Abstract
In an effort to develop an AIDS vaccine that elicits high-frequency cytotoxic-T-lymphocyte (CTL) responses with specificity for a diversity of viral epitopes, we explored two prototype multiepitope plasmid DNA vaccines in the simian-human immunodeficiency virus/rhesus monkey model to determine their efficiency in priming for such immune responses. While a simple multiepitope vaccine construct demonstrated limited immunogenicity in monkeys, this same multiepitope genetic sequence inserted into an immunogenic simian immunodeficiency virus gag DNA vaccine elicited high-frequency CTL responses specific for all of the epitopes included in the vaccine. Both multiepitope vaccine prototypes primed for robust epitope-specific CTL responses that developed following boosting with recombinant modified vaccinia virus Ankara vaccines expressing complete viral proteins. The natural hierarchy of immunodominance for these epitopes was clearly evident in the boosted monkeys. These studies suggest that multiepitope plasmid DNA vaccine-based prime-boost regimens can efficiently prime for CTL responses of increased breadth and magnitude, although they do not overcome predicted hierarchies of immunodominance.
A central role for cytotoxic T lymphocytes (CTL) in containing human immunodeficiency virus (HIV) replication in humans and simian immunodeficiency virus (SIV) replication in monkeys has been suggested by the clear temporal correlation between the partial control of replicating virus during primary infection and the emergence of detectable virus-specific CTL in infected individuals (17). Moreover, monkeys depleted of CD8+ lymphocytes by monoclonal antibody infusion never contained early viral replication and died soon after infection (22). In fact, recent studies with monkeys indicate that vaccine-elicited CTL populations can confer protection against high levels of viral replication and clinical disease progression following simian-HIV (SHIV) or simian immunodeficiency virus (SIV) infection (2, 3, 6).
It is also now assumed that ideal vaccine-elicited HIV-specific CTL responses should recognize the greatest possible diversity of viral epitopes (19). However, studies in HIV-infected humans and SIV-infected monkeys have shown that replicating viruses accrue mutations that allow them to escape from recognition by CTL (12, 20, 21). Importantly, it has also been shown that the immune protection against clinical disease progression observed in monkeys that receive a CTL-based vaccine and are then challenged with an AIDS virus can be lost as viruses emerge with selected mutations that allow them to escape from recognition by CTL (4). It has been postulated that, the greater the diversity of viral epitopes recognized by CTL, the lower the likelihood of emergence of a population of viruses that can escape from recognition by these CTL.
During the course of a viral infection, CTL responses develop a predictable bias in their pattern of epitope recognition. A natural hierarchy of epitope recognition emerges, with most of the CTL response being focused on only a very limited number of epitopes (7, 11). For example, in SHIV- and SIV-infected rhesus monkeys that express the major histocompatibility complex (MHC) class I allele Mamu-A*01, CTL specific for the Gag p11C epitope invariably constitute a dominant response, with the envelope p41A and the polymerase p68A epitopes being subdominant CTL targets (11). Experimental evidence in a variety of systems suggests that this hierarchy develops as a consequence of factors such as the variety of epitope peptide affinities for the relevant MHC class I molecule, the various copy numbers of the epitopes produced by the virus, and differences in epitope processing by the cell's biologic machinery (24). It is conceivable that a vaccine strategy that can bypass the natural hierarchy of epitope bias might result in a particularly broad CTL response that provides longer-lived protection against an infecting AIDS virus.
Vaccine constructs that express a series of minimal epitopes have been explored as a potential strategy for generating immune responses to a variety of viral epitopes (23). Both single (8) and multiepitope (16) constructs have been shown to be immunogenic in mouse models. Limited numbers of studies with nonhuman primates, primarily focusing on the responses to a single dominant epitope, suggest that this approach can elicit detectable CTL responses (1, 14). The diversity and the magnitude of CTL responses generated in primates by multiepitope vaccines carrying epitopes with different degrees of dominance are presently not well understood. The present study was initiated in the Mamu-A*01+ rhesus monkey/SHIV model to systematically evaluate polyepitope vaccine-generated immune responses against both dominant and subdominant epitopes.
Construction of plasmid DNA vaccine.
