SUMMARY
Protein palmitoylation is a reversible lipid modification that regulates membrane tethering for key proteins in cell signaling, cancer, neuronal transmission, and membrane trafficking. Palmitoylation has proven to be a difficult study: Specifying consensuses for predicting palmitoylation remain unavailable, and first-example palmitoylation enzymes—i.e., protein acyltransferases (PATs)—were identified only recently. Here, we use a new proteomic methodology that purifies and identifies palmitoylated proteins to characterize the palmitoyl proteome of the yeast Saccharomyces cerevisiae. Thirty-five new palmitoyl proteins are identified, including many SNARE proteins and amino acid permeases as well as many other participants in cellular signaling and membrane trafficking. Analysis of mutant yeast strains defective for members of the DHHC protein family, a putative PAT family, allows a matching of substrate palmitoyl proteins to modifying PATs and reveals the DHHC family to be a family of diverse PAT specificities responsible for most of the palmitoylation within the cell.
INTRODUCTION
Palmitoylation or protein S-acylation is the thioesterification of fatty acids, usually palmitic acid, to cysteine thiols (for recent reviews on palmitoylation, see Huang and El-Husseini, 2005; Smotrys and Linder, 2004). Like the analogous lipid modifications myristoylation and prenylation, palmitoylation serves to tether proteins to the cytoplasmic surfaces of cellular membranes. Among the proteins that rely on palmitoylation for localized function are many of the key players in cellular signaling, membrane trafficking, cancer, and synaptic transmission, including many G proteins such as Ras- and Rho-like proteins as well as the α and γ subunits of many heterotrimeric G proteins; many nonreceptor tyrosine kinases, e.g., Fyn, Lck, and Yes; and the epithelial nitric oxide synthase (eNOS). Palmitoylation is distinguished from the other two tethering modifications, myristoylation and prenylation, by its reversibility. Reversibility allows for regulation, and, indeed, rapidly cycling palmitoyl modifications have been shown to regulate synaptic plasticity (El-Husseini Ael et al., 2002), β-adrenergic receptor desensitization (Jones et al., 1997; Moffett et al., 2001; Tu et al., 1997; Wedegaertner and Bourne, 1994), and the plasma-membrane-localized activity of H- and N-Ras (Rocks et al., 2005). Palmitoylation also differs from myristoylation or prenylation in that it is often found to modify transmembrane (TM) proteins; thus, the list of palmitoylated proteins also includes many G protein-coupled receptors, viral-envelope proteins, caveolin, and diverse ion channels and ionotropic neurotransmitter receptors. Although the function provided by palmitoylation to such TM proteins is not entirely clear, it may reflect roles in lipid raft and membrane microdomain targeting (Brown and London, 1998; Zacharias et al., 2002).
Given the prevalence and importance of protein palmitoylation, it is surprising how poorly understood its underlying mechanisms remain. With its occurrence in such a wide spectrum of sequence contexts, consensuses that would allow palmitoylation to be predicted from sequence have yet to be defined. The enzymology of palmitoylation also remains poorly defined. The first protein acyltransferases (PATs) were identified only recently, by work in the yeast Saccharomyces cerevisiae (Lobo et al., 2002; Roth et al., 2002). The two identified yeast PATs, Akr1 and Erf2-Shr5, share a common, 50 residue zinc-finger-like sequence, the DHHC cysteine-rich domain, and together point toward the wider DHHC protein family as a possible PAT family. DHHC-family proteins, a collection of polytopic integral membrane proteins, typically share little or no homology beyond their defining DHHC domain; yeast has seven DHHC proteins (including Akr1 and Erf2), while 23 have been identified from the human genome. Recent analyses of various yeast and mammalian DHHC proteins generally support a DHHC protein role in palmitoylation (Ducker et al., 2004; Fukata et al., 2004; Hou et al., 2005; Huang et al., 2004; Keller et al., 2004; Smotrys et al., 2005; Swarthout et al., 2005; Valdez-Taubas and Pelham, 2005). However, the extent of this role is called into question by in vitro demonstrations of DHHC protein-independent palmitoylation (Dietrich et al., 2004; Dietrich and Ungermann, 2004), including apparently bona fide palmitoylations that proceed nonenzymatically in vitro, through direct chemical reaction of acceptor thiols with palmitoyl-CoA (Duncan and Gilman, 1996; Leventis et al., 1997; Veit, 2000). Thus, the overall role of DHHC proteins—and, more generally, of enzymes—in protein palmitoylation remains uncertain.
Palmitoyl proteins typically are identified through in vivo [3H]palmitate labeling, a tedious process involving large amounts of input label, immune precipitation, fluorography, and notoriously long autoradiographic exposures (week- or month-long). With the focus always on individual proteins, the overall scope of palmitoylation’s use in the cell remains untested. Below, we describe a new proteomic method that purifies and identifies palmitoylated proteins. We have applied this method first in the yeast Saccharomyces cerevisiae toward a comprehensive identification of the proteins that comprise the yeast palmitoyl proteome. This approach identifies 12 of the 15 known palmitoyl proteins plus many new palmitoyl proteins. In addition, the application of this approach to mutant yeast strains deficient for different DHHC-family proteins has allowed us to both gauge the impact of DHHC proteins on palmitoylation and match the palmitoyl-protein substrates to cognate modifying DHHC PATs. Our results demonstrate the DHHC protein family to be a family of diverse PAT specificities that mediate the bulk of the protein palmitoylation within the cell.
RESULTS AND DISCUSSION
Purification and Identification of Yeast Palmitoyl Proteins
Our protocol for purifying palmitoyl proteins from complex protein extracts is a proteomic scaling of the acyl-biotinyl exchange (ABE) chemistry of Drisdel and Green (2004). The ABE substitution of biotin for protein palmitoyl modifications consists of three sequential chemical steps: (1) blockade of free thiols with N-ethyl maleimide (NEM), (2) cleavage of palmitoylation thioester linkages with hydroxylamine, and finally (3) the marking of the cysteinyl thiols, newly exposed by hydroxylamine with a thiol-specific biotinylation reagent. Proteins from total yeast membranes were subjected to ABE, and the resulting biotinylated proteins were affinity purified by streptavidin agarose. As a control, half of the protein extract was processed through a parallel protocol that omitted hydroxylamine. The final two purified samples—the experimental +hydroxylamine (EXP) sample and the control +hydroxylamine (CON) sample—were subjected to gel analysis (Figure 1A). Proteins that are common to both samples presumably are purified nonspecifically due either to inappropriate biotinylation or nonspecific streptavidin binding. Proteins unique to the EXP sample (Figure 1A, arrows) should represent purified palmitoyl proteins.
Figure 1. Proteomic Analysis.
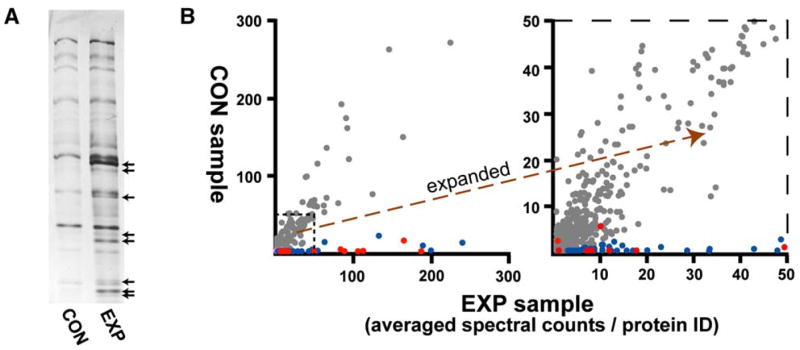
(A) Electrophoretic analysis of purified −hydroxylamine (CON) and +hydroxylamine (EXP) samples. One percent of total purified samples were subjected to SDS-PAGE and silver staining. EXP sample-specific proteins are indicated by arrows.
