SUMMARY
ATPase-facilitated steps during spliceosome function have been postulated to afford opportunities for kinetic proof-reading. Spliceosome assembly requires the ATPase Prp5p, whose activity might thus impact fidelity during initial intron recognition. Using alanine mutations in S. cerevisiae Prp5p we identified a suboptimal intron whose splicing could be improved by altered Prp5p activity, then, using this intron, screened for potent prp5 mutants. These prp5 alleles specifically alter branch region selectivity, with improved splicing in vivo of suboptimal substrates correlating with reduced ATPase activity in vitro for a series of mutants in ATPase motif III (SAT). Because these effects are abrogated by compensatory U2 snRNA mutations or other changes that increase branch region–U2 pairing, these results explicitly link a fidelity event with a defined physical structure, the branch region–U2 snRNA duplex, and provide strong evidence that progression of the splicing pathway requires branch region–U2 snRNA pairing prior to Prp5p-facilitated conformational change.
INTRODUCTION
Both accuracy and flexibility in splice site selection are needed to generate functional mRNAs and to allow for the extensive alternative splicing observed in metazoans, but how such flexibility is achieved is not clear. The spliceosome, a dynamic complex that contains five snRNAs (U1, U2, U4, U5, and U6) and >150 proteins, recognizes three major intron elements during an assembly phase: the 5′ splice site (SS), branch-site region, and 3′SS. The 5′SS and branch region are bound, in part, by base-pairing interactions with U1 and U2 snRNPs, respectively, and the 3′SS by protein factors; the U4/5/6 tri-snRNP joins the complex, and a series of conformational rearrangements and association of additional proteins completes the formation of the catalytic spliceosome comprising a U2/5/6/pre-mRNA core (reviewed in Nilsen, 1998; Staley and Guthrie, 1998).
The DExD/H family of ATPases, including Prp5p, Sub2p, Prp28p, Brr2p, Prp2p, Prp16p, Prp22p, and Prp43p, are thought to facilitate conformational changes within the spliceosome by coupling physical movement with a cycle of ATP binding, hydrolysis, and release of ADP (reviewed in Rocak and Linder, 2004; and see Fig. 1A). Prp16p, a DEAH-box ATPase that facilitates the transition between the 1st and 2nd steps of splicing, was initially identified in a screen for mutants with reduced splicing fidelity (Couto et al., 1987; Burgess et al., 1990; Schwer and Guthrie, 1991). Prp16p mutants with decreased ATPase activity allowed branch site A-to-C mutants to proceed through splicing, rather than being discarded; this effect is thought to be due to a slowed exit from the 1st catalytic step conformation, which allows for increased 1st step catalysis for the BS-C mutant intron before Prp16p-mediated conformational change can result in rejection of unreacted precursor (Konarska and Query, 2005; Villa and Guthrie, 2005). The example of Prp16p-mediated productive progression vs. rejection was predicted to represent a general paradigm for the maintenance of fidelity by ATPases throughout the splicing cycle (Burgess et al., 1990). In this model, rejection of substrates may occur at every ATP-dependent transition, with lowered rates of ATP hydrolysis/conformational change resulting in retention of suboptimal substrates in the productive pathway at any of these points (Burgess and Guthrie, 1993). Further support for this view comes from recent work with the related ATPase Prp22p, which facilitates exit from the 2nd step conformation and thereby release of spliced mRNA: mutant alleles of prp22 improve 2nd step catalysis on suboptimal substrates, presumably by allowing longer dwell time in the 2nd step conformation (Mayas et al., 2006).
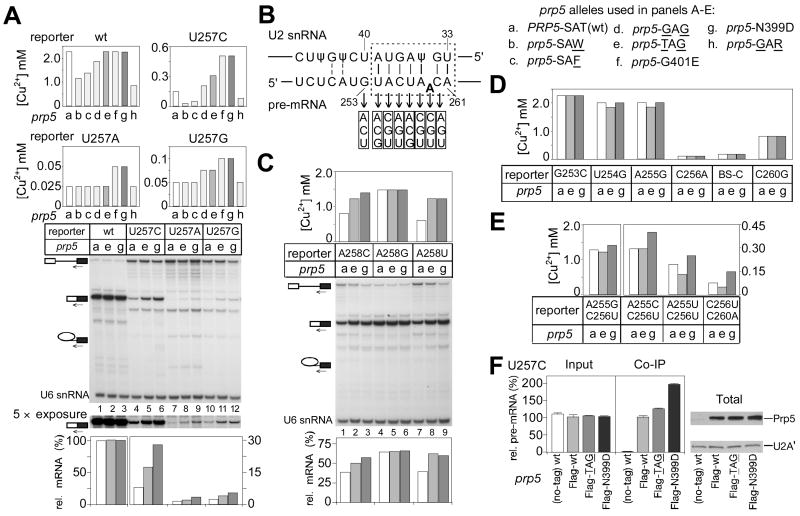
Figure 1. Genetic screen for prp5 alleles that alter splicing substrate selectivity.
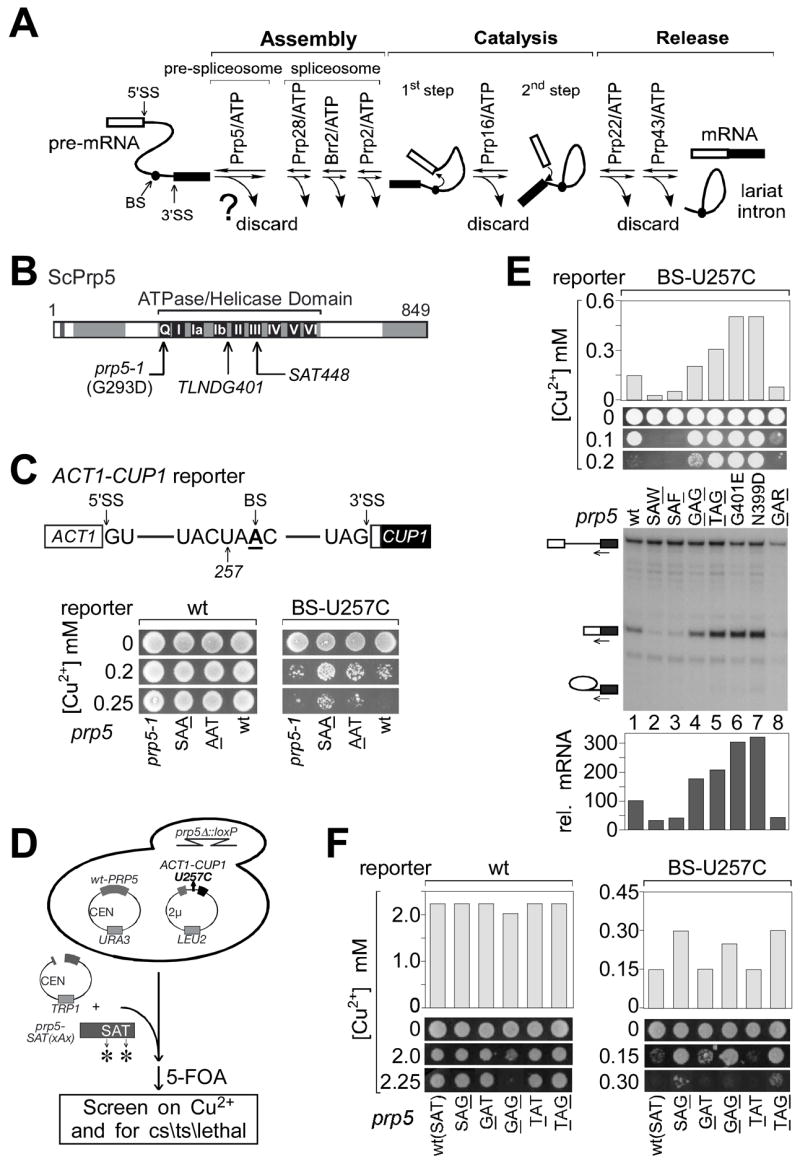
A. ATPase-associated steps during splicing could offer opportunities for kinetic discrimination of suboptimal pre-mRNA substrates (Burgess and Guthrie, 1993).