To explore vaccine strategies that might overcome the natural bias of the immune system to focus CTL responses onto a limited number of dominant epitopes, a multiepitope plasmid DNA vaccine was constructed that encoded a series of peptide fragments of SHIV-89.6 previously shown to be presented to CTL in rhesus monkeys by the MHC class I molecule Mamu-A*01. The 5′-to-3′ order of the epitopes in this vaccine construct was as follows: the control epitope p199A (RYPKTFGWL), HIV-Env p41A (YAPPITGQI), Pol p68A (STPPLVRLV), and Gag p11C (CTPYDINQM), separated one from another with triple alanine spacers (Fig. 1). Such alanine spacers have previously been shown to minimize cleavage bias at epitope junctions that may vary due to the naturally occurring C- and N-terminal sequences of the minimal epitopes themselves (23). The multiepitope construct included a Kozak sequence at the 5′ end, flanking XbaI sites to facilitate its transfer, and a C-terminal myc sequence to facilitate its detection by anti-myc antibodies. DNA-coding sequences were refined to remove potential internal translation initiation sequences and extended tandem or inverted repeats. When not countermanded by these restrictions, codon optimization was also employed in constructing this plasmid DNA vaccine. The synthetic nucleotide sequence was then incorporated into the pV1R DNA vaccine vector (10).
FIG. 1.
Multiepitope sequence encoded by the DNA vaccine construct. Sequences corresponding to the minimal 9-amino-acid-long epitopes p41A, p68A, and p11C, all restricted by the class I allele Mamu-A*01 and derived, respectively, from HIV-1 89.6 envelope, SIVmac239 gag, and SIVmac239 pol, were placed in tandem. Each epitope was flanked on the amino and carboxyl termini by a triple-alanine-spacer sequence. The construct also included an SIV-derived control sequence (p199A) that is not restricted by Mamu-A*01 and a c-myc antibody tag.
Immunogenicity of prototype multiepitope plasmid DNA vaccine.
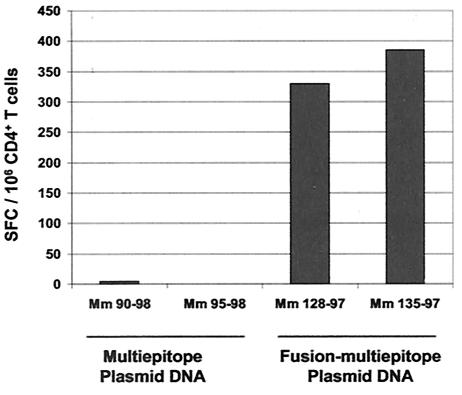
The immunogenicity of this polyepitope vaccine was assessed in a pilot study with two Mamu-A*01+ rhesus monkeys (Mm 90-98 and Mm 95-98) to determine whether the vaccine can generate cytotoxic-T-cell responses of comparable magnitude against all of the epitopes. The animals included in this study were Indian-origin rhesus monkeys selected for their expression of the MHC class I allele Mamu-A*01. A PCR assay employing sequence-specific primers was used to identify Mamu-A*01-positive monkeys (18). Monkeys were maintained in the biologic containment facility of the New England Primate Research Center (Southborough, Mass.) in accordance with the guidelines of the Committee on Animals for the Harvard Medical School and the Guide for the Care and Use of Laboratory Animals. The monkeys were immunized with 8 mg of plasmid DNA delivered by a needle-free Biojector (Bioject Medical Technologies, Inc.) in the left and right quadriceps muscles on a schedule of 0, 4, and 9 weeks. The vaccine-elicited CTL responses specific for the three Mamu-A*01-restricted epitopes and single control epitope were evaluated on a fortnightly schedule following immunization by staining whole blood with Mamu-A*01-peptide tetramer complexes. Tetrameric Mamu-A*01-p11C, -p41A, and -p68A complexes coupled to phycoerythrin were generated as previously described (11). These tetramers were used to stain 100 μl of fresh whole blood on the days noted postvaccination with a cocktail of antibodies that included allophycocyanin-linked anti-CD3 (custom-conjugated FN-18; Beckman Coulter), fluorescein isothiocynate-linked anti-CD8α (SK1; Becton-Dickinson), and phycoerythrin-Texas red-linked anti-CD8αβ (2ST8.5H7; Beckman Coulter) with use of the Immunoprep reagent system and a Q-prep workstation (Beckman Coulter). No tetramer-binding CD8+ T lymphocytes were detected in these sampled fresh peripheral blood cell populations, indicating that high-frequency CTL responses were not generated by these monkeys (Fig. 2). However, low-frequency CTL populations specific for Gag p11C and Env p41A epitopes could be expanded in the lymphocyte population from Mm 95-98 by epitope-peptide stimulation (data not shown). These studies indicated that this polyepitope plasmid DNA vaccine was at best marginally immunogenic in rhesus monkeys as a stand-alone vaccine.
