location of the MP ORF at the 3′end in B3/Rev completely abolished RNA packaging (Fig. 3A), whereas packaging efficiencies approaching near wt level were observed with transcripts of B3/2MP (Fig. 3A). Collectively the data suggest that PE localized in the MP ORF should be positioned in correct context to provide a specific structure or alignment necessary for the recognition of CP subunits. To further substantiate this conjecture, a set of four independent deletion variants of B3 was constructed and the packaging efficiency of these deleted RNAs was examined (Fig. 3B). When the distance between the PE and the NE (i.e., 3′ TLS) was reduced by 436 nt (as in variant B3/ΔPS; Fig. 3B), the packaging efficiency was indistinguishable from that of wt B3 (Fig. 3B). A significant drop in the packaging efficiency, as much as 50%, was clearly evident with an increased deletion, as exemplified by variant B3/ΔPX (Fig. 3B). This bias was even more pronounced for the larger deletions (B3/ΔPH and B3/ΔPK; Fig. 3B) resulting in the complete elimination of packaging. These observations suggest that a minimal distance between the PE and the NE is required either to maintain the structural features of the PE (Fig. 3B) or to provide optimal cis-interaction between PE and the NE.
The PE in conjunction with the NE promotes assembly of nonviral sequence into BMV virions.
The data presented above has demonstrated that packaging of B3 into virions requires PE and NE. If these are the only two elements required for packaging RNA into BMV virions, then any unrelated RNA sequence engineered to have these two elements should be directed into BMV virions. To substantiate this hypothesis, three chimeras were constructed. Two chimeras, B3/Xe 5′PE and B3/Xe 3′PE (Fig. 4A), are the derivatives of the assembly defective B3/Xe (Fig. 1A) whereas the third chimera, B3/Xe.1, is derivative of B3/ΔMP-BC (see Materials and Methods). When in vitro assembly assays were performed with radiolabeled transcripts of all three chimeras, efficient assembly of BMV virions was observed in each case (Fig. 4A). The presence of each chimera in BMV virions with the expected size was confirmed by agarose gel analysis (Fig. 4B).
In vivo evidence that packaging of B4 is coupled to that of its progenitor B3.
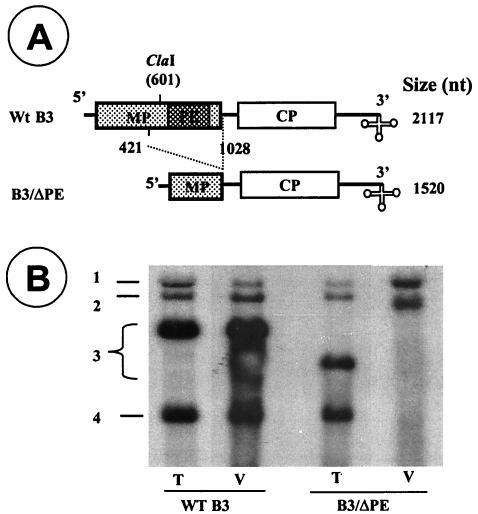
Since B4 is derived from the 3′ half of B3 by internal initiation on progeny minus strand, it retains the 3′ TLS but lacks the PE of B3. To verify the in vitro function of PE in vivo and to get an insight as to how B4 copackages with B3, a variant clone of B3 referred to as B3/ΔPE was constructed by deleting a central 591-nt region of the MP ORF (Fig. 5A). The rationale for deleting such a large region is to distinguish the replicated B3/ΔPE progeny from those of wt B3. Barley protoplasts were transfected with a mixture containing transcripts of wt B1and B2 and B3/ΔPE. In BMV, deletion of the MP ORF does not interfere with amplification of B3 or production of B4 (10). Consequently analysis of total RNA from transfected protoplasts revealed that B3/ΔPE replicated efficiently and synthesized wt levels of B4 (Fig. 5B). However, when virions were purified from the same batch of transfected protoplasts and their RNA content analyzed by Northern blots, a profile distinct from that of total RNA was observed. Interestingly virions of B3/ΔPE packaged only progeny B1 and B2 (Fig. 5B). As expected, B3 was not packaged into virions due to the lack of the PE. Progeny B4 could have been found in B3/ΔPE virions, had the packaging of B4 been independent of B3. Taken together these results suggest that B4 cannot be packaged independently into virions and its packaging is coupled to that of B3.
FIG. 5.