(B) Graphical depiction of MS analysis. For each identified protein, averaged and normalized EXP sample spectral counts from four wild-type samples (x axis) is plotted against averaged, normalized CON sample spectral counts (y axis). Shown at right is an expanded view of the indicated portion. Known palmitoyl proteins are indicated in red; the new candidate palmitoyl proteins are blue. Proteins with substantial representations from both the EXP and CON samples are gray. Note that 3 of the 15 known palmitoyl proteins fail to cluster with other known palmitoyl proteins, being detected either by low spectral count numbers (Ykt6) or from both EXP and CON samples (Hem14 and Tub1). See Table S1 for supporting MS/MS data.
MudPIT (multi-dimensional protein identification technology), a robust, tandem-MS-based proteomic methodology capable of identifying thousands of proteins per run (Link et al., 1999; Washburn et al., 2001), was used to identify EXP and CON sample proteins. Candidate palmitoyl proteins were defined as proteins with exclusive, or substantially higher, EXP versus CON sample representations. To compare the relative abundance of each protein in the two samples, we relied on per-protein spectral counts—i.e., the number of peptide identifications (including redundant identifications) linked to each protein identification—as a rough metric of relative protein abundance (Liu et al., 2004). Combined data from MudPIT analyses of four paired EXP-CON samples (see Table S1 in the Supplemental Data available with this article online) are graphically represented in Figure 1B: For each protein, averaged EXP sample spectra counts are plotted on the x axis against averaged CON sample spectra counts on the y axis. The vast majority of the identified proteins show substantial representations in both the CON and EXP samples (Figure 1B, gray dots); these proteins tend to be highly abundant proteins (e.g., ribosomal proteins, chaperonins, etc.; Ghaemmaghami et al., 2003) that likely contaminate both samples nonspecifically. All 15 known yeast palmitoyl proteins were detected (Figure 1B, red dots); 12 of the 15 clustered near the x axis with significantly higher EXP versus CON sample representations. The other proteins clustering along the x axis with the known palmitoyl proteins (Figure 1B, blue dots) are investigated below as new palmitoyl-protein candidates.
Proteins were ranked according to both their EXP sample abundance and their EXP/CON spectra count ratio (the 70 most abundant proteins with EXP/CON ratios greater than 5.5 are listed and described in Table S2). To cull false positives, proteins were individually tested for palmitoylation by either the ABE protocol or the standard [3H]palmitate in vivo labeling protocol (example tests are shown in Figure S1, with overall results of this testing summarized in Table S2). Many of the false positives appear to be statistical anomalies, i.e., contaminant proteins that fail to be subtracted due to chance CON sample underdetection. As the ABE chemistry detects palmitoylation through detection of the thioester linkage, another false-positive class are proteins that use thioester linkages for biochemistries other than palmitoylation. Indeed, the two proteins most prominently detected in the proteomic analysis, Lat1 and Pdx1 (Table S2), both fall into this category; both are subunits of the mitochondrial pyruvate dehydrogenase complex, and both utilize covalently bound lipoic-acid prosthetic groups to transiently accept acetyl moieties in thioester linkage (for pyruvate decarboxylation). Pdx1 is robustly labeled by the ABE protocol (data not shown) but shows no hint of in vivo labeling by [3H]palmitate (Figure S1). Three other prominently detected proteins, Gcv3, Acp1, and Ubc1, also use thioesters in their enzymatic mechanism and thus also likely fall into this same false-positive category (Table S2). Given the positive ABE detection of these thioester-using impostors, we have relied predominantly on in vivo [3H]palmitate labeling as the primary test for validating palmitoylation; palmitoylation was confirmed for 26 of the candidate proteins by this test (Figure S1). Testing results for all of the top 70 candidate proteins are summarized in Table S2.
New Palmitoyl Proteins
Overall, our proteomic analysis detected 47 palmitoyl proteins: 12 of the 15 known palmitoyl proteins plus 35 new palmitoyl proteins (Figure 2). The new palmitoyl proteins span a wide range of cellular functions (for protein descriptions, see Table S2), including three G proteins (Gpa2, Rho2, and Rho3), three Golgi-localized mannosyltransferases (Mnn1, Mnn10, and Mnn11), the plasma-membrane-localized phosphatases Psr1 and Psr2, the phosphoinositol-4-kinase Lsb6, and the yeast sphingosine kinase homolog Lcb4. Also prominently detected as palmitoylated were many SNARE proteins (mediators of vesicular fusion), with 8 of the 23 yeast SNARE proteins being detected as palmitoylated, as well as a number of amino acid permeases (AAPs, the plasma-membrane-localized mediators of amino acid import). Many of the new palmitoyl proteins have cysteines positioned in typical palmitoylation contexts—e.g., proximal to motifs for either N-terminal myristoylation or C-terminal prenylation—or, for TM proteins, cysteines near predicted TM domain (TMD) boundaries (Figure 2). Strikingly, all 13 palmitoyl proteins within the single-TMD category, a group which includes the eight SNARE proteins, have cysteines that map cytoplasmically adjacent to their TMD (Figure 2).
Figure 2. Yeast Palmitoyl Proteins.
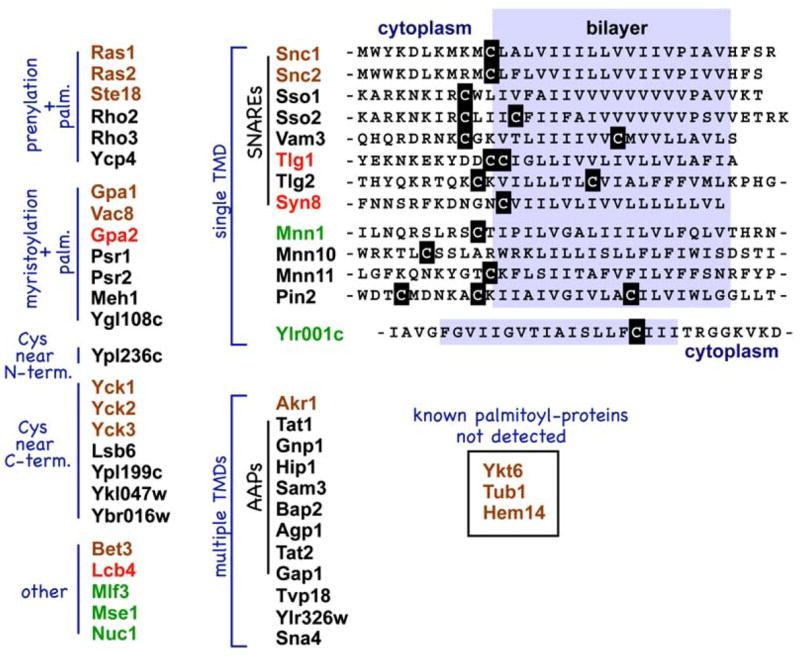
Palmitoyl proteins, both new and known, detected by the MS analysis are classed by presence or absence of predicted TMDs and by positioning of likely palmitoyl-accepting cysteines. Known palmitoyl proteins are shown in brown; new palmitoyl proteins are shown in black. For each of the listed proteins, Table S2 provides a full summary of the evidence supporting palmitoylation. Proteins lacking strong independent confirmation of palmitoylation are shown in green. Four proteins for which palmitoylation has been confirmed in recent published reports (Harashima and Heitman, 2005; Kihara et al., 2005; Valdez-Taubas and Pelham, 2005) are shown in red. At right, TMD-proximal sequences are shown for the indicated proteins.