FIG. 2.
Kinetics of the epitope-specific CTL responses detected in freshly isolated peripheral blood of monkeys immunized with the multiepitope or the fusion-multiepitope plasmid DNA immunogen. The upper panels show tetramer-specific CD8+-T-lymphocyte responses of the two monkeys (Mm 90-98 and Mm 95-98) that received three inoculations (weeks 0, 4, and 9) of the multiepitope plasmid DNA. The lower panels show tetramer-specific CD8+-T-lymphocyte responses of the two monkeys (Mm 128-97 and Mm 135-97) that received three inoculations (weeks 0, 4, and 8) of the fusion-multiepitope plasmid DNA. Peripheral blood was assessed for p11C-, p41A-, and p68A-tetramer binding to CD8+ CD3+ lymphocytes. Differences in the time to peak of epitope-specific responses following multiple DNA inoculations in the fusion multiepitope group of immunized monkeys may be due to the inherent biologic variability between the two animals. −, data not available.
Immunogenicity of fusion-multiepitope plasmid DNA vaccine.
We next considered the possibility that the multiepitope sequence may require presentation in a different immunologic context to optimally generate responses against the constituent epitope peptides. Since we have previously shown that a codon-optimized SIV gag DNA immunogen in this same plasmid vector is immunogenic, we reasoned that presenting the DNA encoding the Mamu-A*01-restricted epitopes in the context of that same SIV gag DNA should provide an appropriately immunogenic context for this vaccine. We therefore constructed a new plasmid DNA vaccine comprised of the same multiepitope sequence fused to the coding region of the carboxyl terminus of the full-length gag gene. However, to insure that a vaccine-elicited p11C-specific CTL response represented an immune response induced by the multiepitope sequence rather than p11C in the context of the gag gene product, we deleted the internal p11C coding sequence of the gag gene. The fusion of the multiepitope sequence with the gag gene was accomplished by inserting the short multiepitope sequence as a tag just before the stop codon of the full-length gag gene with a deletion of p11C.
As in the pilot study of the prototype multiepitope vaccine construct, two naïve Mamu-A*01+ rhesus monkeys (Mm 128-97 and Mm 135-97) were vaccinated with 8 mg of inocula of the fusion-multiepitope DNA vaccine by intramuscular route with a Biojector, each monkey receiving three inoculations at monthly intervals. Circulating CD8+ T lymphocytes binding the Mamu-A*01-p11C, -p41A, and -p68A tetramers were measured in both freshly isolated whole blood and epitope peptide-stimulated peripheral blood mononuclear cells (PBMC). In contrast to the simple multiepitope vaccine construct, the fusion-multiepitope vaccine construct elicited readily detectable populations of CD8+ tetramer-binding T lymphocytes as early as 2 weeks following the first plasmid DNA inoculation (Fig. 2). In fact, peripheral blood CD8+ T lymphocytes specific for each of the Mamu-A*01-restricted epitopes were generated by this vaccine at levels comparable to those generated by vaccines expressing full-length proteins in earlier studies (10). Moreover, fresh blood tetramer staining to assess peptide-specific CD8+-T-lymphocyte responses in circulating PBMC (Fig. 2) indicated that the fusion-multiepitope vaccine elicited readily detectable CTL responses against all three epitopes without the usual marked bias toward the dominant p11C epitope (11).
