Abstract
In the absence of a hepatitis C virus (HCV) culture system, the use of a Semliki Forest virus replicon expressing genes encoding HCV structural proteins that assemble into HCV-like particles provides an opportunity to study HCV morphogenesis. Using this system, we showed that the HCV core protein constitutes the budding apparatus of the virus and that its targeting to the endoplasmic reticulum by means of the signal sequence of E1 protein is essential for budding. In addition, the aspartic acid at position 111 in the HCV core protein sequence was found to be crucial for virus assembly, demonstrating the usefulness of this system for mapping amino acids critical to HCV morphogenesis.
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis and may lead to liver cirrhosis and hepatocellular carcinoma. With an estimated 170 million people worldwide chronically infected with HCV, this disease has emerged as a serious global health problem since the cloning of the viral genome in 1989 (8). Indeed, it has been predicted that chronic HCV infection will lead to an increase in the prevalence of hepatocellular carcinoma in the United States over the next 2 to 3 decades (39). HCV is a small, enveloped, positive-strand RNA virus belonging to the genus Hepacivirus of the Flaviviridae family. Its genome of approximately 9,600 nucleotides contains, at both the 5′ and 3′ ends, untranslated regions (UTRs) which flank a single open reading frame encoding a single polyprotein precursor of about 3,000 amino acids (aa) (13). The viral polyprotein can be broadly divided into two regions: the N-terminal one-third encodes the structural components of the virion, including the putative nucleocapsid or core protein and two envelope proteins (E1 and E2), and the remaining two-thirds encode the nonstructural proteins (22). These nonstructural proteins (NS2 through NS5B) have various functions, particularly in HCV genome replication, during the life cycle of the virus. They form a cytoplasmic membrane-associated complex similar to that formed by related positive-strand RNA viruses. The nonstructural proteins are separated from the structural proteins by the short hydrophobic polypeptide p7, the function of which is unknown (22). Translation to generate HCV polyprotein is initiated via an internal ribosome entry site located within the 5′ UTR. During translation, the mature viral products are generated from the polyprotein by a series of cleavage events. The structural components are produced by cellular protease-mediated cleavages, whereas processing of the nonstructural proteins requires virus-encoded proteases (41). The core protein is produced at the N-terminal end of the polyprotein and is followed by the signal sequence of the E1 envelope glycoprotein. The signal sequence targets the nascent polypeptide chain to the endoplasmic reticulum (ER), allowing the translocation of E1 into the ER lumen, which is essential for the membrane-dependent processing of the core protein (23, 36). In the ER lumen, cleavage by a signal peptidase liberates the N-terminal end of E1, leaving the 191 aa of the core protein anchored by the signal peptide (30). The core protein is then processed by the intramembrane protease SPP, resulting in release of the 173 N-terminal aa of the core protein from the ER (31, 43). This 173-aa core protein is the mature form found in the viral particle (47). HCV polyprotein organization and processing have been well characterized, but little is known about HCV structure and assembly due to the lack of an efficient in vitro culture system for this virus. A significant advance in HCV research was made with the development of subgenomic HCV RNAs, consisting of sequences encoding nonstructural proteins flanked by the 5′ and 3′ UTRs, which self-replicate in the Huh7 hepatoma cell line (5, 26). However, recently developed systems for the expression of the full-length HCV polyprotein in the context of these self-replicating RNAs have not allowed the production of virus particles (34). Using a recombinant Semliki Forest virus (SFV) replicon expressing genes encoding the three HCV structural proteins (core, E1, and E2), Blanchard et al. have demonstrated the assembly of these proteins into HCV-like particles in mammalian cells (3, 4). However, most of these HCV-like particles seem to display an abortive budding process (3, 4). Our attempts to purify HCV-like particles from the cell lysate gave disappointing results. We believe that, in the absence of a membrane completely coating the HCV-like particles, these particles are unstable and may be damaged by the purification procedures. A similar hypothesis was put forward in a recent report describing the in vitro assembly of alphavirus core-like particles (32). Also, our efforts to obtain HCV-like particles in hepatoma cells such as the Huh7 cell line were unsuccessful, since the SFV vector is less efficient in these cells.