SNARE Proteins
Eight of the twenty-three yeast SNARE proteins are identified here as palmitoylated (Figure 2); all eight have juxta-TMD cysteines as likely palmitoyl acceptors (Figure 2). The 15 SNAREs not detected by our analysis conspicuously lack such cysteines. While palmitoylation had been previously demonstrated for two of the eight palmitoylated SNAREs—namely for the redundant plasma-membrane v-SNAREs Snc1 and Snc2 (Couve et al., 1995)—this widespread yeast SNARE palmitoylation reported both here and in a recent paper (Valdez-Taubas and Pelham, 2005) was not anticipated. SNARE palmitoylation likely is not a yeast-limited phenomenon: Cursory inspection of mammalian SNARE sequences (Bock et al., 2001) finds a high proportion (15 of 36) also with cysteines cytoplasmically adjacent to their membrane-anchoring TMD. Thus, some function for SNARE palmitoylation has been conserved through evolution. Palmitoylation may serve to target SNAREs to lipid rafts or, alternatively, could regulate or participate more directly in the membrane fusion event itself.
Though all eight SNAREs with juxta-TMD cysteines were detected by our analysis, one palmitoylated yeast SNARE, Ykt6 (Dietrich et al., 2005; Fukasawa et al., 2004), was missed. Ykt6 is an atypical SNARE: It lacks a TMD and instead relies entirely on C-terminal lipidation, dual prenylation-palmitoylation, for a regulated anchoring to the donor bilayer; at any given point in time, only a fraction of the Ykt6 protein is palmitoylated and hence membrane anchored (Dietrich et al., 2005). Our poor detection of Ykt6 (low-level detection in only one of the four MudPIT runs; Table S1) may indicate low fractional palmitoylation for Ykt6.
Amino Acid Permeases
AAPs, polytopic membrane proteins with 12 predicted TMDs, are the plasma-membrane-localized importers of amino acids; 25 AAPs with different transport specificities are identifiable from the yeast genome. Intriguingly, the four AAPs detected as palmitoylated in our initial analysis—i.e., Tat1, Gnp1, Sam3, and Hip1—all have the identical C-terminal sequence Phe-Trp-Cys (Figure 3A). A fifth AAP, Bap2, also with the C-terminal Phe-Trp-Cys sequence, was identified by the MS analysis at a somewhat lower significance level (ranking below the top 70 candidates at position 90). Cys-to-Ser mutation of the C-terminal cysteines for Tat1, Gnp1, Hip1, and Bap2 fully abolishes palmitoylation (Figure 3B), indicating service of the C-terminal cysteines as the sole palmitoyl acceptors. Six additional AAPs not detected by our MS analysis also have C-terminal cysteines: Agp1, Bap3, Gap1, and Tat2 have the C-terminal Phe-Trp-Cys sequence, while Mmp1 ends with Phe-Phe-Cys and Mup3 with Cys-Leu-Cys (Figure 3A); the 14 other yeast AAPs lack C-terminal cysteines. The lack of MS detection for these other AAPs with C-terminal cysteines may reflect the low expression expected for many AAPs in the rich culture conditions used for preparation of the proteomic extracts; indeed, as shown below, with forced AAP expression from the GAL1 promoter, we do find additional members of this AAP sequence class to be palmitoylated. As suggested above for the SNARE proteins, palmitoylation also may serve to regulate function and/or sorting of many yeast AAPs.
Figure 3. Palmitoylation Acceptor Sites.
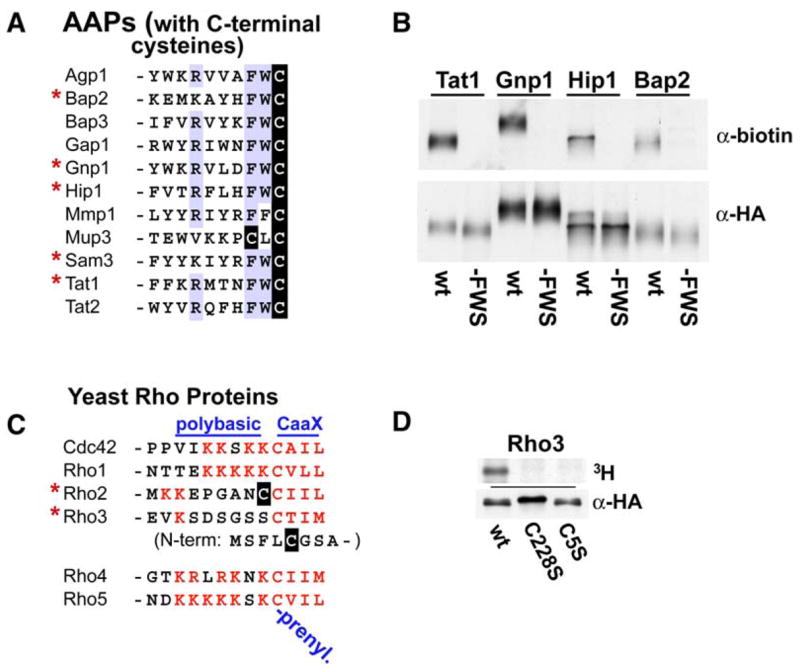
(A) Consensus elements for the 11 amino acid permeases (AAPs) with C-terminal cysteines. The five AAPs identified as palmitoylated by the proteomic analysis are indicated with asterisks.
(B) AAP C-terminal Cys is required for palmitoylation. For the indicated AAPs, palmitoylation of both the wild-type AAP (wt) and the C-terminal Cys-to-Ser mutant version (−FWS) was assessed using a scaled-down version of the acyl-biotinyl exchange (ABE) protocol (Supplemental Experimental Procedures). To facilitate this analysis, AAPs were N-terminally FLAG/HA epitope tagged and overexpressed from the GAL1 promoter (2 hr expression period). Following ABE, AAPs were anti-FLAG immunoprecipitated and then blotted for biotinylation (α-biotin; indicative of palmitoylation) or overall recovery (α-HA).
(C) Membrane tethering sequences of yeast Rho proteins. The C-terminal 13 amino acid residues of the six yeast Rho proteins are shown. CaaX prenylation motifs and polybasic domains are red, and likely palmitoyl-accepting cysteines are highlighted in black.
(D) N-terminal Rho3 palmitoylation requires prior C-terminal prenylation. Cells expressing wild-type or the two indicated mutant Rho3 proteins from the GAL1 promoter (2 hr expression period) were labeled with [3H]palmitic acid. Immunoprecipitated Rho3 was analyzed for both label incorporation (top panel) and Rho3 protein recovery (bottom panel). The 4× HA/FLAG epitope tag used for this analysis (for immunoprecipitation and for Western detection) was inserted internally within the RHO3 ORF, between codons Ala217 and Thr218 (14 codons from the RHO3 stop codon), to avoid disrupting N- and C-terminal lipidation sites.