In vivo evidence demonstrating that B4 is incompetent for autonomous packaging in vivo. (A) The structure of wt B3 and its variant referred to as B3/ΔPE designed to lack a region encompassing the PE is shown. The lengths of wt B3 and its variant transcripts are shown. (B) Autoradiograph of an agarose gel showing profiles of total (T) and virion RNA (V) recovered from barley protoplasts transfected with all three wt BMV transcripts or a mixture containing wt B1and B2 and a B3/ΔPE variant. Total and virion RNA isolated, respectively, from 105 and 106 transfected protoplasts were denatured with glyoxal and subjected to agarose gel analysis. Following electrotransfer, the blot was hybridized with a 32P-labeled riboprobe complementary to the highly conserved 3′ TLS sequence. The positions of genomic and subgenomic RNA are shown.
DISCUSSION
In this work, in vitro assembly assays mapped both essential and dispensable regions for packaging B3 into icosahedral virions. It was demonstrated that the “packaging core” of B3 is bipartite consisting of NE and a specific cis-acting PE (Fig. 1A and Fig. 2). The highly conserved 3′ TLS functions as a NE for CP subunits whereas the essential component of the PE is a 187-nt sequence localized within the nonstructural MP (Fig. 2). It was also observed that the position of the MP ORF (containing the PE) with respect to the 3′TLS appears to profoundly influence B3 packaging (Fig. 3A). Finally, barley protoplast experiments provided the first genetic evidence that B4 is incompetent for efficient autonomous packaging into virions and prior packaging of B3 is obligatory to promote copackaging of B4 into the same virion (Fig. 5B).
Packaging signal in B3 is bipartite.
The distribution of four BMV RNAs into three identical capsids (20) and the high degree of specificity exhibited by the BMV CP (5, 16) suggest its interaction with viral RNA is specific. However nonspecific interactions were observed between the CP and BMV RNA acting separately or synergistically on the same RNA molecule. Duggal and Hall (7) demonstrated that the interaction between CP and 3′ TLS is nonspecific leading to virion assembly by compaction of the RNA through neutralization of the negative charge on the phosphate backbone. It was previously proposed that, in icosahedral single-strand RNA viruses, binding of structural proteins to a specific RNA structure form an assembly initiation complex, which nucleates the subsequent addition of CP subunits to complete a mature virion (31). Our recent observation that TLS-less BMV RNAs cannot assemble into virions (4) suggest a role for this domain in nucleating a higher-order arrangement of CP dimers that serves as an intermediate on the encapsidation pathway. Thus, TLS functioning as a NE might promote the formation of pentamers of dimers via a number of mechanisms. For example, the N-terminal region of BMV CP contains an arginine-rich motif (ARM) that is conserved among plant and nonplant viruses (21) and implicated in RNA binding during encapsidation. By binding to the basic residues of the N terminus, RNA (specifically 3′TLS or host tRNAs) might neutralize the charge repulsion between noncovalent CP dimers to form pentamers of dimers. The fact that no empty virions are found in vivo favors the hypothesis that plant or viral TLS acts as a scaffold for nucleating CP dimers to form pentamers of dimers. Furthermore the nonspecific interaction between the CP and the viral TLS (as well as host tRNAs) may serve to stabilize these pentamers or productive complexes of pentamers in favor of nonproductive aggregates that fail to associate with a genomic RNA and progress to full virions. In doing so the nonspecific interactions could stimulate encapsidation in vivo at early times when the concentrations of viral components are comparatively low, or serve to chaperone productive capsid formation late in the infection when high concentrations of CP could lead to nonproductive aggregates.
By contrast to nonspecific interactions, RNA features that specifically interact with CP and are involved in the assembly of icosahedral virions have been described for retroviruses (1), Sindbis virus (28) and TCV (26, 27). Such specific interactions are envisioned to play a significant role in selectively removing viral RNAs from the pool of heterologous host RNAs present in the cytoplasm (5, 9). Therefore we propose that selective recognition of a domain specific for each of the four BMV RNAs occurs after or in concert with TLS/CP interaction. This specificity filter together with size constraints dictated by the capsid size may aid in segregating four RNAs into three identical capsids. Thus, the PE in B3 identified in this study could be considered as a selective domain for CP interaction and facilitate specific packaging of B3. The existence of similar specific domains for B1 and B2, distinct from that of B3, are envisioned to exist.
Mechanism of B3 and B4 copackaging.
The results presented here also invoke important questions concerning B3 and B4 copackaging. Virions containing only B4 can be assembled efficiently in vitro (4). However analysis of RNA distribution in purified virions obtained from natural infections revealed that 94% of virions contained one molecule each of B3 and B4 whereas only 6% contained three molecules of B4 suggesting that in vivo B4 is incompetent for efficient autonomous assembly (12). This study identified that B3 packaging is mediated by a signal specifically encoded within the MP gene that is absent in B4 and its deletion not only debilitated packaging of B3 but also B4 when analyzed in vivo (Fig. 5B). Thus, the question that remains to be addressed is how do B3 and B4 copackage into a single virion?