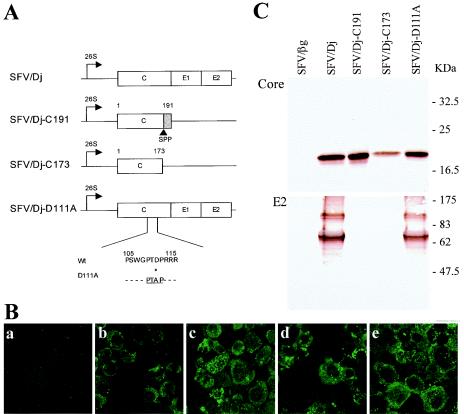
Although the abortive budding of the HCV-like particles limits the interest of our model, this system may constitute an original tool for studying the minimum requirements for HCV morphogenesis in a cellular context (3, 4). The HCV core protein self-assembles in cell-free assays (21). We therefore investigated whether production of the core protein alone, with or without the E1 signal sequence, leads to the morphogenesis of virus-like particles. HCV core sequences were subcloned from the HCV cDNA clone Dj (genotype 1a) containing the C-E1-E2 sequence (clone Dj6.4; GenBank accession number AF529293), which has been described elsewhere (4). The Dj-C191 construct, encoding the 191-aa core protein, and the Dj-C173 construct, encoding the 173-aa core protein (Fig. 1A), were amplified by PCR using PfuTurbo DNA polymerase (Stratagene, La Jolla, Calif.). We introduced a stop codon into each of the antisense primers at the 3′ end of the core protein-coding region. These PCR products were inserted into the BamHI site of the expression vector pSFV1 (Life Technologies, Rockville, Md.) under the control of the SP6 RNA polymerase promoter and upstream from the 5′ SFV UTR (Fig. 1A).
FIG. 1.
Description of the SFV RNAs encoding HCV structural proteins and expression in BHK-21 cells. (A) RNAs transcribed from the four SFV DNA constructs. The SFV/Dj construct is our original Dj6.4 clone, encoding HCV core-E1-E2, and is described elsewhere (4). The horizontal line in each construct indicates the segment of the SFV genome. The position of the subgenomic SFV promoter is indicated (26S). Boxes show the inserted genes, and the shaded box in the SFV/Dj-C191 construct indicates the signal sequence targeting the E1 envelope glycoprotein to the ER. SPP, signal peptide peptidase processing between residues 173 and 174. The SFV/Dj-D111A construct was produced by introducing a D111-to-A substitution in the original SFV/Dj construct, i.e., introducing a viral L domain, PTAP. (B) Immunofluorescence staining, using a monoclonal anti-core antibody, of BHK-21 cells electroporated with SFV RNA β-Gal (a), Dj (b), Dj-C191 (c), Dj-C173 (d), and Dj-D111A (e). This assay was performed by following a standard procedure (35), using the mouse monoclonal anti-HCV core antibody MAb 1856 (Virostat, Portland, Maine). (C) Western blotting of these cells with monoclonal anti-core and anti-E2 antibodies. For this standard assay (4), the anti-core MAb 1856 gave poor results. We therefore carried out immunoblotting with the human monoclonal anti-core antibody B12.F8 (11) and the mouse monoclonal anti-E2 (10). Size markers (in kilodaltons) were obtained from New England Biolabs (Beverly, Mass.).
The incomplete HCV virus-like particle budding process, despite its occurrence at the ER membrane, is reminiscent of the situation in late (L)-domain mutant viruses, in which the failure of the final budding step results in virions remaining permanently tethered to the host cell membrane (29). The L domains are short sequences such as PTAP, PPXY, and YXXL that have been identified in the Gag proteins of a number of retroviruses and in the matrix proteins of rhabdoviruses and filoviruses. These domains play a critical role in the pinching off of virus particles from the plasma membrane (12, 29). We therefore investigated whether the introduction of an L domain into the HCV core sequence would lead to the complete pinching off of the HCV virus-like particles. An L-domain sequence was introduced into the core protein sequence of the Dj clone by mutation, i.e., conversion of the aspartic acid at position 111 into an alanine (PTD111P to PTA111P). This site-directed mutagenesis was performed with a QuikChange PCR mutagenesis kit (Stratagene), using the Dj DNA as a template, to yield mutant Dj-D111A (Fig. 1A). DNA sequencing was used to check that the original sequence was conserved in all constructs and that the D111A mutation had been correctly introduced.