Rho Proteins
Two of the six yeast Rho protein actin regulators, Rho2 and Rho3, are identified as being palmitoylated (Figure 2). The C-terminal sequence of Rho2, -Cys-Cys-Ile-Ile-Leu, is typical of dually prenylated-palmitoylated proteins, having a likely palmitoyl-accepting cysteine proximal (in this case, adjacent) to a C-terminal -CaaX prenylation consensus in which C represents cysteine, aa represents any two aliphatic amino acids, and X represents the amino acid specifying farnesylation or geranylgeranylation (Figure 3C). The other five yeast Rho proteins also have -CaaX prenylation consensuses but lack proximal cysteines for palmitoylation (Figure 3C). However, four of the five have proximal polybasic sequences that are thought to stabilize membrane attachments of proteins with single-lipid tethers through electrostatic interaction with anionic phospholipid head groups (Resh, 1999). The exception is Rho3, which lacks both proximal cysteines and polybasic sequences (Figure 3C). Looking farther afield within the Rho3 sequence to account for its palmitoylation, one does find a cysteine at the Rho3 N terminus that maps to a unique Rho3 N-terminal extension (Figure 3C). Mutation of this cysteine (i.e., C5S) does indeed abolish Rho3 palmitoylation (Figure 3D). Furthermore, Rho3 palmitoylation also is abolished by mutation of the C-terminal prenyl acceptor (i.e., C228S) (Figure 3D), consistent with the dogma for dually prenylated-palmitoylated proteins that palmitoylation requires prior prenylation (Dunphy and Linder, 1998; el-Husseini Ael and Bredt, 2002; Smotrys and Linder, 2004). Thus, our results indicate that Rho3 is a unique example of a dually prenylated-palmitoylated protein—prenylated at its C terminus and palmitoylated at its N terminus. Simultaneous tethering of both N and C termini could be accommodated by the conserved Rho protein structure: Both N and C termini emerge from the same face of the folded protein and are spatially proximal (Ihara et al., 1998; Wei et al., 1997). While the majority of mammalian Rho/Rac-family actin regulators appear to tether to membranes exclusively through C-terminal lipidations and polybasic sequences, a few, like yeast Rho3, have cysteine-containing N-terminal sequences suggestive of possible dual N-terminal tethering: most notably RhoJ, but also RhoU, Rnd1, Rnd2, and Rnd3.
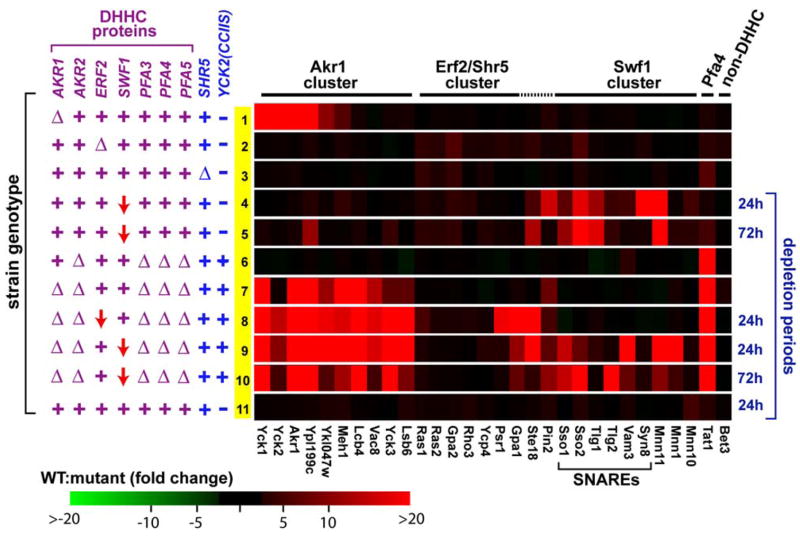
Analysis of DHHC PAT-Deficient Strains
We and others have suggested that the DHHC protein family may function as a diverse family of PAT specificities (Lobo et al., 2002; Roth et al., 2002). To assess overall DHHC protein involvement in palmitoylation, we have applied our proteomic method to mutant yeast strains either singly or multiply deficient for the seven yeast DHHC proteins. Proteins relying on particular DHHC proteins for their palmitoylation were expected to be lost from the palmitoyl proteomes of the DHHC protein-deficient strain. Palmitoyl-proteome profiles deriving from MudPIT analyses of nine different DHHC mutant yeast strains are graphically summarized in Figure 4. Palmitoyl proteins underrepresented in the mutant strain profiles relative to wild-type are shaded red: Proteins fully lost (i.e., >20-fold underrepresentations relative to wild-type) are bright red, while proteins having more subtle underrepresentations are indicated with corresponding intermediate red shadings (Figure 4). To improve the statistical quality of this analysis, focus was limited to just the 30 most prominently detected palmitoyl proteins, i.e., those detected by the highest spectra count values.
Figure 4. Palmitoyl-Proteome Profiles of Different DHHC Protein-Deficient Strains.
For the 30 top-ranking palmitoyl proteins, EXP sample representations (based on normalized spectral counts; see Table S3 for supporting MS/MS data) from the different mutant strains were compared to averaged EXP representations derived from the four wild-type MS/MS runs. The relevant genotypes of the different mutant strains are indicated at left: wild-type alleles (+), deletion alleles (Δ), or depletion alleles (red downward arrow). Proteins with 20-fold or greater mutant strain underrepresentations are depicted as red, with intermediate levels of underrepresentation converted to intermediate shadings of red as described (Supplemental Experimental Procedures). Similarly, proteins with mutant sample overrepresentations are depicted by shadings of green. (Subtle green shadings are faintly apparent for only a few boxes within this matrix.) For strains utilizing GAL1-driven ERF2 or SWF1 depletion alleles, the depletion time period (period of growth on glucose medium) is indicated at right. The row numbers in yellow at left allow the results from individual strains to be referenced from the text. An EXP sample from the isogenic wild-type parent strain was analyzed as a control (row 11). Note that some of the profiled strains harbor the akr1-suppressing lys2Δ::YCK2(CCIIS) allele (as indicated). The substantial Yck2 palmitoylation that is found to persist in some of the profiled akr1Δ strains (e.g., row 7) likely reflects ongoing Akr1-independent palmitoylation of the Yck2(CCIIS) mutant.
Substrates for the Akr1, Erf2-Shr5, and Swf1 PATs
Strains individually deficient for Akr1, Swf1, or Erf2 were profiled first. For akr1Δ cells (Figure 4, row 1), significant underrepresentations were found for six proteins: the two known Akr1 substrates, Yck1 and Yck2 (Roth et al., 2002); Akr1 itself; and Meh1, Ypl199c, and Ykl047w. While the loss of Akr1 from the profile is trivial (cells are akr1Δ), Meh1, Ypl199c, and Ykl047w may represent new Akr1 substrates; indeed, all three, when individually tested, were found to depend on Akr1 for palmitoylation (Figure 5A). In addition, two other palmitoyl proteins that ranked somewhat lower in our proteomic analysis, Sna4 and Ypl236c, also showed Akr1-dependent palmitoylation (Figure 5A). While comparison of these putative Akr1 substrates reveals no obvious consensus motif, there are some loose commonalities: Most, but not all, are soluble proteins that tether to bilayers solely through either N- or C-terminal palmitoylation (Figure 5A).
Figure 5. DHHC PAT-Dependent Palmitoylation.
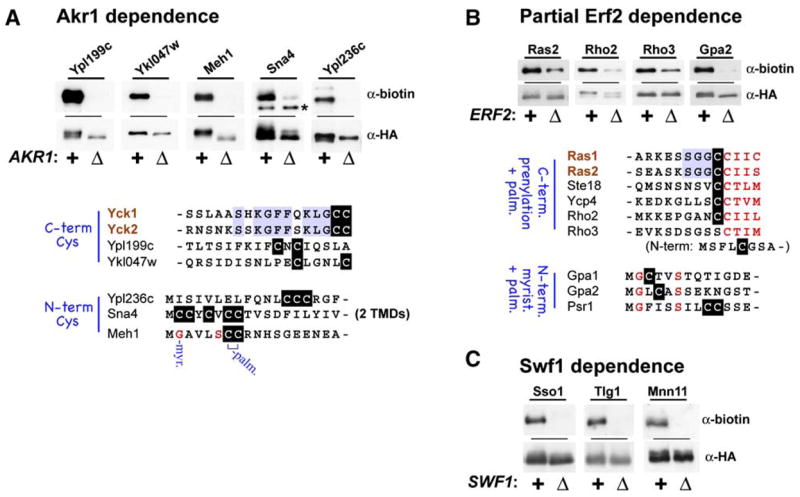
For the indicated proteins, palmitoylation in the wild-type strain context (+) was compared with that in the indicated DHHC gene-deletion strain context (Δ): akr1Δ (A), erf2Δ (B), or swf1Δ (C). Palmitoylation was assessed by the small-scale ABE protocol as described for Figure 3B. The tested proteins were expressed from the GAL1 promoter and were either N- or C-terminally tagged with the dual HA/FLAG epitope tag (see Table S2). For (A) and (B), putative Akr1 and Erf2-Shr5 substrates are listed and sequences surrounding likely palmitoyl-accepting cysteines are shown. Note that Sna4 and Meh1 are unique among the putative Akr1 substrates in that Sna4 has two predicted TMDs and Meh1 is predicted to be heterolipidated: myristoylated in addition to palmitoylated. The asterisk marks a nonspecific band that crossreacts with the anti-biotin IgG.