B. Schematic of S. cerevisiae Prp5p, indicating conserved motifs within its ATPase/helicase domain. Regions mutated in this study and the prp5-1 mutation are indicated.
C. (Upper) Schematic of ACT1-CUP1 pre-mRNA reporter used for monitoring splicing effects in vivo (Lesser and Guthrie, 1993). (Lower) Growth on copper of strains carrying one reporter mutant, U257C, is slightly improved by alanine mutations in the SAT motif of Prp5p.
D. Strategy of genetic screen for more-potent prp5 alleles that alter splicing of the U257C reporter, and for alleles conferring conditional and lethal growth defects. The selected prp5 alleles are listed in Table 1.
E. Analysis of seven prp5 alleles that alter splicing of U257C. (Upper) Graph of maximum copper concentration that allows cell growth. (Middle) Cell growth on selected copper plates, as indicated. (Lower) Analysis of in vivo RNA levels for ACT1-CUP1 reporter by primer extension. Primer complementary to the 3′ exon was used to monitor levels of pre-mRNA, mRNA, and lariat intermediate (indicated by icons).
F. Glycine at position T448 (SAG) is the critical change within motif III that confers improved splicing upon the U257C reporter.
Prp5p, originally identified as a mutant in RNA biogenesis (rna5; Hartwell, 1967) and later shown to be a splicing factor (Lustig et al., 1986), is required early in spliceosome assembly (Ruby et al., 1993). PRP5 interacts genetically with numerous U2 snRNP components: PRP9, -11, and -21 (homologs of SF3a), CUS1 (homolog of SF3b145), CUS2, and U2 snRNA stem-loop IIa (Ruby et al., 1993; Perriman et al., 2003 and references therein). The ATPase activity of S. pombe Prp5p is required for stable association of U2 snRNP with pre-mRNA, and both fission yeast and human Prp5p contain distinct domains that separately interact with U1 and U2 snRNPs (Xu et al., 2004), contributing to the formation of pre-spliceosome (U1/U2/pre-mRNA) complexes. Additional in vitro experiments suggest that Prp5p and the SF3a proteins facilitate an ATP-dependent alteration in U2 structure, and that Prp5p is required for the branch-pairing region of U2 snRNA to become accessible to the intron branch region (Wiest et al., 1996; Abu Dayyeh et al., 2002). Depletion of Cus2p allows U2–intron binding in the absence of ATP, although Prp5p is still required, suggesting that one result of Prp5p activity is removal of Cus2p (Perriman and Ares, 2000).
Given that Prp5p facilitates stable binding of introns to U2 snRNP, altered Prp5p activity might change substrate selectivity at an early stage in the splicing pathway. To search for alleles of S. cerevisiae prp5 that would improve splicing of suboptimal substrates, we first identified an intron sensitive to altered Prp5p activity and then used this suboptimal intron to screen for prp5-SAT mutants that alter substrate selectivity. This strategy identified a variety of prp5 mutants that modulate splicing fidelity of introns mutated in the branch region that pairs with U2 snRNA; analogously, the prp5 alleles also modulate splicing of wild-type (wt) introns in the presence of U2 alleles mutated in the branch-pairing region. Either hyper-stabilized U2–branch region pairing or compensatory changes between U2 alleles and mutant intron branch regions result in loss of splicing alteration by the prp5 alleles, providing strong evidence that a lowered stability of the branch region–U2 duplex is the critical feature suppressed by prp5 alleles. Together, these results demonstrate that Prp5p activity modulates intron selectivity and indicate that this modulation occurs at the time of establishment of branch region–U2 snRNA pairing.
RESULTS
Genetic screen for prp5 alleles that alter splicing substrate selectivity
To obtain alleles of prp5 that would modulate the efficiency of spliceosome assembly on suboptimal substrates, we performed two rounds of screening in S. cerevisiae – the first to identify suboptimal intron substrates that would be affected by prp5 mutants, and the second, using such a substrate, to screen for more-potent prp5 alleles. DExH/D ATPase domains contain highly conserved motifs; motif III, SAT, is thought to be required for coupling of ATP hydrolysis to conformational change and therefore we considered mutants in SAT to be good candidates to alter Prp5p’s activity. We tested two such mutants, prp5-S446A (AAT) and -T448A (SAA), as well as the originally-isolated prp5-1 allele (a G293D mutation in the Q motif; Abu Dayyeh et al., 2002) (Fig. 1B). To analyze splicing effects in vivo, we employed ACT1-CUP1 reporters (Lesser and Guthrie, 1993), which allow for monitoring of splicing efficiency by conferring copper resistance to yeast deleted of chromosomal CUP1 genes (Fig. 1C). Fifteen ACT1-CUP1 reporters, representing mutations at the 5′SS, branch region, and 3′SS, were tested for growth on copper in the presence of these three prp5 alleles, in comparison to wt-PRP5. One mutant reporter, carrying a U257C mutation in the branch region, showed weak but detectably improved growth on copper in the presence of prp5-AAT and prp5-SAA, but not prp5-1 (Fig. 1C), indicating that mutation of SAT in Prp5p can improve splicing of suboptimal introns, and that the U257C reporter would be suitable for screens for more-potent prp5 alleles.
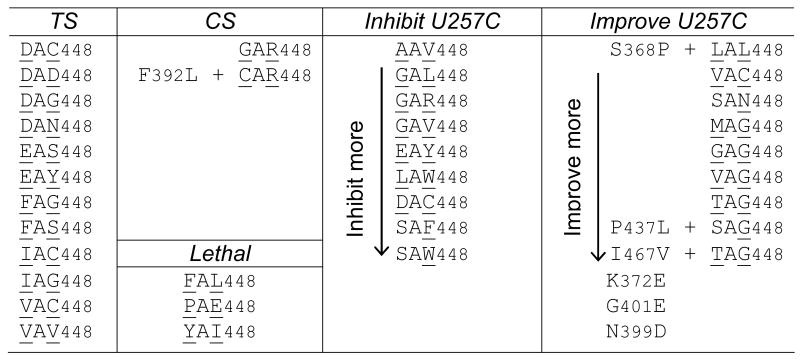
A library of prp5 alleles randomized at the Ser and Thr residues of SAT (see Experimental Procedures) was screened for temperature-sensitive (ts), cold-sensitive (cs), or lethal phenotypes, and for improvement or exacerbation of the growth defect on copper of a strain carrying the U257C reporter (Fig. 1D). We obtained numerous ts alleles, most of which contain bulky amino acids at the randomized positions; two cs alleles, both of which contain Arg instead of Thr in SAT; and numerous lethal alleles, three of which are described here (Table 1). Because ts alleles did not detectably alter splicing of suboptimal introns, they are not discussed here further.
Table 1.
Selected prp5 alleles that have growth phenotypes or alter splicing of BS-U257C reporter.
|
Two classes of mutant alleles were obtained that significantly improved copper growth for U257C (Table 1): those in the SAT motif, for which the most common feature is Gly at position T448; and those with mutations outside SAT, within the sequence T397LNDGKL403, close to ATPase motif Ib (Fig. 1B), likely generated during PCR in construction of the mutant library. Alleles obtained from the screen for exacerbation of the U257C growth defect were predominantly characterized by an aromatic group (Trp or Phe) at position T448 (Table 1).