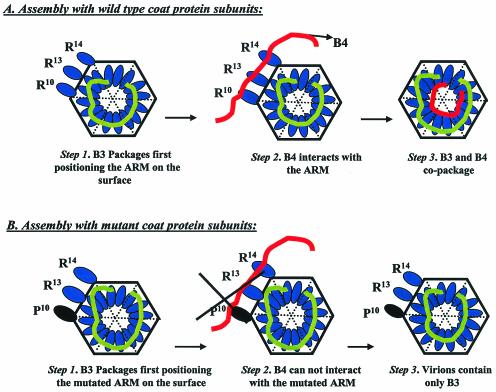
We hypothesize that B3 and B4 copackage into a single virion either sequentially or in a concerted manner involving RNA-RNA interactions. The first model envisages (Fig. 6) that B3 and B4 copackage sequentially in a fashion similar to that proposed for bacteriophage φ6 (15). This model predicts that binding of wt CP subunits with NE and PE of B3 results in prior packaging of B3 into a single virion (Fig. 6A, step 1). This provides an architectural feature to the virion such that the basic N-ARM of the CP is displayed on the surface of the virion (Fig. 6A, step 1), similar to that observed for icosahedral virions of flock house virus (2). Such surface conformation allows specific interaction of B4 with the required arginine residues (Fig. 6A, step 2), promoting copackaging of B3 and B4 into a single virion (Fig. 6A, step 3). As observed previously in vivo (3), a mutation in the N-ARM (e.g., R10→P10; Fig. 6B, step 1) will not affect prior packaging of B3 but B4 interaction is disrupted (Fig. 6B, step 2). As a consequence the virion will contain only B3 (Fig. 6B, step 3).
FIG. 6.
A schematic model for the sequential packaging of B3 and B4 into a single virion. (A) In step 1, prior packaging of B3 occurs with wt CP subunits positioning the N-ARM on the surface of the virion. In step 2, a specific interaction between B4 and arginine residues of N-ARM occurs. In step 3, this results in B3 and B4 copackaging into the same virion. (B) In step 1, as in panel A, B3 is packaged into the virions by mutant CP subunits. In step 2, however, mutation of a crucial arginine residue disrupts B4 interaction with the N-ARM. Consequently, in step 3 the virion contains only B3 as exemplified by previous in vivo analysis (3).
Alternatively, the underlying principle for the concerted packaging model derives from our observations that deletion of PE not only blocked B3 packaging but also B4. Thus, the PE could be acting in trans to form a complex with B4 resulting in copackaging. We predict that this complex formation is mediated through either RNA-RNA interactions or the CP subunits (up on binding to PE) function directly or indirectly as chaperones to promote B3 and B4 copackaging. Experiments are in progress to verify these two models.
Acknowledgments
We thank George Grantham for excellent technical assistance and Theo Dreher, Shou-Wei Ding, and Dan Gallie for helpful comments on the manuscript.
This research was supported in part by grant GM064465 from the National Institutes of Health (to A.L.N.R.).
REFERENCES
- 1.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 2.Bothner, B., A. Schneemann, D. Marshall, V. Reddy, J. E. Johnson, and G. Siuzdak. 1999. Crystallographically identical virus capsids display different properties in solution. Nat. Struct. Biol. 6:114-116. [DOI] [PubMed] [Google Scholar]
- 3.Choi, Y. G., and A. L. N. Rao. 2000. Molecular studies on bromovirus capsid protein: VII. Selective packaging of BMV RNA4 by specific N-terminal arginine residues. Virology 275:207-217. [DOI] [PubMed] [Google Scholar]
- 4.Choi, Y. G., T. W. Dreher, and A. L. N. Rao. 2002. tRNA elements mediate the assembly of an icosahedral RNA virus. Proc. Natl. Acad. Sci. USA 99:655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuillel, M., M. Herzog, and L. Hirth. 1979. Specificity of in vitro reconstitution of bromegrass mosaic virus. Virology 95:146-153. [DOI] [PubMed] [Google Scholar]
- 6.Dreher, T. W., A. L. N. Rao, and T. C. Hall. 1989. Replication in vivo of mutant brome mosaic virus RNAs defective in aminoacylation. J. Mol. Biol. 206:425-438. [DOI] [PubMed] [Google Scholar]
- 7.Duggal, R., and T. C. Hall. 1993. Identification of domains in brome mosaic virus RNA-1 and coat protein necessary for specific interaction and encapsidation. J. Virol. 67:6406-6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fosmire, J. A., K. Hwan, and S. Makino. 1992. Identification and characterization of a coronavirus packaging signal. J. Virol. 66:3522-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox, J. M., J. E. Johnson, and M. J. Young. 1994. RNA/protein interactions in icosahedral virus assembly. Semin. Virol. 5:51-60. [Google Scholar]