The yeast Ras PAT, like the recently identified mammalian H- and N-Ras PAT (Swarthout et al., 2005), appears to be composed of two subunits: the DHHC protein Erf2 and the non-DHHC protein Shr5 (Lobo et al., 2002). While Erf2-Shr5 mediates palmitoylation of Ras2, in the absence of this PAT—i.e., in erf2Δ, shr5Δ, or erf2Δ shr5Δ cells—substantial residual Ras2 palmitoylation continues (Bartels et al., 1999; Lobo et al., 2002), indicating that Erf2-Shr5-independent mechanisms also are capable of Ras2 palmitoylation. Consistent with this, our profiles of the erf2Δ and shr5Δ cell palmitoyl proteomes find Ras2, as well as the homologous and functionally redundant Ras1, to be underrepresented but not fully lost (Figure 4, rows 2 and 3, subtle red shadings). In erf2Δ cells, underrepresentations of 4.5- and 4-fold were found for Ras1 and Ras2, respectively, while 4.5- and 3-fold underrepresentations were found for shr5Δ cells (see Table S3 for supporting MS data). Similar 3- to 6-fold underrepresentations in erf2Δ and/or shr5Δ cells also are apparent for other palmitoyl proteins (Figure 4, rows 2 and 3), suggesting that these proteins also may partially rely on Erf2-Shr5 for palmitoylation; indeed, for Gpa2, Rho2, and Rho3, Erf2-dependent palmitoylation has been confirmed (Figure 5B). Interestingly, all of the nine proteins implicated to date as likely Erf2-Shr5 substrates are predicted to be heterolipidated, being either C-terminally prenylated-palmitoylated (Ras1, Ras2, Ste18, Ycp4, and Rho2), N-terminally myristoylated-palmitoylated (Gpa1, Psr1, Gpa2), or (in the case of Rho3) C-terminally prenylated and N-terminally palmitoylated (Figure 5B).
While none of the seven DHHC genes is individually required for yeast cell viability, a significant growth impairment was found to be associated with the swf1Δ allele (A.F.R. and N.G.D., unpublished data). Therefore, rather than profiling a swf1Δ strain, we opted instead to profile a GAL1-SWF1 strain that has the chromosomal regulatory sequences upstream of the SWF1 ORF replaced by the regulatable GAL1 promoter. The GAL1-SWF1 allele is conditional: GAL1-SWF1 cells grow normally on galactose medium and show impaired growth following a prolonged period of Swf1 depletion instituted through glucose-mediated repression of the GAL1 promoter. Following depletion periods of 24 hr or 72 hr, underrepresentations were found for many of the top palmitoyl proteins in the GAL1-SWF1 strain. Consistent with a recent report that implicated Swf1 in SNARE protein palmitoylation (Val-dez-Taubas and Pelham, 2005), significant underrepresentations were seen for many of the palmitoylated SNARE proteins (Figure 4, rows 4 and 5). Other proteins showing evidence of Swf1-dependent palmitoylation include the three mannosyltransferases, Mnn1, Mnn10, and Mnn11, as well as the yeast prion induction protein Pin2. Like the SNAREs, these other proteins all have juxta-TMD mapping cysteines (Figure 2), suggesting a Swf1 preference for substrates of this type. For the Sso1 and Tlg1 SNARE proteins and the mannosyltransferase subunit Mnn11, Swf1-dependent palmitoylation has been confirmed in individual tests (Figure 5C).
Pfa4: A New PAT for AAP Palmitoylation
Given the precedent of Erf2-Shr5-independent palmitoylation for Ras2 (Bartels et al., 1999; Lobo et al., 2002), we were concerned that many other palmitoyl proteins might also rely on the actions of multiple, overlapping PATs. Much of our analysis, therefore, has concentrated on strains with multiple DHHC gene deletions. Thus, an akr2Δ pfa3Δ pfa4Δ pfa5Δ strain deleted for four previously uncharacterized DHHC genes was profiled. Only one of the top palmitoyl proteins was found to fully drop out, this being the tryptophan permease Tat1 (Figure 4, row 6). To analyze the individual contributions of the four DHHC proteins deficient in this strain, Tat1 palmitoylation was assessed in strains individually deleted for the four DHHC genes. While the akr2Δ, pfa3Δ, or pfa5Δ mutations were without effect, in pfa4Δ cells, Tat1 palmitoylation was fully abolished (Figure 6A). To see whether Pfa4 might play a more general role in AAP palmitoylation, eight additional AAPs were examined for palmitoylation in wild-type and in pfa4Δ cells (Figure 6B). This survey included, in addition to three AAPs that were shown above to palmitoylated (Gnp1, Hip1, and Bap2), five AAPs of unknown palmitoylation status. Three of the five, Tat2, Agp1, and Gap1, have C-terminal Phe-Trp-Cys sequences (Figure 3A), while the other two, Lyp1 and Can1, do not. The results nicely extend prior results: The three Phe-Trp-Cys-containing AAPs are found to be palmitoylated, while the two lacking this motif are not (Figure 6B). Furthermore, for all eight of the palmitoylated AAPs, palmitoylation is found to be fully Pfa4 dependent (Figure 6B). Thus, like SNARE protein palmitoylation, AAP palmitoylation also has a dedicated PAT, Pfa4. These palmitoylated AAPs represent the first Pfa4 substrates and allow Pfa4 to be grouped with Akr1, Erf2-Shr5, Swf1, and the recently characterized Pfa3 (Smotrys et al., 2005) as a member of the yeast DHHC PAT family; five of the seven yeast DHHC proteins now have been directly linked to palmitoylation.
Figure 6. Pfa4-Dependent AAP Palmitoylation.
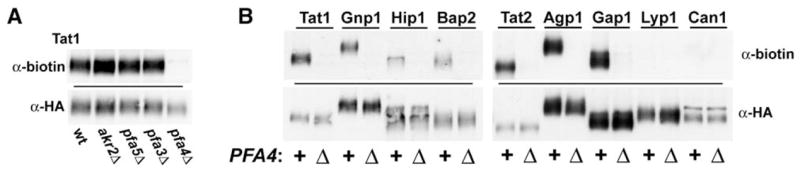
Palmitoylation of the indicated AAPs was assessed by the small-scale ABE protocol as described for Figure 3B.
(A) Deconvolution of the DHHC protein requirement for Tat1 palmitoylation.
(B) Pfa4-dependent palmitoylation for the Phe-Trp-Cys-containing AAPs.