Seven representative prp5 alleles were chosen for further analyses: four that improved U257C growth on copper (two within the SAT motif, G446AG448 and T446AG448, and two outside SAT, G401E and N399D), two that exacerbated U257C growth on copper (SAW448 and SAF448), and one cs allele (G446AR448). The alleles that improved U257C growth on copper also yielded increased levels of U257C mRNA, relative to wt-PRP5 (Fig. 1E, cf. lanes 4–7 to 1) and did not affect the wt reporter (except for prp5-GAG, which slightly inhibited the wt reporter; data not shown, and Fig. 2A). The other alleles, prp5-SAW, -SAF and -GAR, inhibited splicing of all reporters tested, including the wt reporter (Fig. 1E, 2A, and data not shown). In all cases, altered levels of mRNA detected by primer extension correlated with altered growth in the presence of copper.
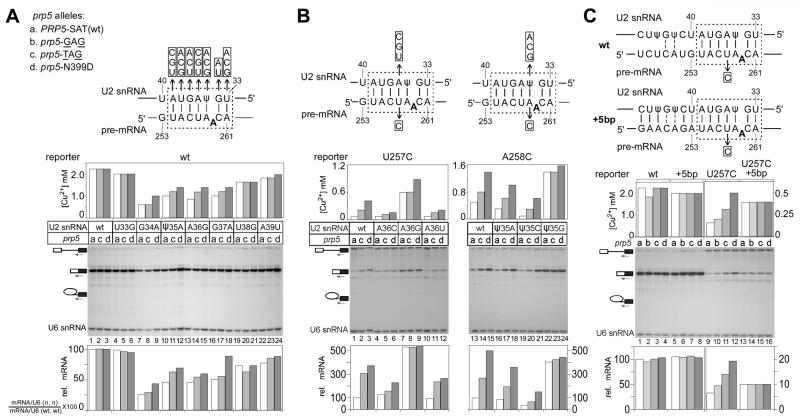
Figure 2. prp5 alleles modulate substrate selectivity primarily at two positions in the branch-flanking region.
A. Growth in the presence of copper (upper) and analysis of mRNA levels in vivo (lower) indicate that prp5 alleles improve splicing of U257A and U257G reporters, as well as the previously-tested U257C mutant. A longer exposure is provided to more clearly show mRNA levels. Primer complementary to U6 snRNA was used in the same reaction as a loading control. Quantitations: the mRNA level in each lane as determined by phosphorimaging was normalized to the U6 snRNA loading control, then divided by the normalized mRNA level in the strain containing wt-PRP5 and wt ACT1-CUP1 reporter to give relative mRNA levels.
B. Schematic of base-pairing interactions between U2 snRNA and the branch region of ACT1-CUP1 pre-mRNA, indicating branch region mutations used in this figure and Fig. S1.
C. prp5-TAG and -N399D alleles improve growth on copper and in vivo mRNA level of reporters mutated at position A258 in the branch region.
D. prp5 alleles do not improve growth on copper for all single mutations that are outside of branch region U257 and A258. One mutant at each position is shown; for other mutants at these positions, see Fig. S1.
E. Reporters carrying double mutations at branch region positions other than U257 and A258 also are improved by the prp5-N399D allele.
F. prp5 alleles increase the steady-state levels of suboptimal branch region pre-mRNA present in splicing complexes in vivo. (Left) Quantitative-RT-PCR analysis of anti-Flag-Prp5p-purified complexes indicates an increased association of U257C pre-mRNAs with splicing complexes by prp5-TAG and prp5-N399D alleles. The level of pre-mRNA was normalized to U6 snRNA. Each bar is from a triplet assay; error bars represent standard deviations. (Right) Visualization of total Flag-Prp5p and U2A’ (Lea1p) by western blot indicates that levels of Prp5-TAG and Prp5-N399D proteins are similar to wt-Prp5p.
Because the selected SAT alleles that improved U257C splicing contained mutations at both randomized positions (S446 and T448), we made additional prp5 mutants (SAG, GAT, and TAT) to deconvolute the effects of these positions. prp5-SAG improved U257C growth on copper (Fig. 1F) to a level indistinguishable from that of the prp5-TAG mutant, and slightly better than did the prp5-GAG mutant. In contrast, prp5-GAT and -TAT did not improve U257C growth on copper. Thus, Gly at position T448 (SAG) is the critical amino acid change that confers improved splicing upon the U257C intron reporter.
prp5 alleles modulate substrate selectivity primarily at two positions in the branch-flanking region
To investigate the nucleotide and positional specificity of the altered intron selectivity exhibited by prp5 alleles, we first tested other nucleobases at position U257; all four prp5 alleles that improved splicing of the U257C reporter also improved splicing of the U257G reporter, and the strongest two alleles improved U257A (Fig. 2A). Thus, defects associated with all mutations at U257 can be suppressed by prp5 alleles.
Second, we tested other positions within the branch region from G253 to C260, testing all three mutations at every position, using one prp5 allele from each class that improved U257C splicing (Fig. 2B). prp5-TAG and -N399D alleles improved overall splicing for A258C and A258U mutants, compared to wt-PRP5 (Fig. 2C, lanes 1–3, 7–9). Splicing of the A258G reporter was not improved by prp5 alleles (Fig. 2C, lanes 4–6); however, the A258G reporter yields near-wt levels of mRNA in the presence of wt-PRP5, consistent with an A258G–ψ35 wobble base-pair providing near-wt branch region–U2 duplex stability. In contrast, no improvement was detectable for any mutations from G253 to C256, at the branch site (A259), or at position C260 (Fig. 2D, Fig. S1, and data not shown).
No 5′ or 3′SS mutations tested were improved by prp5-TAG and -N399D alleles; however, all were exacerbated by prp5-SAW and -SAF, as were branch region mutants (data not shown). Although Prp5-SAW and -SAF mutant proteins were present at reduced levels, perhaps partly explaining their inhibitory phenotypes, alleles that improved U257C splicing were present at indistinguishable levels from wt-Prp5p (data not shown and Fig. 2F), implicating protein function rather than steady-state levels in improvement of splicing.
These data suggest that effects of the prp5 alleles are specific to two positions within the branch region, 257 and 258. One possibility is that branch region–U2 snRNA stability is critical and that these two positions are more important in pairing with U2 snRNA than are flanking positions; alternatively, it is possible that prp5 alleles selected in our screen have positional specificity because of the U257C reporter used in their selection, but that other prp5 alleles could have a different specificity. This latter possibility seems unlikely, as additional screens for prp5-SAT mutants using C256A or C260G reporters (whose splicing was not detectably altered by the previously-selected prp5 alleles) as well as with 5′SS mutant reporters U2A and A3G did not yield prp5 alleles that improved splicing.