To clarify why a weakly immunogenic multiepitope DNA sequence can elicit robust CD8+-T-cell responses when configured as a fusion construct with a DNA sequence encoding a large immunogenic protein, we assessed Gag-specific CD4+-T-cell responses in PBMC of the monkeys vaccinated with either the stand-alone multiepitope vaccine or the fusion-multiepitope vaccine (Fig. 3). To evaluate the SIV Gag-specific CD4+-T-cell responses in the vaccinated monkeys, we assessed gamma interferon (IFN-γ) production by CD8+-lymphocyte-depleted PBMC in an ELISPOT assay following exposure to a pool of overlapping 15-amino-acid peptides spanning the entire SIV Gag protein as previously described (4). An evaluation of ELISPOT responses of such CD4+-T-cell-enriched PBMC of the vaccinated monkeys demonstrated that substantial Gag-specific CD4+-T-cell responses were generated in the monkeys immunized with the fusion-multiepitope construct but not the multiepitope construct alone. This observation suggests that insufficient CD4+-T-lymphocyte help may have been generated in monkeys immunized with the multiepitope vaccine construct alone to induce robust CD8+-T-lymphocyte responses to multiple CTL epitopes.
FIG. 3.
Multiepitope and fusion-multiepitope plasmid DNA immunogen elicited Gag-specific CD4+-T-lymphocyte IFN-γ ELISPOT responses. Two weeks following the third DNA inoculation, CD8+-lymphocyte-depleted PBMC from the vaccinated monkeys were exposed to a pool of 15-amino-acid-long peptides spanning the SIV Gag protein and were assessed for IFN-γ production.
Multiepitope DNA vaccine priming for live recombinant vector-boosted CTL responses.
If a plasmid DNA immunogen is used in HIV vaccination, it will likely be as a priming step for a live recombinant vector booster to potentiate vaccine-elicited CTL and/or antibody responses. We therefore sought to determine the efficiency of CTL priming induced by these multiepitope plasmid DNA immunogens for a recombinant poxvirus vector booster. We boosted each of the plasmid DNA-primed monkeys with two modified vaccinia virus Ankara (MVA) recombinants, one carrying the full-length SIV Gag-Pol proteins and the other the HIV-1 89.6 Env protein, constructed as described previously (5, 9). These SIV and HIV genes encode the three Mamu-A*01-restricted epitopes included in the multiepitope plasmid DNA immunogens. Following a fourth and final DNA immunization and an identical 10-month resting period, each of the DNA-primed monkeys was inoculated with 108 PFU of both of the recombinant MVA constructs by the intramuscular route. We then compared the immune responses elicited in these four DNA-primed, recombinant MVA-boosted monkeys to those of a group of previously naïve Mamu-A*01+ rhesus monkeys that were immunized only with the two recombinant MVA immunogens.
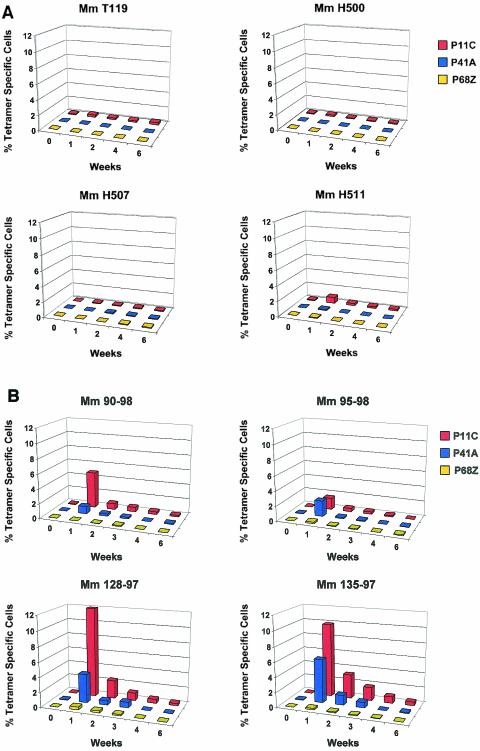
Vaccine-elicited CTL responses were monitored in these monkeys by quantitating tetramer-binding CD8+ T lymphocytes in freshly isolated peripheral blood samples (Fig. 4 and 5). The control monkeys that received only the recombinant MVA immunogens developed low-frequency CTL responses that were predominantly focused on the dominant p11C Gag epitope (Fig. 5A). Very-low-frequency and sporadic CTL responses were detected with specificity for the nondominant p41A and p68A epitopes. However, the monkeys that received priming immunizations with the multiepitope plasmid DNA vaccines developed readily demonstrable CTL responses specific for subdominant epitopes (Fig. 5B). The two monkeys that were immunized with the fusion-multiepitope construct had consistently higher-frequency responses to all three epitopes than did the two monkeys immunized with the simple multiepitope plasmid construct. These augmented epitope-specific CTL responses in the fusion multiepitope DNA-primed monkeys following the poxvirus booster suggest that an optimal DNA priming step may lead to augmented responses to the booster (compare Fig. 5B to Fig. 2).