Overlapping DHHC PAT Functionality
Though all seven DHHC gene deletions are individually nonessential, strain inviabilities are encountered with cumulative DHHC gene deletion. For instance, the viable akr2Δ pfa3Δ pfa4Δ pfa5Δ strain can sustain introduction of the akr1Δ allele only in the presence of the akr1 suppressor lys2Δ::YCK2(CCIIS). The Yck2(CCIIS) mutant expressed from lys2Δ::YCK2(CCIIS) has the Yck2 C-terminal, palmitoyl-accepting Cys-Cys dipeptide replaced by the five C-terminal residues of Ras2, i.e., Cys-Cys-Ile-Ile-Ser; like Ras2, Yck2(CCIIS) is farnesylated, palmitoylated, and trafficked to the plasma membrane independently of Akr1 function (Roth et al., 2002). lys2Δ::YCK2(CCIIS) restores casein kinase activity to the akr1Δ cell plasma membrane and thereby suppresses a variety of akr1-associated phenotypes, including its associated growth defect (Y. Feng and N.G.D., unpublished data).
Analysis of the 5-fold-deleted akr1Δ akr2Δ pfa3Δ pfa4Δ pfa5Δ strain finds significant underrepresentations for many of the top palmitoyl proteins (Figure 4, row 7). In addition to the expected loss of Akr1 and Pfa4 substrates, underrepresentations also are seen for many proteins that retain significant palmitoylation in both the akr1Δ and the akr2Δ pfa3Δ pfa4Δ pfa5Δ parental strains (Figure 4, rows 1 and 6). For these proteins, namely Lcb4, Vac8, Yck3, and Lsb6, Akr1 likely acts redundantly with one (or several) of the other DHHC proteins deleted, i.e., with either Akr2, Pfa3, Pfa4, or Pfa5. A recent report found palmitoylation of the sphingosine kinase Lcb4 to partially require Akr1 function; Lcb4 palmitoylation was reduced by about 60%–80% in akr1Δ cells (Kihara et al., 2005). Close inspection of our data finds a subtle, 2.6-fold reduction for Lcb4 representation in the akr1Δ cell sample (Figure 4, row 1 and Table S3). Building on this result, the present finding of abolished Lcb4 palmitoylation in the 5-fold-deleted strain (Figure 4, row 7), indicates that, in addition to Akr1, one or more of four other DHHC proteins—i.e., Akr2, Pfa3, Pfa4, and/or Pfa5—also can participate in Lcb4 palmitoylation. Similarly, a recent report found the palmitoylation of the vacuolar armadillo repeat protein Vac8 to partially depend on Pfa3 (Hou et al., 2005; Smotrys et al., 2005); coupling this to the current MS result (Figure 4, row 7) implicates Akr1 as the PAT responsible for the residual Vac8 palmitoylation that persists in pfa3Δ cells.
Our finding that Vac8 palmitoylation is fully DHHC PAT dependent contrasts with a prior report that found Vac8 palmitoylation to be fully DHHC PAT independent in vitro (Dietrich et al., 2004): In vitro, the atypical SNARE Ykt6 suffices to mediate Vac8 palmitoylation; indeed, just the 140 residue Ykt6 longin domain, which was also found to have binding affinity for palmitoyl-CoA and palmitic acid, suffices to promote palmitoylation (Dietrich et al., 2004). However, a caveat accompanying all in vitro demonstrations of protein palmitoylation is the well-known capacity of palmitoyl-CoA for direct chemical modification of substrate cysteines; indeed, in vitro, nonenzymatic palmitoylations have been demonstrated for a number of different proteins (Duncan and Gilman, 1996; Leventis et al., 1997; Veit, 2000). Consistent with view in which Ykt6 acts to augment a basal level of direct chemical palmitoylation, significant Vac8 palmitoylation is reported even in the absence of added Ykt6 (Dietrich et al., 2004). Efforts to investigate Ykt6 participation in palmitoylation in vivo are hampered by the YKT6 requirement for yeast cell viability.
Deficiencies for Swf1 and Erf2 could be introduced into the 5-fold-deleted akr1Δ akr2Δ pfa3Δ pfa4Δ pfa5Δ strain only as conditional-depletion alleles. Two strains were constructed by introducing either the GAL1-SWF1 or the GAL1-ERF2 depletion allele into the 5-fold-deleted strain. Profiles of the resulting GAL1-SWF1 6-fold-deficient strain (Figure 4, rows 9 and 10) yielded essentially the result expected for a simple summation of the previous GAL1-SWF1 strain result (Figure 4, rows 4 and 5) with the 5-fold-deleted strain result (Figure 4, row 7). In contrast, analysis of the GAL1-ERF2 6-fold-deficient strain found that three proteins, namely Gpa1, Psr1, and Ste18, that had shown partial underrepresentations in the erf2Δ strain (Figure 4, row 2) were now fully lost from the 6-fold-defi-cient strain profile (Figure 4, row 8). Thus, overlapping PAT substrate specificities are again implicated—in this instance, an overlap of Erf2-Shr5 with one (or several) of the five other DHHC activities deleted in this strain. The five other putative Erf2-Shr5 substrates, namely Ras1, Ras2, Gpa2, Rho3, and Ycp4, continue to show ongoing, residual palmitoylation in the 6-fold-deficient strain context (Figure 4, row 8), similar to that seen in erf2Δ and shr5Δ cells (Figure 4, rows 2 and 3).
The DHHC PAT Family
The DHHC proteins have emerged only recently as a possible family of PATs (Lobo et al., 2002; Roth et al., 2002). While the past year has seen a variety of different mammalian and yeast DHHC proteins linked to different palmitoylations (Ducker et al., 2004; Fukata et al., 2004; Hou et al., 2005; Huang et al., 2004; Keller et al., 2004; Smotrys et al., 2005; Swarthout et al., 2005; Valdez-Taubas and Pelham, 2005), the overall extent of the role of this family in palmitoylation has also been called into question by in vitro demonstrations of DHHC protein-independent palmitoylation (Dietrich et al., 2004; Dietrich and Ungermann, 2004), including apparently bona fide palmitoylations that proceed in vitro fully nonenzymatically, through direct chemical reaction of acceptor thiols with palmitoyl-CoA (Duncan and Gilman, 1996; Leventis et al., 1997; Veit, 2000). Our analysis of multiply DHHC protein-deficient strains, including two strains each deficient for six of the seven yeast DHHC proteins, highlights DHHC protein participations in the palmitoylation of 29 of the 30 palmitoyl proteins surveyed by this analysis (Figure 4), indicating that the DHHC PATs clearly mediate the bulk of the palmitoylation within the yeast cell.
Nonetheless, for some of the proteins, substantial palmitoylation persists even in strains multiply deficient for the DHHC proteins. For instance, five of the putative Erf2-Shr5 substrates, namely Ras1, Ras2, Rho3, Gpa2, and Ycp4, show reduced palmitoylation in erf2Δ cells that is not further diminished with disruption of five of the six other DHHC functions (Figure 4, row 8). The Erf2-Shr5-independent activity responsible for the residual palmitoylation of these five palmitoyl proteins remains unclear. Unfortunately, due to technical obstacles, we have been unable to profile strains fully deficient for all seven DHHC proteins; thus, the residual palmitoylation could still possibly reflect overlapping DHHC PAT action. However, it is also possible that the residual palmitoylation of these partial Erf2-Shr5 substrates is DHHC protein independent, mediated either by yet to be discovered non-DHHC PATs or perhaps by nonenzymatic mechanisms.