To test the possibility that splicing of reporters mutated at positions other than position 257 and 258 might be improved by prp5 alleles in the context of more-strongly inhibited spliceosome assembly, we tested reporters mutated at multiple positions in the branch region. Double mutations at A255+C256 or at C256+C260 were slightly improved by the strongest prp5-N399D allele, although not detectably improved or slightly exacerbated by the prp5-TAG allele (Fig. 2E). Taken together, these data suggest that alteration of substrate selection by prp5 alleles is strongest at two positions, U257 and A258, but that other branch-region positions can also be affected.
prp5 alleles led to an increased association of suboptimal branch region pre-mRNA with spliceosomal complexes. Prp5p associates with pre-mRNA in complexes with U1 and U2 snRNPs in mammals and S. pombe (Xu et al., 2004), suggesting that its association with pre-mRNA is a good indicator of pre-spliceosome formation (U1/U2/pre-mRNA complexes). We used Ch-IP-like conditions to monitor Prp5/U1/U2/pre-mRNA complexes in vivo, using Flag-tagged Prp5p, in situ formaldehyde crosslinking, and anti-Flag chromatography, followed by quantitative-RT-PCR of associated pre-mRNA. prp5-TAG or prp5-N399D led to increased association of U257C pre-mRNA with such complexes relative to wt-PRP5 (Fig. 2F), whereas wt-pre-mRNA was not increased (data not shown), suggesting that prp5 alleles enable increased incorporation of suboptimal branch region pre-mRNA into splicing complexes.
prp5 alleles improve splicing of wt intron in the presence of U2 snRNA mutants
The combination of wt intron and U2 snRNA with a mutated branch site-binding region (Fig. 3A and Fig. S1) should result in a similar duplex alteration as would branch region mutants in the presence of wt-U2 snRNA. All single base mutations of U2 snRNA from U33 to A39 were constructed and tested (excepting G34C, which was extremely dominant negative). Except for A39G and mutations at position U33, all U2 snRNA mutants were lethal as the sole copy (data not shown), consistent with previous reports and confirming the importance of the U2 branch-pairing region (Parker et al., 1987; Miraglia et al., 1991; Pascolo and Séraphin, 1997). Therefore, further experiments using these U2 mutants were carried out in the presence of a plasmid-borne copy of wt-U2 snRNA. In all cases, the presence of the mutant U2 allele resulted in reduced growth on copper and lower mRNA levels of the wt reporter, compared to two copies of wt-U2 (shown for one mutant allele at each position in U2 in Fig. 3A; other mutants shown in Fig. S1). In comparison to wt-PRP5, prp5-TAG and -N399D alleles improved growth in the presence of copper and increased mRNA levels for strains carrying wt reporter and U2 snRNAs mutated at positions G34, ψ35, A36, and G37 to varying extents, whereas little or no difference was detectable in strains carrying U2 snRNAs mutated at positions U33, U38, or A39 (which also are the U2 mutants within the branch-pairing region that have the least detrimental effects on cell growth as second copies)(Fig. 3A and Fig. S1). [note: some mutants within the branch-pairing region might reduce pseudouridylation at position 35, as has been described for positions near to ψ42 (Yang et al., 2005); such a possibility does not appear to affect the present analysis, as we do not detect changes in splicing of branch region mutant reporters in the absence of the position 35 pseudouridine synthase, Pus7p (D. Smith, M. Konarska, CQ, unpublished data).]
Figure 3. Suboptimal branch region–U2 snRNA base-pairing is the critical feature suppressed by prp5 alleles.
A. prp5 alleles improve splicing of a wt intron in the presence of U2 snRNA mutants.
B. prp5 alleles provide little or no additional improvement to the partial suppression by compensatory changes in the U2 branch-pairing region of defects due to U257C and A258C mutations.
C. Additional potential base-pairs between U2 snRNA and the intron branch region partially suppress the U257C defect; prp5 alleles provide no additional improvement.
A–C: (upper) Schematic of base-pairing interactions between U2 snRNA and the intron branch region, indicating U2 snRNA and ACT1-CUP1 reporter mutations used below. (middle) Graphical representation of maximum copper concentration that allows cell growth. (lower) Analysis by primer extension of in vivo RNA levels for ACT1-CUP1 reporters as in Fig. 1E. For panels A and B, all tested strains carried both wt-U2 and mutant U2 snRNA alleles on CEN plasmids; all strains were otherwise isogenic and contained disrupted chromosomal loci of U2 snRNA and PRP5. mRNA quantitations are as in Fig. 2A except divided by the normalized mRNA level in the strain containing wt-PRP5 and wt-U2 snRNA.
Our prp5 alleles improved splicing of wt reporters in the presence of U2 snRNA alleles carrying mutations at positions 34–37 of the branch-pairing region. These data, together with the improved splicing of branch region mutant reporters in the presence of wt-U2, suggest that a property of the branch region–U2 duplex is a critical limiting feature of the defect that can be suppressed by prp5 alleles.
Restored base-pairing results in loss of enhanced splicing by prp5 alleles
To compare the importance of duplex stability and nucleobase identity of the branch region–U2 duplex, we tested branch region mutants combined with compensatory mutants in U2 snRNA. It is known that the presence of a mutant U2 molecule containing compensatory changes improves splicing of introns mutated at some positions within the branch region relative to the presence of wt-U2 alone (Parker et al., 1987; Wu and Manley, 1989; Zhuang and Weiner, 1989); however, such suppressor U2 snRNAs do not fully restore wt splicing, at least in part because branch region intron mutants are defective at multiple steps in the splicing pathway. Thus, we anticipated that such compensatory changes in branch region–U2 pairing would partially improve overall splicing – if duplex stability were critical for the observed improvement by prp5 alleles, then the presence of prp5 alleles would improve splicing to little or no greater extent; by contrast, if some other feature, such as base identity, were critical for the improvement by prp5 alleles, then additional improvement would be observed.
As expected, splicing of the U257C reporter was improved in the presence of U2-A36G, but not in the presence of other U2-A36 alleles (Fig. 3B, cf. lane 7 to 1, 4, 10); similarly, splicing of the A258C reporter was improved in the presence of U2-ψ35G, but not other U2-ψ35 alleles (cf. lane 22 to 13, 16, 19). Importantly, the prp5-TAG allele did not result in improved splicing of the U257C reporter in the presence of U2-A36G (cf. lane 7 to 8) or of the A258C reporter in the presence of U2-ψ35G (cf. lane 22 to 23). The prp5-N399D allele resulted in slightly improved growth on copper in both cases (~15–30% improvement with the compensatory U2 allele compared to ~300% improvement with a wt or non-compensatory U2 allele), but little difference in mRNA levels; the slight discordance between growth on copper and mRNA levels can be explained by effects of the mutant U2 alleles on splicing of endogenous (wt) introns, which would be improved by the prp5-N399D allele (Fig. 3A), resulting in an overall healthier strain (data not shown). These results also dismiss an alternative explanation for suppression of U2 mutants – that prp5 alleles might enhance the mutation, eliminating the mutant U2 from a mixed population of wt and mutant U2 snRNAs. The lack of effect of prp5 mutants in the strains with compensatory U2–branch region changes eliminates this possibility for suppression of mutants at U2 positions 35 and 36.
We also tested the effects of increased base-pairing by extension of the branch region–U2 duplex. Reporters containing either 5 or 2 additional potential base-pairs with U2 snRNA were prepared in combination with either an otherwise-wt branch region or the U257C mutation. The +5bp change did not significantly affect splicing of an otherwise-wt intron (Fig. 3C, cf. lanes 5–8 to 1–4), but did improve splicing of the U257C mutant (Fig. 3C, cf. lanes 13 to 9). In the presence of the +5bp mutation, prp5-TAG or -N399D alleles resulted in no further improvement in U257C splicing (cf. lanes 9–12 to 13–16). Similar results were observed in the presence of +2bp mutations (data not shown). In contrast, U257C reporters carrying different (non-complementary) mutations at the same five positions showed significant improvement in the presence of prp5 alleles; and, U257C reporters carrying the +2bp mutations did not show improved growth on copper in the presence of U2 alleles mutated at the corresponding two positions (data not shown), consistent with the effect of the +5 and +2 mutations being enhanced pairing with U2.