- 10.French, R., and P. Ahlquist. 1987. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J. Virol. 61:1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frolova, E., I. Frolov, and S. Schlesinger. 1997. Packaging signals in alphaviruses. J. Virol. 71:248-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hull, R. 1976. The behavior of salt-labile plant viruses in gradients of cesium sulphate. Virology 75:18-25. [DOI] [PubMed] [Google Scholar]
- 13.Matthews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 14.McBride, M. S., and A. T. Panganiban. 1997. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J. Virol. 71:2050-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mindich, L. 1999. Precise packaging of the three genomic segments of the double stranded RNA of bacteriophage variant φ6. Microbiol. Mol. Biol. Rev. 63:149-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osman, F., Y. G. Choi, G. L. Grantham, and A. L. N. Rao. 1998. Molecular studies on bromovirus capsid protein. V. Evidence for the specificity of brome mosaic virus encapsidation using chimera of brome mosaic and cucumber mosaic viruses expressing heterologous coat proteins. Virology 251:438-448. [DOI] [PubMed] [Google Scholar]
- 17.Patton, J. T., and E. Spencer. 2000. Genome replication and packaging of segmented double-stranded RNA viruses. Virology 277:217-225. [DOI] [PubMed] [Google Scholar]
- 18.Qiao, X., J. Qiao, and L. Mindich. 1997. Stoichiometric packaging of the three genomic segments of double-stranded RNA bacteriophage φ6. Proc. Natl. Acad. Sci. USA 94:4074-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu, F., and T. J. Morris. 1997. Encapsidation of turnip crinkle virus is defined by a specific packaging signal and RNA size. J. Virol. 71:1428-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao, A. L. N. 2001. Bromoviruses, p. 155-158. In O. C. Maloy and T. D. Murray (ed.), Encyclopedia of plant pathology. John Wiley & Sons, Toronto, Canada.
- 21.Rao, A. L. N., and G. L. Grantham. 1996. Molecular studies on bromovirus capsid protein: II. Functional analysis of the amino terminal arginine rich motif and its role in encapsidation, movement and pathology. Virology 226:294-305. [DOI] [PubMed] [Google Scholar]
- 22.Rao, A. L. N., T. W. Dreher, L. E. Marsh, and T. C. Hall. 1989. Telomeric function of the tRNA-like structure of brome mosaic virus RNA. Proc. Natl. Acad. Sci. USA 86:5335-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook., J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Schneemann, A., and D. Marshall. 1998. Specific encapsidation of nodavirus RNAs is mediated through the C terminus of capsid precursor protein alpha. J. Virol. 72:8738-8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorger, P. G., P. G. Stockley, and S. C. Harrison. 1986. Structure and assembly of turnip crinkle virus II. Mechanism of reassembly in vitro. J. Mol. Biol. 191:639-658. [DOI] [PubMed] [Google Scholar]
- 26.Wei, N., L. A. Heaton, T. J. Morris, and S. C. Harrison. 1990. Structure and assembly of turnip crinkle virus. VI. Identification of coat protein binding sites on the RNA. J. Mol. Biol. 214:85-95. [DOI] [PubMed] [Google Scholar]
- 27.Wei, N., and T. J. Morris. 1991. Interactions between viral coat protein and specific binding region on turnip crinkle virus RNA. J. Mol. Biol. 222:437-443. [DOI] [PubMed] [Google Scholar]
- 28.Weiss, B., U. Geigenmuller-Gnirke, and S. Schlesinger. 1994. Interactions between sindbis viral RNAs and 68 amino acid derivative of the viral capsid protein further define the capsid binding site. Nucleic Acids Res. 22:780-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao, X., J. Fox, N. Olson, T. S. Baker, and M. J. Young. 1995. In vitro assembly of cowpea chlorotic mottle virus from coat protein expressed in Escherichia coli and in vitro transcribed viral cDNA. Virology 207:486-494. [DOI] [PubMed] [Google Scholar]
- 30.Zimmern, D. 1977. The nucleotide sequence at the origin for assembly on tobacco mosaic virus RNA. Cell 11:463-482. [DOI] [PubMed] [Google Scholar]
- 31.Zlotnick, A., R. Aldrich, J. M. Johnson, P. Ceres, and M. J. Young. 2000. Mechanism of capsid assembly for an icosahedral plant virus. Virology 277:450-456. [DOI] [PubMed] [Google Scholar]