One protein, Bet3, shows a palmitoylation that is notably undiminished in any of the different DHHC mutant strains (Figure 4). Bet3 is a component of the TRAPP tethering complex for ER-to-Golgi trafficking, and its palmitoylation is noteworthy in other regards as well. The Bet3 palmitoyl modification was discerned from recent crystallographic structural analyses (Kim et al., 2005; Turnbull et al., 2005). Interestingly, the attached acyl chain, rather than being disposed as a possible tether, was found instead to be oriented inwards, being entirely buried within a close-fitting hydrophobic pocket of the protein. Furthermore, mutation of the cysteinyl acceptor site affects neither Bet3 functionality nor Bet3 capacity for peripheral association with its resident Golgi membranes. The acylated Bet3 for the two structural studies that reported palmitoylation was purified from either yeast (Turnbull et al., 2005) or overproducing E. coli (Kim et al., 2005). For E. coli-produced Bet3, three forms were reported: a nonacylated form as well as forms having either the 16-carbon palmitoyl or the 14-carbon myristoyl moiety attached to Cys80 in thioester linkage. The finding of protein acylation occurring in E. coli is a bit surprising: Bacteria lack both DHHC proteins as well as any evidence for protein acyl modifications similar to protein palmitoylation that is prevalent in eukaryotes. Might Bet3 palmitoylation be autocatalytic? This could explain both the acylation of Bet3 in bacteria and also the DHHC PAT-independent palmitoylation that we now document for Bet3 (Figure 4). Palmitoyl-CoA, we note, if fitted into the deep acyl binding pocket of Bet3, would be nicely positioned for direct reaction with the acceptor cysteine that locates at the mouth of the hydrophobic pocket on the Bet3 surface.
Overall, we find that substrates of the DHHC PATs tend to group by obvious common features. For instance, the putative Swf1 substrates—a group that includes the palmitoylated SNAREs as well as Mnn1, Mnn10, Mnn11, and Pin2—all have cysteines that map cytoplasmically adjacent to TMDs (Figure 2). Likewise, we identify Pfa4 as a new member of the DHHC PAT family that is apparently devoted to palmitoylation of the AAPs, a family of plasma-membrane transporters with 12 TMDs and a conserved C-terminal Phe-Trp-Cys palmitoylation site (Figure 3A and Figure 6B). Erf2-Shr5 substrates all appear to be heterolipidated, being either prenylated or myristoylated in addition to being palmitoylated (Figure 5B), and Akr1 substrates tend to be hydrophilic proteins that tether to membranes solely through N- or C-terminal palmitoyl modifications (Figure 5A). We conclude that the DHHC protein family is a family of both discrete and overlapping PAT specificities—a diverse range of specificities that together mediate the bulk of the diverse palmitoylations that occur within the eukaryotic cell.
EXPERIMENTAL PROCEDURES
Strains
Strains isogenic to LRB759 (Panek et al., 1997) were utilized throughout, except for the testing of putative Erf2 substrates (Figure 5B) and the DHHC protein dependence of AAP palmitoylation (Figure 6), which utilized Yeast Deletion Consortium strains (ResGen/Invitrogen) (see Table S4 for complete strain list). The construction of the multiply DHHC gene-deleted strains is described in the Supplemental Experimental Procedures.
Purification of Palmitoyl Proteins for MS Analysis
Protein extracts derived from total membranes of 60 × 109 log-phase yeast cells grown in YEPD were subjected to the three chemical treatment steps of the acyl-biotinyl exchange (ABE) protocol (see Supplemental Experimental Procedures). Protein was transferred between chemical treatment steps by collecting proteins at the conclusion of one treatment step by chloroform-methanol precipitation (Wessel and Flugge, 1984), which was followed by a 10 min, 37°C resolubilization and denaturation in SB (4% SDS, 50 mM Tris, 5 mM EDTA [pH 7.4]) and then a 4-fold dilution into the chemicals comprising the next treatment step. Proteins biotinylated by ABE were bound to streptavidin agarose, washed with buffer, and eluted through cleavage of the cysteine-biotin disulfide linkage with 1% β-mercaptoethanol.
Mass Spectrometry/MudPIT
Purified palmitoyl proteins were reduced with 5 mM TCEP (Pierce), alkylated with 10 mM iodoacetamide, and proteolyzed with endo-LysC and trypsin. Peptides resolved by the in-line 2D-chromatographic separation steps that characterize MudPIT (Link et al., 1999; Washburn et al., 2001) were introduced as they eluted from the microcapillary columns over an 8 hr period directly into a LTQ ion tandem mass spectrometer (Thermo Finnigan). Peptide identifications were made by SEQUEST (Eng et al., 1994) correlation of tandem mass spectrometry data to theoretical spectra derived from the translated Saccharomyces Genome Database (SGD; 05/23/03 release) (Cherry et al., 1998). For additional detail, see Supplemental Experimental Procedures.
Testing Palmitoylation for Individual Proteins
To confirm candidate protein palmitoylation and also DHHC protein dependencies, epitope-tagged versions of individual proteins were tested for palmitoylation. ORFs for the tested proteins were amplified from yeast genomic DNA to GAL1-driven expression plasmids based on pRS316 (CEN/ARS/URA3) that attached either a HA/FLAG dual epitope tag to the N terminus or a 3× HA/FLAG tag to the C terminus. Following a period of galactose-induced expression, palmitoylation was assessed either through analysis of in vivo [3H]palmitate incorporation (Roth et al., 2002) or with a scaled-down version of the ABE proteomic protocol. For additional detail, see Supplemental Experimental Procedures.
Supplementary Material
Supplemental Data
Supplemental Data include Supplemental Experimental Procedures, one figure, four tables, and Supplemental References and can be found with this article online at http://www.cell.com/cgi/content/full/125/5/1003/DC1/.
Acknowledgments
We thank Ed Ziff (New York University School of Medicine) for early insights leading up to this work, Charlie Boone (University of Toronto) for critiquing the manuscript, and Anthony Cimini and Amina Ahmed (Wayne State University) for constructing some of the plasmids used in this work. This work was supported by NIH grants GM65525 (N.G.D.) and RR11823 (J.R.Y.).