Furthermore, alleles of prp5 and of the DEAH-ATPase prp16, required for the transition from the 1st to the 2nd step conformation (Schwer and Guthrie, 1991), exhibit different phenotypes in combination with both U257C and U257C+extended base-pairing reporters. First, whereas prp5 alleles showed strong improvement of U257C splicing, prp16 alleles improve this reporter only slightly (consistent with the notion that U257C is strongly suboptimal in spliceosome assembly as well as at least somewhat impaired for the 1st step). Second, whereas prp5 alleles show no additional improvement of the U257C+5bp reporter, the prp16–101 allele strongly improved this reporter (Fig. S2) (suggesting that extended pairing with U2 improves spliceosome assembly, affecting the same event as Prp5p, whereas prp16 mutants improve the 1st step, a distinct event from that improved by increased pairing to U2).
Collectively, these data indicate that the positive effects of prp5 alleles are superseded by enhanced stability of the branch region–U2 duplex. From these results we conclude that the effects of the prp5 alleles occur at the same time in the spliceosome assembly pathway as does formation of the duplex.
Deletion of CUS2 does not alter splicing fidelity
Deletion of CUS2, which encodes a U2 snRNP protein, suppresses some conditionally lethal prp5 defects in vivo and allows for formation of pre-mRNA–U2 snRNP complexes in vitro in the absence of ATP (Perriman and Ares, 2000; Perriman et al., 2003). These observations led to the proposal that Cus2p antagonizes the action of Prp5p, and that a result of Prp5p ATPase activity may be removal of Cus2p from the forming spliceosome (Perriman et al., 2003). If the interaction between Prp5p and Cus2p impacts the same event in which prp5 alleles alter intron selectivity, then cus2 deletion should also alter intron selectivity, but in a manner opposite to prp5 alleles. We tested the effect of cus2 deletion on a variety of intron mutants; in the presence of wt-PRP5, there was no detectable difference in splicing or growth on copper in the presence or absence of Cus2p (Fig. 4D and data not shown). Likewise, cus2 deletion did not detectably alter the effects on splicing of a variety of intron mutants by prp5-TAG and -N399D alleles that do not alter overall cell growth (Fig. 4E and data not shown). Thus, cus2 deletion affects a different event(s) from that in which prp5 alleles alter selectivity of branch-region mutants.
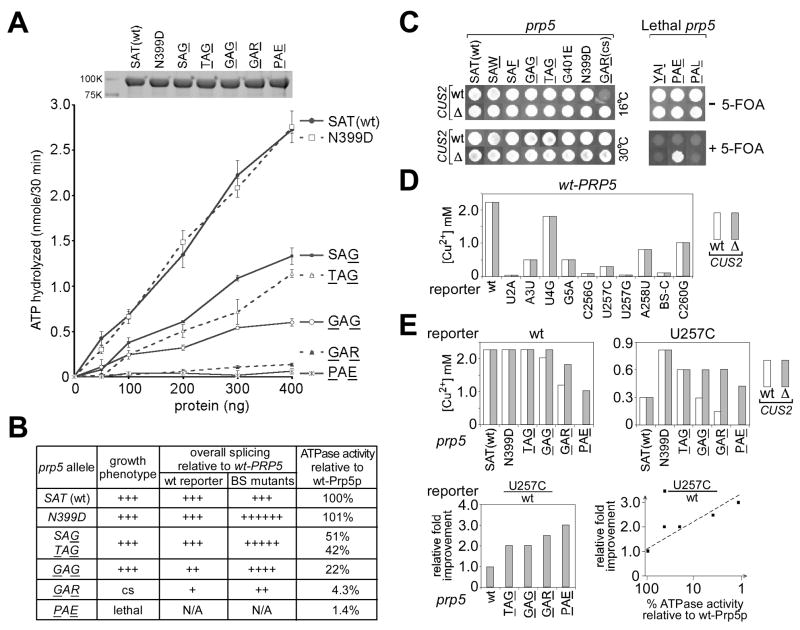
Figure 4. ATP hydrolysis by prp5-SAT alleles correlates with their phenotypes.
A. In vitro ATPase assay of Prp5p mutants using His6-tagged proteins purified from E. coli (Coomassie blue-stained gel, upper insert). All values, with standard errors, are averaged from at least two parallel reactions.
B. Relative phenotypes and ATPase activities of Prp5p mutants. Calculation of relative ATPase activities was based on the least squares fitted lines in panel A.
C. CUS2 deletion suppresses the cs phenotype of prp5-GAR (Left) and rescues the otherwise-lethal phenotype of prp5-PAE (Right).
D. CUS2 deletion does not alter splicing fidelity. In the presence of wt-PRP5, growth on copper plates of strains carrying a variety of intron mutants did not change when CUS2 was deleted.
E. In the absence of CUS2, the ATPase activity of prp5-SAT mutants correlates with ability to improve splicing of suboptimal intron substrates. Deletion of CUS2 improves growth of strains carrying prp5 alleles with low ATPase activity, but not prp5 alleles with relatively high ATPase activity, indicated by wt and U257C reporters (upper). In the absence of CUS2, lowered ATPase activity of prp5-SAT mutants correlated with stronger improved splicing of the U257C mutant. Relative improvement for each prp5 allele was calculated by normalizing to the growth of wt reporter (lower left) and plotted against the ATPase activity given in Fig. 4B (lower right).
ATP hydrolysis by prp5-SAT alleles correlates with their phenotypes
The SAT motif in the ATPase domain of DExD/H proteins is thought to be required to couple ATP hydrolysis with remodeling of RNA-RNA or RNA-protein complexes (reviewed in Rocak and Linder, 2004). In vitro assays for S. pombe Prp5p and other proteins in the DExH/D family, e.g. Prp22p, have shown that mutations in the SAT motif change both ATP hydrolysis and functional activity (Schwer and Meszaros, 2000; Xu et al., 2004).
ATP hydrolysis by mutant Prp5p proteins was measured in vitro using recombinant His6-tagged proteins purified from E. coli (Fig. 4A). The Prp5-N399D mutant hydrolyzed ATP at nearly the same rate as wt-Prp5p, consistent with the mutated residue being outside a highly conserved ATPase motif. In contrast, Prp5-SAG, -TAG, and -GAG mutants exhibited lower ATPase activities (51%, 42%, and 22% compared to wt, respectively); and Prp5-GAR (cs) and -PAE (lethal) mutants showed extremely low ATPase activities (4.3% and 1.4%, respectively) (Fig. 4A, B).
Consistent with previous reports, deletion of CUS2 suppressed the cold sensitivity of prp5-GAR, allowing growth at 16°C (Fig. 4C, left), and rescued one lethal allele, prp5-PAE (Fig. 4C, right), allowing for the severely ATPase-defective prp5-PAE allele to be tested for alteration of branch region usage in vivo. Although in the absence of Cus2p both prp5-GAR and -PAE alleles inhibited splicing of the wt reporter, they improved splicing of the branch region mutants improved by other prp5 alleles, e.g. U257C (Fig. 4E and data not shown); thus, prp5 alleles with reduced ATPase activity, even though not selected from the screen for improvement of U257C splicing, nevertheless yield improved splicing of branch-region mutants.
Because deletion of CUS2 bypassed a growth defect of prp5 mutants (that does not involve the event that alters branch region fidelity), strains without CUS2 allowed for a more direct comparison of ATPase effects to altered fidelity. ATP hydrolysis by mutant Prp5-SAT proteins inversely correlated with their splicing phenotype: prp5-SAT alleles with the lowest ATPase activity exhibited the greatest relative improvement of U257C splicing (Fig. 4E, lower).