FIG. 4.
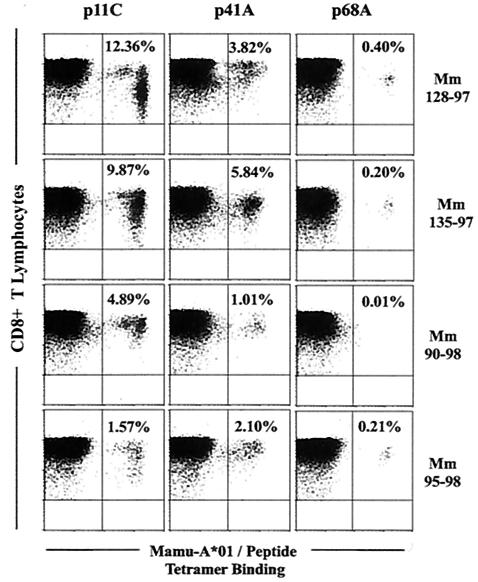
Epitope-specific CD8+-T-lymphocyte responses in peripheral blood 1 week following recombinant MVA boosting of monkeys primed with multiepitope and fusion-multiepitope plasmid DNA vaccines. Freshly isolated peripheral blood was assessed for p11C-, p41A-, and p68A-tetramer binding to CD8+ CD3+ lymphocytes. Monkeys Mm 90-98 and Mm 95-98 were primed with the multiepitope, and monkeys Mm128-97 and Mm135-97 were primed with the fusion-multiepitope plasmid DNA vaccine.
FIG. 5.
Kinetics of epitope-specific CTL responses as measured by peptide/tetramer binding to freshly isolated peripheral blood lymphocytes of monkeys that were vaccinated with either a recombinant MVA-priming/recombinant MVA-boosting (A) or a plasmid DNA-priming/recombinant MVA-boosting (B) regimen. Mm 90-98 and Mm95-98 were primed with the simple multiepitope DNA vaccine, while Mm128-97 and Mm135-97 were primed with the fusion-multiepitope DNA vaccines. Whole blood was obtained at the indicated time points following recombinant MVA boosting and was assessed for p11C-, p41A-, and p68A-tetramer binding to CD8+ CD3+ lymphocytes.
Interestingly, while the usual dominance hierarchy of epitopes was not apparent in the freshly stained peripheral blood of monkeys immunized with the fusion-multiepitope plasmid DNA vaccine constructs, this situation changed following recombinant MVA boosting. The monkeys that were primed with either the multiepitope or the fusion-multiepitope plasmid DNA vaccine constructs developed robust CTL responses specific for both the dominant and subdominant epitopes after recombinant MVA boosting. However, the usual dominance hierarchy seen during natural infection was readily apparent in three of the four monkeys, with the p11C epitope-specific CTL population detected at approximately a two-to-fourfold-higher frequency than CTL specific for the subdominant p41A epitope (Fig. 4). We cannot rule out the possibility that we may be able to alter these epitopic dominance patterns by boosting with recombinant poxvirus vaccines expressing increased copy numbers of the nondominant epitopes.
While vaccine constructs comprised of a series of defined CTL epitopes have been studied in mouse models (13, 15, 23), their utility in primates has not been extensively explored. The results of the present study suggest that a plasmid DNA encoding only a limited number of CTL epitopes is unlikely to be viable as a stand-alone vaccine. Even when enormous amounts of the multiepitope plasmid DNA vaccine were administered to rhesus monkeys, minimal CTL responses were generated.
Interestingly, however, a plasmid DNA vaccine construct containing the poorly immunogenic multiepitope DNA sequence fused to a gene encoding a large protein proved quite immunogenic. We cannot formally rule out the possibility that the poor immunogenicity of the prototype multiepitope construct was due to inefficient processing of the epitope peptides and that the processing of the fusion-multiepitope construct was improved. However, since we showed that DNA encoding the larger fusion protein induced CD4+-T-lymphocyte responses, it is likely that Gag-specific T-cell helper responses elicited by this larger protein augmented the CTL responses to the epitopes encoded by the multiepitope sequence. These vaccine-elicited CTL responses were in fact comparable in magnitude to those elicited in monkeys by plasmid DNA vaccines encoding full-length viral proteins (10).