References
- Bartels DJ, Mitchell DA, Dong X, Deschenes RJ. Erf2, a novel gene product that affects the localization and palmitoylation of Ras2 in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6775–6787. doi: 10.1128/mcb.19.10.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock JB, Matern HT, Peden AA, Scheller RH. A genomic perspective on membrane compartment organization. Nature. 2001;409:839–841. doi: 10.1038/35057024. [DOI] [PubMed] [Google Scholar]
- Brown DA, London E. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, et al. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couve A, Protopopov V, Gerst JE. Yeast synaptobrevin homologs are modified posttranslationally by the addition of palmitate. Proc Natl Acad Sci USA. 1995;92:5987–5991. doi: 10.1073/pnas.92.13.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich LE, Ungermann C. On the mechanism of protein palmitoylation. EMBO Rep. 2004;5:1053–1057. doi: 10.1038/sj.embor.7400277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich LE, Gurezka R, Veit M, Ungermann C. The SNARE Ykt6 mediates protein palmitoylation during an early stage of homotypic vacuole fusion. EMBO J. 2004;23:45–53. doi: 10.1038/sj.emboj.7600015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich LE, Peplowska K, LaGrassa TJ, Hou H, Rohde J, Ungermann C. The SNARE Ykt6 is released from yeast vacuoles during an early stage of fusion. EMBO Rep. 2005;6:245–250. doi: 10.1038/sj.embor.7400350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drisdel RC, Green WN. Labeling and quantifying sites of protein palmitoylation. Biotechniques. 2004;36:276–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- Ducker CE, Stettler EM, French KJ, Upson JJ, Smith CD. Huntingtin interacting protein 14 is an oncogenic human protein: palmitoyl acyltransferase. Oncogene. 2004;23:9230–9237. doi: 10.1038/sj.onc.1208171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JA, Gilman AG. Autoacylation of G protein alpha subunits. J Biol Chem. 1996;271:23594–23600. doi: 10.1074/jbc.271.38.23594. [DOI] [PubMed] [Google Scholar]
- Dunphy JT, Linder ME. Signalling functions of protein palmitoylation. Biochim Biophys Acta. 1998;1436:245–261. doi: 10.1016/s0005-2760(98)00130-1. [DOI] [PubMed] [Google Scholar]
- el-Husseini Ael D, Bredt DS. Protein palmitoylation: a regulator of neuronal development and function. Nat Rev Neurosci. 2002;3:791–802. doi: 10.1038/nrn940. [DOI] [PubMed] [Google Scholar]
- El-Husseini Ael D, Schnell E, Dakoji S, Sweeney N, Zhou Q, Prange O, Gauthier-Campbell C, Aguilera-Moreno A, Nicoll RA, Bredt DS. Synaptic strength regulated by palmitate cycling on PSD-95. Cell. 2002;108:849–863. doi: 10.1016/s0092-8674(02)00683-9. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Fukasawa M, Varlamov O, Eng WS, Sollner TH, Rothman JE. Localization and activity of the SNARE Ykt6 determined by its regulatory domain and palmitoylation. Proc Natl Acad Sci USA. 2004;101:4815–4820. doi: 10.1073/pnas.0401183101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M, Fukata Y, Adesnik H, Nicoll RA, Bredt DS. Identification of PSD-95 palmitoylating enzymes. Neuron. 2004;44:987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Harashima T, Heitman J. Gα subunit Gpa2 recruits kelch repeat subunits that inhibit receptor-G protein coupling during cAMP-induced dimorphic transitions in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:4557–4571. doi: 10.1091/mbc.E05-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H, Subramanian K, LaGrassa TJ, Markgraf D, Dietrich LE, Urban J, Decker N, Ungermann C. The DHHC protein Pfa3 affects vacuole-associated palmitoylation of the fusion factor Vac8. Proc Natl Acad Sci USA. 2005;102:17366–17371. doi: 10.1073/pnas.0508885102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, El-Husseini A. Modulation of neuronal protein trafficking and function by palmitoylation. Curr Opin Neurobiol. 2005;15:527–535. doi: 10.1016/j.conb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Huang K, Yanai A, Kang R, Arstikaitis P, Singaraja RR, Metzler M, Mullard A, Haigh B, Gauthier-Campbell C, Gutekunst CA, et al. Huntingtin-interacting protein HIP14 is a palmitoyl transferase involved in palmitoylation and trafficking of multiple neuronal proteins. Neuron. 2004;44:977–986. doi: 10.1016/j.neuron.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Ihara K, Muraguchi S, Kato M, Shimizu T, Shirakawa M, Kuroda S, Kaibuchi K, Hakoshima T. Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue. J Biol Chem. 1998;273:9656–9666. doi: 10.1074/jbc.273.16.9656. [DOI] [PubMed] [Google Scholar]
- Jones TL, Degtyarev MY, Backlund PS., Jr The stoichiometry of G alpha(s) palmitoylation in its basal and activated states. Biochemistry. 1997;36:7185–7191. doi: 10.1021/bi9628376. [DOI] [PubMed] [Google Scholar]
- Keller CA, Yuan X, Panzanelli P, Martin ML, Alldred M, Sassoe-Pognetto M, Luscher B. The gamma2 subunit of GABA(A) receptors is a substrate for palmitoylation by GODZ. J Neurosci. 2004;24:5881–5891. doi: 10.1523/JNEUROSCI.1037-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Kurotsu F, Sano T, Iwaki S, Igarashi Y. Long-chain base kinase Lcb4 Is anchored to the membrane through its palmitoylation by Akr1. Mol Cell Biol. 2005;25:9189–9197. doi: 10.1128/MCB.25.21.9189-9197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Sohn EJ, Seo J, Lee KJ, Lee HS, Hwang I, White-way M, Sacher M, Oh BH. Crystal structure of bet3 reveals a novel mechanism for Golgi localization of tethering factor TRAPP. Nat Struct Mol Biol. 2005;12:38–45. doi: 10.1038/nsmb871. [DOI] [PubMed] [Google Scholar]
- Leventis R, Juel G, Knudsen JK, Silvius JR. Acyl-CoA binding proteins inhibit the nonenzymic S-acylation of cysteinyl-containing peptide sequences by long-chain acyl-CoAs. Biochemistry. 1997;36:5546–5553. doi: 10.1021/bi963029h. [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Lobo S, Greentree WK, Linder ME, Deschenes RJ. Identification of a Ras palmitoyltransferase in Saccharomyces cerevi-siae. J Biol Chem. 2002;277:41268–41273. doi: 10.1074/jbc.M206573200. [DOI] [PubMed] [Google Scholar]
- Moffett S, Rousseau G, Lagace M, Bouvier M. The palmitoylation state of the beta(2)-adrenergic receptor regulates the synergistic action of cyclic AMP-dependent protein kinase and beta-adrenergic receptor kinase involved in its phosphorylation and desensitization. J Neurochem. 2001;76:269–279. doi: 10.1046/j.1471-4159.2001.00005.x. [DOI] [PubMed] [Google Scholar]
- Panek HR, Stepp JD, Engle HM, Marks KM, Tan PK, Lemmon SK, Robinson LC. Suppressors of YCK-encoded yeast casein kinase 1 deficiency define the four subunits of a novel clathrin AP-like complex. EMBO J. 1997;16:4194–4204. doi: 10.1093/emboj/16.14.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- Roth AF, Feng Y, Chen L, Davis NG. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J Cell Biol. 2002;159:23–28. doi: 10.1083/jcb.200206120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smotrys JE, Linder ME. Palmitoylation of intracellular signaling proteins: regulation and function. Annu Rev Biochem. 2004;73:559–587. doi: 10.1146/annurev.biochem.73.011303.073954. [DOI] [PubMed] [Google Scholar]
- Smotrys JE, Schoenfish MJ, Stutz MA, Linder ME. The vacuolar DHHC-CRD protein Pfa3p is a protein acyltransferase for Vac8p. J Cell Biol. 2005;170:1091–1099. doi: 10.1083/jcb.200507048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem. 2005;280:31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- Tu Y, Wang J, Ross EM. Inhibition of brain Gz GAP and other RGS proteins by palmitoylation of G protein alpha subunits. Science. 1997;278:1132–1135. doi: 10.1126/science.278.5340.1132. [DOI] [PubMed] [Google Scholar]
- Turnbull AP, Kummel D, Prinz B, Holz C, Schultchen J, Lang C, Niesen FH, Hofmann KP, Delbruck H, Behlke J, et al. Structure of palmitoylated BET3: insights into TRAPP complex assembly and membrane localization. EMBO J. 2005;24:875–884. doi: 10.1038/sj.emboj.7600565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez-Taubas J, Pelham H. Swf1-dependent palmitoylation of the SNARE Tlg1 prevents its ubiquitination and degradation. EMBO J. 2005;24:2524–2532. doi: 10.1038/sj.emboj.7600724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit M. Palmitoylation of the 25-kDa synaptosomal protein (SNAP-25) in vitro occurs in the absence of an enzyme, but is stimulated by binding to syntaxin. Biochem J. 2000;345:145–151. [PMC free article] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wedegaertner PB, Bourne HR. Activation and depalmitoylation of Gs alpha. Cell. 1994;77:1063–1070. doi: 10.1016/0092-8674(94)90445-6. [DOI] [PubMed] [Google Scholar]
- Wei Y, Zhang Y, Derewenda U, Liu X, Minor W, Nakamoto RK, Somlyo AV, Somlyo AP, Derewenda ZS. Crystal structure of RhoA-GDP and its functional implications. Nat Struct Biol. 1997;4:699–703. doi: 10.1038/nsb0997-699. [DOI] [PubMed] [Google Scholar]
- Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane micro-domains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data
Supplemental Data include Supplemental Experimental Procedures, one figure, four tables, and Supplemental References and can be found with this article online at http://www.cell.com/cgi/content/full/125/5/1003/DC1/.