DISCUSSION
We have used a series of genetic screens in S. cerevisiae to isolate potent prp5 alleles that modulate splicing fidelity. Whereas it was previously thought that improved assembly of splicing complexes on suboptimal introns was achieved through accessory binding proteins and that the role of Prp5p ATPase was to facilitate access of the intron branch region to U2 snRNA, we show that decreased Prp5p ATPase activity in a series of SAT mutants results in improved splicing of introns with suboptimal branch regions. We propose that Prp5p-facilitated conformational change competes with branch region–U2 snRNA pairing to control progression of the splicing pathway, such that pairing followed by conformational change results in productive splicing, but conformational change prior to pairing results in an abortive path. Thus, reducing the rate of ATP hydrolysis or otherwise impairing the ATPase domain reduces the requirement for branch region complementarity to U2. This mechanism of altering the required stringency of branch region–U2 pairing may allow for the reduced complementarity between U2 snRNA and the branch region of many introns in metazoans.
Prp5p affects spliceosome assembly at the time of branch region–U2 pairing
Many ATPases are required during spliceosome assembly, catalysis, and disassembly, presumably facilitating conformational changes by coupling physical movement to ATP hydrolysis. Prp5p is required early in spliceosome assembly, at the time of stable U2 snRNP binding, although its exact role in U2 snRNP–intron interaction is unknown. Based on the paradigm of Prp16p as a modulator of fidelity at the time of exit from the 1st step conformation (Burgess et al., 1990), we hypothesized that alleles of prp5 that modulate spliceosome assembly on suboptimal introns should be obtainable. Using a library of prp5 mutants randomized in the Ser and Thr codons of the SAT motif of the ATPase domain, we identified a collection of mutants (both SAT and non-SAT mutants) that improve overall splicing of introns mutated at the two positions preceding the branch site (UACUAAC).
To ascertain whether these effects were linked to the branch region–U2 snRNA duplex, we tested combinations of reporters and U2 mutants. Mutants in the branch-pairing region of U2 decreased overall splicing of a wt intron, and this defect was suppressed by prp5 alleles, suggesting that it is a property of the altered duplex contributing to poor splicing that prp5 alleles suppress. Two possibilities for this property are duplex stability and base identity. At both positions within the branch region that are strongly affected by prp5 alleles, we observed improvement in overall splicing in the presence of the compensatory U2 snRNA allele, but importantly the presence of prp5 alleles no longer improved splicing, or did so to a much reduced extent. Similarly, the presence of additional pairing improved overall splicing of the branch region mutants, and the presence of prp5 alleles did not provide added improvement (whereas such reporters were further improved by alleles of prp16). Both of these results suggest that prp5 alleles compensate for a reduced stability of the branch region–U2 duplex (and not base identity). That mutants in the middle portion of the duplex both impair splicing and are suppressed by prp5 alleles to a much greater extent than those near the ends of the duplex is consistent with thermodynamic measurements of duplex stabilities, which indicate that mismatches near the ends of short duplexes have significantly lower destabilizing effects than those in the middle (Kierzek et al., 1999). These data explicitly link the fidelity effects of prp5 alleles to a defined physical structure, the branch region–U2 duplex.
A recent genome-wide analysis of alteration of splicing by mutations in core spliceosomal components revealed a large group of introns whose splicing was inhibited by the prp5-1 allele at a non-permissive temperature (Pleiss et al., 2007). Notably, there were no introns whose splicing was improved by prp5-1, consistent with this being a loss-of-function mutation. It is likely that our prp5-SAW and -SAF alleles (and most ts alleles) are analogous to prp5-1, in that they inhibit wt intron splicing (Fig. 2A).
Prp5p has been proposed to remove the U2 snRNP protein Cus2p (Perriman et al., 2003) and to increase accessibility of the branch-pairing region of U2 snRNA to the intron (Wiest et al., 1996; Abu Dayyeh et al., 2002). The failure of cus2 deletions to alter branch region selectivity in a manner opposite to that of prp5 alleles argues strongly that any event(s) in which Prp5p activity is opposed by Cus2p is distinct from that in which Prp5p affects the stringency of branch region usage. In addition, the ability of cus2Δ strains to suppress cell growth defects of cs or lethal prp5 alleles allowed us to test their effects on splicing. These independently-isolated, ATPase-defective alleles yield the strongest relative improvement to the splicing of branch-flanking mutants (Fig. 4E), providing strong evidence that the improved splicing of branch region mutants is inversely correlated with the ATPase activity of Prp5p SAT mutants.
Competition between Prp5p activity and branch region–U2 pairing controls progression of spliceosome assembly
All prp5-SAT mutants that improved splicing of branch region mutants exhibited reduced ATPase activity, which presumably results in a reduced ability to promote conformational change. In other studied DExH/D proteins, reduced ATPase activity results in reduced helicase activity in vitro; however, helicase activity can also be impaired without reduced ATPase activity (Schwer and Meszaros, 2000). It is possible that the selected non-SAT prp5 mutants (N399D and G401E) are in this latter category of reduced activity in facilitating conformational change, or that they may inhibit interactions within the spliceosome, increasing non-productive hydrolysis events.
The simplest explanation for the ability of reduced-activity prp5 alleles to improve overall splicing when branch region–U2 pairing is suboptimal is a model in which these alleles allow longer time for inefficient duplex formation. In such a scenario, wt introns and U2 snRNA would pair efficiently, and subsequent ATP hydrolysis by Prp5p would facilitate a conformational change in U2 snRNP that ‘locks’ U2 snRNP onto the intron (Fig. 5A). Formation of the branch region–U2 duplex might even contribute to stimulation of Prp5p ATPase, by an induced fit conformational change. By contrast, a near-complementary branch region–U2 structure would pair inefficiently (slower kon and faster koff; both rates are altered by the extent of base-pairing (Morrison and Stols, 1993)) and wt-Prp5p would hydrolyze ATP prior to duplex formation, resulting in a similar, but abortive, conformational change in the U2 snRNP–intron complex (Fig. 5B). The slowed activity of mutant prp5 alleles (either because of decreased ATP hydrolysis or because of non-productive cycles of hydrolysis) would allow more time for an inefficient duplex to form, after which a productive rearrangement in U2 structure could be achieved (Fig. 5C). Such rearrangement is likely to involve the stable deposition of the SF3a/b proteins onto the intron region surrounding the branch site (Gozani et al., 1996) and also might involve the U2 stem IIa/IIc structure, recently reported to alter spliceosome progression at multiple points (Hilliker et al., 2007; Perriman and Ares, 2007), although it is not known whether these competing structures affect the fidelity of branch region usage at the Prp5p-mediated event.
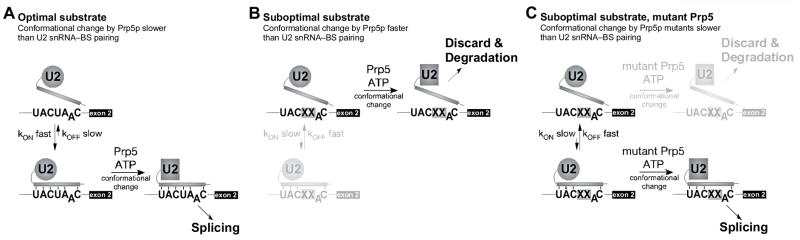
Figure 5. Schematic of outcomes of Prp5p-mediated spliceosomal transition, depending on relative rates of branch region–U2 snRNA pairing and conformational change by Prp5p.
A. Productive pathway of splicing, where kon is fast and koff is slow with wild-type substrate, and therefore duplex formation is faster than the rate of conformational change by Prp5p.