Although the multiepitope plasmid DNA construct without the large protein did not elicit detectable CTL responses, immunization with this construct nonetheless primed for robust secondary epitope-specific CTL responses that were detected following recombinant MVA boosting. However, the monkeys that were immunized with the fusion-multiepitope plasmid DNA construct prior to recombinant MVA boosting generated impressive CTL responses to all three Mamu-A*01-restricted epitopes that were greater in magnitude than those seen in the monkeys that received the plasmid DNA encoding only the multiepitope sequence. Both multiepitope plasmid DNA immunogens therefore primed for secondary epitope-specific responses.
A rationale for exploring the use of multiepitope DNA immunogens as components of an HIV vaccine is that they may be capable of increasing the breadth of CTL epitope-specific priming. This presumably would occur through increasing immune responses to nondominant CTL epitopes. The present studies suggest that multiepitope plasmid DNA vaccines can efficiently prime for CTL responses of increased breadth but when employed in concert with established boosting regimens do not overcome usual hierarchies of immunodominance. Overcoming the proclivity of the immune system to focus CD8+-T-cell responses on a limited number of epitopes therefore remains a major challenge for the development of CTL-based vaccines for AIDS.
Acknowledgments
We gratefully acknowledge Bernard Moss and Linda S. Wyatt for making available the MVA constructs used in this study. We thank Carol I. Lord, Christine E. Nickerson, and Wenyu Lin for technical assistance and Keith A. Reimann and Karen Hershberger for helpful discussions.
We also acknowledge support from NIH grants CA-50139, AI-35351, and AI-85343.
REFERENCES
- 1.Allen, T. M., T. U. Vogel, D. H. Fuller, B. R. Mothe, S. Steffen, J. E. Boyson, T. Shipley, J. Fuller, T. Hanke, A. Sette, J. D. Altman, B. Moss, A. J. McMichael, and D. I. Watkins. 2000. Induction of AIDS virus-specific CTL activity in fresh, unstimulated peripheral blood lymphocytes from rhesus macaques vaccinated with a DNA prime/modified vaccinia virus Ankara boost regimen. J. Immunol. 164:4968-4978. [DOI] [PubMed] [Google Scholar]
- 2.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2002. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Vaccine 20:1949-1955. [DOI] [PubMed] [Google Scholar]
- 3.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 4.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 5.Barouch, D. H., S. Santra, M. J. Kuroda, J. E. Schmitz, R. Plishka, A. Buckler-White, A. E. Gaitan, R. Zin, J. H. Nam, L. S. Wyatt, M. A. Lifton, C. E. Nickerson, B. Moss, D. C. Montefiori, V. M. Hirsch, and N. L. Letvin. 2001. Reduction of simian-human immunodeficiency virus 89.6P viremia in rhesus monkeys by recombinant modified vaccinia virus Ankara vaccination. J. Virol. 75:5151-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 7.Berzofsky, J. A. 1991. Mechanisms of T cell recognition with application to vaccine design. Mol. Immunol. 28:217-223. [DOI] [PubMed] [Google Scholar]
- 8.Ciernik, I. F., J. A. Berzofsky, and D. P. Carbone. 1996. Induction of cytotoxic T lymphocytes and antitumor immunity with DNA vaccines expressing single T cell epitopes. J. Immunol. 156:2369-2375. [PubMed] [Google Scholar]