B. Alternative outcomes for suboptimal substrates (X’s indicate a suboptimal intron branch-site-flanking region). kon is slow and koff is fast in spliceosomes formed on suboptimal substrates. In the case of wild-type Prp5p, conformational change occurs prior to duplex formation, resulting in Prp5p-mediated transition prior to branch region–U2 snRNA pairing (upper path).
C. In the presence of mutations of the Prp5p ATPase domain, the rate of conformational change is also slowed, resulting in longer opportunity to achieve branch region–U2 snRNA pairing and allowing progression of the productive assembly pathway for suboptimal substrates (lower path).
Common features of mechanisms that modulate fidelity
It is now clear that at least three spliceosomal ATPases impact the splicing of suboptimal introns. In each case, it is thought that mutant alleles allow more time for a limiting event preceding a conformational change. prp5 alleles improve splicing of branch region mutants by allowing more time for duplex formation with U2 snRNA; prp16 alleles improve substrates suboptimal for 1st step catalysis (e.g. BS-C) by allowing longer dwell time in the 1st step conformation (Konarska and Query, 2005; Villa and Guthrie, 2005); prp22 alleles improve splicing of substrates suboptimal for 2nd step catalysis (e.g. 3′SS mutants) by allowing longer dwell time in the 2nd step conformation (Mayas et al., 2006). Such common features suggest that other spliceosomal ATPases – as proposed (Burgess and Guthrie, 1993) – will similarly affect a preceding event, in each case limiting for different sets of (suboptimal) intron features or other spliceosomal interactions. The strategy used here, of making weak mutants in an ATPase, screening for substrates that can be improved, then screening for potent alleles of the ATPase, provides a path for identification of additional fidelity events and better understanding of ATPase function.
One prediction of the kinetic proof-reading model is that organisms that routinely use weaker splice signals might have reduced ATPase activities. For example, compared to the highly conserved UACUAAC sequence in S. cerevisiae where the UA positions preceding the branch site adenosine are 100% conserved (Grate and Ares, 2002), these positions are 21% non-U and 47% non-purine, respectively, in experimentally-determined human branch regions (Kol et al., 2005, and references therein), and the A position preceding the branch site is 6% non-purine in predicted S. pombe branch regions (Wood et al., 2002). A reduced activity of Prp5 in such systems could facilitate splicing of introns with such reduced branch region–U2 complementarity. Consistent with this, we detect ~60–70% reduction in ATPase activity in Prp5p proteins derived from S. pombe or human relative to S. cerevisiae Prp5p (YZX & CQ, unpublished data). Another prediction of the model is that modulation of Prp5 activity could affect alternative splicing in higher eukaryotes in situations where there is competition between different branch sites. Indeed, RNAi knock-down screens in Drosophila identified several core components of the spliceosome (including Prp5p and other ATPases) that changed alternative splicing patterns (Park et al., 2004). We propose that lowered Prp5p activity would increase use of otherwise-skipped branch+3′ splice sites containing non-consensus branch regions (in particular at the UACUAAC positions), and that higher Prp5p activity would increase skipping of such suboptimal branch+3′SS regions.
EXPERIMENTAL PROCEDURES
Strains and plasmids
S. cerevisiae strains are listed in Table 2. ACT1-CUP1 reporter plasmids (Lesser and Guthrie, 1993) and U2 snRNA plasmids were prepared by overlapping PCR and in vivo gap repair cloning.
Table 2.
Yeast strains
| Strain | Genotype |
|---|---|
| yYZX02 | MATa, ade2 cup1Δ::ura3 his3 leu2 lys2 prp5Δ::loxP trp1, pRS316-PRP5 (PRP5 URA3 CEN ARS) |
| yYZX06 | MATa, ade2 cup1Δ::ura3 his3 leu2 lys2 prp5Δ::loxP trp1, cus2Δ::loxP pRS316-Prp5 (PRP5 URA3 CEN ARS) |
| yYZX11 | MATa, ade2 cup1Δ::ura3 his3 leu2 lys2 prp5Δ::loxP trp1, snr20 Δ::loxP pRS314-PRP5 (PRP5 TRP1 CEN ARS), pRS316-U2 (SNR20 URA3 CEN ARS) |
S. cerevisiae strains used in this study were derived from the above three strains, containing plasmid-borne alleles of prp5 and U2 snRNA (snr20) as indicated in the figures.
Screen of randomized prp5-SAT mutants
A pool of two-step PCR products covering S. cerevisiae PRP5 gene from +952 to +1473 was amplified using ExTaq DNA polymerase (Takara), in which the 6 nts encoding Ser and Thr at prp5-S446AT448 were randomized. pRS314(TRP)-PRP5, digested by NheI and BsmI, was co-transformed with the above PCR products for gap repair recombination in strain yYZX02 containing U257C ACT1-CUP1 reporter. Transformants were replicated to 5-FOA plates to lose the URA-marked wt-PRP5 plasmid; colonies were either picked for testing growth at different temperatures or were replicated to copper plates to select prp5 mutants that altered splicing. Selected prp5 mutant plasmids were isolated from yeast cells, identified by sequencing, and transformed back to yeast strains to confirm their phenotypes.
Copper assays
Cultures were grown to mid-log phase in –Leu or –His–Leu medium, diluted to 0.2 OD, and equal volumes were dropped onto –Leu plates containing 0, 0.025, 0.05, 0.075, 0.1, 0.125, 0.15, 0.2, 0.25, 0.3, 0.4, 0.5, 0.6, 0.8, 1.0, 1.2, 1.4, 1.5, 1.6, 1.8, 2.0, 2.25, or 2.5 mM CuSO4 (Lesser and Guthrie, 1993). Plates were scored and photographed after 3 days at 30°C.
RNA analysis
Primer extensions were carried out as described (Siatecka et al., 1999), using YAC6 5′-GGCACTCATGACCTTC, complementary to exon 2 of the reporter, and yU6-61 5′-GAACTGCTGATCATCTCTG, complementary to U6 snRNA as a loading control. Extension products were separated in 7% polyacrylamide/8 M urea gels, visualized by autoradiography, and quantified using phosphorimaging and ImageQuant 5.0.
Quantitative RT-PCR
Sequence encoding 3xFLAG was appended to the 5′-end of prp5 ORFs and expressed in yYZX02. Complexes were cross-linked in vivo using 1% formaldehyde, quenched using 0.38 M glycine. Glass-bead cell lysates were sonicated and applied to anti-FLAG M2 affinity gel (Sigma). After washing, bead-bound material was released by pronase digestion and cross-links reversed with heat treatment; total RNA was isolated followed by DNase I digestion. RNA was detected using an ABI PRISM 7900HT real-time PCR system (Applied Biosystems) with QuantiTect SYBR Green (Qiagen). Primers for ACT1-CUP1 pre-mRNA: 5′-TCTTTTATTTGCTACTGTGTCTCATGT and 5′-TTAATTCGCTGAACCCGGTA; for U6 snRNA: 5′-CGAAGTAACCCTTCGTGGAC and 5′-TCTCTTTGTAAAACGGTTCATCC.
Expression of recombinant ScPrp5p and ATPase assays
His6-tagged Prp5 proteins were expressed in E. coli, purified by affinity binding to Ni-NTA agarose (Qiagen), and dialyzed against buffer D [20 mM HEPES-KOH, pH 7.9, 0.2 mM EDTA, 100 mM KCl, 0.5 mM dithiothreitol, 1 mM PMSF, 20% glycerol]. ATPase assays were carried out at 30°C for 30 min in the presence of poly(A) RNA as previously described (Xu et al., 2004).
Supplementary Material
Figure S1. Summary of growth on copper of strains carrying either intron branch region mutants, or wt reporter in the presence of U2 snRNA mutants. prp5-TAG and -N399D alleles improve overall splicing of wt reporter in the presence of U2 snRNA alleles mutated at positions G34,ψ 35, A36, and G37 (upper), and improve splicing of intron branch region mutants at two positions, U257 and A258 (lower).
Figure S2. prp5 and prp16 alleles have distinct effects on splicing of suboptimal branch region mutants.
(Upper) Graph of maximum copper concentration that allows cell growth. (Lower) Cell growth on selected copper plates, as indicated. prp5-TAG and -N399D alleles improve splicing of branch region mutant U257C, but provide no additional improvement to branch region mutant (U257C+5bp) that contains extended pairing to U2 snRNA. In contrast, the prp16-101 allele provides only slight improvement to the U257C reporter, but provides significant additional improvement to the U257C+5bp mutant.
Acknowledgments
We are grateful to David McPheeters (CWRU) for wt-U2 snRNA plasmids, and Magda Konarska, Duncan Smith, and Jon Warner for helpful discussions and critical readings of the manuscript. This work was supported by NIH grant GM57829 to C.C.Q. and by a Cancer Center Support (core) grant from the NCI to AECOM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu Dayyeh BK, Quan TK, Castro M, Ruby SW. Probing interactions between the U2 small nuclear ribonucleoprotein and the DEAD-box protein, Prp5. J Biol Chem. 2002;277:20221–20233. doi: 10.1074/jbc.M109553200. [DOI] [PubMed] [Google Scholar]
- Burgess S, Couto JR, Guthrie C. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell. 1990;60:705–717. doi: 10.1016/0092-8674(90)90086-t. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Guthrie C. Beat the clock: paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem Sci. 1993;18:381–384. doi: 10.1016/0968-0004(93)90094-4. [DOI] [PubMed] [Google Scholar]
- Couto JR, Tamm J, Parker R, Guthrie C. A trans-acting suppressor restores splicing of a yeast intron with a branch point mutation. Genes Dev. 1987;1:445–455. doi: 10.1101/gad.1.5.445. [DOI] [PubMed] [Google Scholar]
- Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- Grate L, Ares M., Jr Searching yeast intron data at Ares lab Web site. Methods Enzymol. 2002;350:380–392. doi: 10.1016/s0076-6879(02)50975-7. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967;93:1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker AK, Mefford MA, Staley JP. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev. 2007;21:821–834. doi: 10.1101/gad.1536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzek R, Burkard ME, Turner DH. Thermodynamics of single mismatches in RNA duplexes. Biochemistry. 1999;38:14214–14223. doi: 10.1021/bi991186l. [DOI] [PubMed] [Google Scholar]
- Kol G, Lev-Maor G, Ast G. Human-mouse comparative analysis reveals that branch-site plasticity contributes to splicing regulation. Hum Mol Genet. 2005;14:1559–1568. doi: 10.1093/hmg/ddi164. [DOI] [PubMed] [Google Scholar]
- Konarska MM, Query CC. Insights into the mechanisms of splicing: more lessons from the ribosome. Genes Dev. 2005;19:2255–2260. doi: 10.1101/gad.1363105. [DOI] [PubMed] [Google Scholar]
- Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in S. cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig AJ, Lin RJ, Abelson J. The yeast RNA gene products are essential for mRNA splicing in vitro. Cell. 1986;47:953–963. doi: 10.1016/0092-8674(86)90810-x. [DOI] [PubMed] [Google Scholar]
- Mayas RM, Maita H, Staley JP. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miraglia L, Seiwert S, Igel AH, Ares M., Jr Limited functional equivalence of phylogenetic variation in small nuclear RNA: yeast U2 RNA with altered branchpoint complementarity inhibits splicing and produces a dominant lethal phenotype. Proc Natl Acad Sci USA. 1991;88:7061–7065. doi: 10.1073/pnas.88.16.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison LE, Stols LM. Sensitive fluorescence-based thermodynamic and kinetic measurements of DNA hybridization in solution. Biochemistry. 1993;32:3095–3104. doi: 10.1021/bi00063a022. [DOI] [PubMed] [Google Scholar]
- Nilsen TW. RNA-RNA interactions in nuclear pre-mRNA splicing. In: Simons RW, Grunberg-Manago M, editors. RNA Structure and Function. New York: Cold Spring Harbor Laboratory Press; 1998. pp. 279–307. [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci USA. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Siliciano PG, Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987;49:229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Pascolo E, Séraphin B. The branchpoint residue is recognized during commitment complex formation before being bulged out of the U2 snRNA-pre-mRNA duplex. Mol Cell Biol. 1997;17:3469–3476. doi: 10.1128/mcb.17.7.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman R, Ares M., Jr ATP can be dispensable for prespliceosome formation in yeast. Genes Dev. 2000;14:97–107. [PMC free article] [PubMed] [Google Scholar]
- Perriman R, Barta I, Voeltz GK, Abelson J, Ares M., Jr ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc Natl Acad Sci USA. 2003;100:13857–13862. doi: 10.1073/pnas.2036312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman RJ, Ares M., Jr Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev. 2007;21:811–820. doi: 10.1101/gad.1524307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript Specificity in Yeast Pre-mRNA Splicing Revealed by Mutations in Core Spliceosomal Components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- Ruby SW, Chang TH, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- Schwer B, Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349:494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- Schwer B, Meszaros T. RNA helicase dynamics in pre-mRNA splicing. EMBO J. 2000;19:6582–6591. doi: 10.1093/emboj/19.23.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siatecka M, Reyes JL, Konarska MM. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev. 1999;13:1983–1993. doi: 10.1101/gad.13.15.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Villa T, Guthrie C. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes Dev. 2005;19:1894–1904. doi: 10.1101/gad.1336305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiest DK, O’Day CL, Abelson J. In vitro studies of the Prp9.Prp11.Prp21 complex indicate a pathway for U2 small nuclear ribonucleoprotein activation. J Biol Chem. 1996;271:33268–33276. doi: 10.1074/jbc.271.52.33268. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Wu J, Manley JL. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989;3:1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Xu YZ, Newnham CM, Kameoka S, Huang T, Konarska MM, Query CC. Prp5 bridges U1 and U2 snRNPs and enables stable U2 snRNP association with intron RNA. EMBO J. 2004;23:376–385. doi: 10.1038/sj.emboj.7600050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, McPheeters DS, Yu YT. Psi35 in the branch site recognition region of U2 small nuclear RNA is important for pre-mRNA splicing in S. cerevisiae. J Biol Chem. 2005;280:6655–6662. doi: 10.1074/jbc.M413288200. [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Weiner AM. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989;3:1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Summary of growth on copper of strains carrying either intron branch region mutants, or wt reporter in the presence of U2 snRNA mutants. prp5-TAG and -N399D alleles improve overall splicing of wt reporter in the presence of U2 snRNA alleles mutated at positions G34,ψ 35, A36, and G37 (upper), and improve splicing of intron branch region mutants at two positions, U257 and A258 (lower).
Figure S2. prp5 and prp16 alleles have distinct effects on splicing of suboptimal branch region mutants.
(Upper) Graph of maximum copper concentration that allows cell growth. (Lower) Cell growth on selected copper plates, as indicated. prp5-TAG and -N399D alleles improve splicing of branch region mutant U257C, but provide no additional improvement to branch region mutant (U257C+5bp) that contains extended pairing to U2 snRNA. In contrast, the prp16-101 allele provides only slight improvement to the U257C reporter, but provides significant additional improvement to the U257C+5bp mutant.