- 9.Earl, P. L., W. Sugiura, D. C. Montefiori, C. C. Broder, S. A. Lee, C. Wild, J. Lifson, and B. Moss. 2001. Immunogenicity and protective efficacy of oligomeric human immunodeficiency virus type 1 gp140. J. Virol. 75:645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egan, M. A., W. A. Charini, M. J. Kuroda, J. E. Schmitz, P. Racz, K. Tenner-Racz, K. Manson, M. Wyand, M. A. Lifton, C. E. Nickerson, T. Fu, J. W. Shiver, and N. L. Letvin. 2000. Simian immunodeficiency virus (SIV) gag DNA-vaccinated rhesus monkeys develop secondary cytotoxic T-lymphocyte responses and control viral replication after pathogenic SIV infection. J. Virol. 74:7485-7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egan, M. A., M. J. Kuroda, G. Voss, J. E. Schmitz, W. A. Charini, C. I. Lord, M. A. Forman, and N. L. Letvin. 1999. Use of major histocompatibility complex class I/peptide/β2M tetramers to quantitate CD8+ cytotoxic T lymphocytes specific for dominant and nondominant viral epitopes in simian-human immunodeficiency virus-infected rhesus monkeys. J. Virol. 73:5466-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans, D. T., D. H. O'Connor, P. Jing, J. L. Dzuris, J. Sidney, J. da Silva, T. M. Allen, H. Horton, J. E. Venham, R. A. Rudersdorf, T. Vogel, C. D. Pauza, R. E. Bontrop, R. DeMars, A. Sette, A. L. Hughes, and D. I. Watkins. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270-1276. [DOI] [PubMed] [Google Scholar]
- 13.Hanke, T., T. J. Blanchard, J. Schneider, C. M. Hannan, M. Becker, S. C. Gilbert, A. V. Hill, G. L. Smith, and A. McMichael. 1998. Enhancement of MHC class I-restricted peptide-specific T cell induction by a DNA prime/MVA boost vaccination regime. Vaccine 16:439-445. [DOI] [PubMed] [Google Scholar]
- 14.Hanke, T., R. V. Samuel, T. J. Blanchard, V. C. Neumann, T. M. Allen, J. E. Boyson, S. A. Sharpe, N. Cook, G. L. Smith, D. I. Watkins, M. P. Cranage, and A. J. McMichael. 1999. Effective induction of simian immunodeficiency virus-specific cytotoxic T lymphocytes in macaques by using a multiepitope gene and DNA prime-modified vaccinia virus Ankara boost vaccination regimen. J. Virol. 73:7524-7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanke, T., J. Schneider, S. C. Gilbert, A. V. Hill, and A. McMichael. 1998. DNA multi-CTL epitope vaccines for HIV and Plasmodium falciparum: immunogenicity in mice. Vaccine 16:426-435. [DOI] [PubMed] [Google Scholar]
- 16.Ishioka, G. Y., J. Fikes, G. Hermanson, B. Livingston, C. Crimi, M. Qin, M. F. del Guercio, C. Oseroff, C. Dahlberg, J. Alexander, R. W. Chesnut, and A. Sette. 1999. Utilization of MHC class I transgenic mice for development of minigene DNA vaccines encoding multiple HLA-restricted CTL epitopes. J. Immunol. 162:3915-3925. [PubMed] [Google Scholar]
- 17.Kuroda, M. J., J. E. Schmitz, W. A. Charini, C. E. Nickerson, M. A. Lifton, C. I. Lord, M. A. Forman, and N. L. Letvin. 1999. Emergence of CTL coincides with clearance of virus during primary simian immunodeficiency virus infection in rhesus monkeys. J. Immunol. 162:5127-5133. [PubMed] [Google Scholar]
- 18.Kuroda, M. J., J. E. Schmitz, W. A. Charini, C. E. Nickerson, C. I. Lord, M. A. Forman, and N. L. Letvin. 1999. Comparative analysis of cytotoxic T lymphocytes in lymph nodes and peripheral blood of simian immunodeficiency virus-infected rhesus monkeys. J. Virol. 73:1573-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMichael, A., and T. Hanke. 2002. The quest for an AIDS vaccine: is the CD8+ T-cell approach feasible? Nat. Rev. Immunol. 2:283-291. [DOI] [PubMed] [Google Scholar]
- 20.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493-499. [DOI] [PubMed] [Google Scholar]
- 21.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, et al. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453-459. [DOI] [PubMed] [Google Scholar]
- 22.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 23.Toes, R. E., R. C. Hoeben, E. I. van der Voort, M. E. Ressing, A. J. van der Eb, C. J. Melief, and R. Offringa. 1997. Protective anti-tumor immunity induced by vaccination with recombinant adenoviruses encoding multiple tumor-associated cytotoxic T lymphocyte epitopes in a string-of-beads fashion. Proc. Natl. Acad. Sci. USA 94:14660-14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yewdell, J. W., and J. R. Bennink. 1999. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 17:51-88. [DOI] [PubMed] [Google Scholar]