The electroporation of BHK-21 cells with RNA produced by transcription of these constructs was performed as previously described (4). For the control, recombinant β-galactosidase (β-Gal) RNA, encoding the β-Gal protein, was synthesized from the expression vector pSFV3 (Life Technologies). Using immunocytochemistry, we observed no major differences between BHK-21 cells transfected with our various SFV-HCV constructs and stained with a monoclonal anti-core antibody (Fig. 1B). More than 90% of the cells gave a positive result with an intense, cytoplasmic granular pattern of fluorescence, with very little staining of the nucleus. This is consistent with the results of other reports showing a similar subcellular distribution of C191 and C173 core proteins (23, 47), although other studies have reported changes in the distribution of the core protein, from the cytoplasm to the nucleus, if the C terminus is truncated (24, 37, 45). Cells transfected with the β-Gal RNA gave no fluorescent signal. As reported previously (4), a monoclonal antibody directed against the HCV core protein detected by Western blotting two species in BHK-21 cells transfected with the SFV/Dj construct: a major band at approximately 20 kDa and a minor band 19 kDa in size (Fig. 1C). The other constructs gave similar banding patterns, except for the Dj-C173 construct, for which only the larger band was detected. Contrary to what was previously thought (4), these two species do not correspond to the immature and mature forms of the HCV core protein, because expression from the Dj-C173 construct led to the detection of the larger species. This indicates that the smaller species is probably a degradation product of the HCV mature core protein. Such a degradation product was also reported in other studies in which large amounts of HCV core protein were produced (1, 9). Our observations are also consistent with the results of a recent study showing that the production of HCV structural proteins from an SFV vector in BHK-21 cells leads to the detection of a single HCV core species corresponding to the mature form of the core protein (31). The lack of detection of the smaller species with the Dj-C173 construct probably resulted from the smaller amount of HCV core protein produced with this construct than with the others. The lower levels of core protein produced from the Dj-C173 construct were confirmed in repeat experiments. The HCV core protein is a substrate of the ubiquitin-proteasome pathway (38), and this may reflect higher levels of proteolysis of the core protein in the absence of the E1 signal sequence. The monoclonal anti-E2 antibody detected a major band at 70 kDa, corresponding to the glycosylated form of the E2 protein, for both Dj and Dj-D111A (Fig. 1C). A minor band was also detected at 100 kDa. This band may correspond to an E1-E2 heterodimer or to the product of an incomplete cleavage at the site between E1 and E2, as previously described (4). The C191 and C173 constructs gave no signal with the anti-E2 antibody, and cells transfected with the β-Gal RNA gave no bands with either the anti-core or anti-E2 antibody.
HCV core protein was quantified in the cell culture supernatant or cell lysate of BHK-21 cells transfected with the various constructs by means of a commercially available enzyme immunoassay (Total HCV core antigen assay; Ortho Clinical Diagnostics, Raritan, N.J.), performed according to the manufacturer's instructions (Table 1). The smallest amount of intracellular core protein (23 ng per million cells) was obtained with the Dj-C173 construct, whereas the largest amounts were obtained with the Dj and Dj-C191 constructs (112 and 125 ng per million cells, respectively). This was consistent with the results described above for the Western blotting. In contrast, HCV core protein levels were considerably lower, in the range of 1 to 2.5 ng per million cells, in the extracellular medium. This sharp contrast between intracellular and extracellular levels suggests that the release of core protein into the cell medium is more likely to result from cellular lysis than efficient secretion. The highly cytopathic nature of the SFV expression system is consistent with this hypothesis.
TABLE 1.
HCV core protein quantification
| Construct | Amt of core protein (ng/million cells)
|
|
|---|---|---|
| Intracellulara | Extracellularb | |
| β-Gal | Negativec | Negative |
| Dj | 112 | 1.9 |
| Dj-C191 | 125 | 1.0 |
| Dj-C173 | 23 | 1.5 |
| Dj-D111A | 72 | 2.5 |
Cell lysates were tested at a dilution of 1:1,000, and overflow samples were then tested after 1:5,000 dilution.
Cell culture supernatants were tested undiluted, and overflow samples were then tested after 1:10 dilution (all dilutions were made with fresh culture medium).
Below the cutoff value (2.5 pg/ml).
The ultrastructural changes in the BHK-21 cells transfected with the various constructs were studied with electron microscopy techniques extensively described elsewhere (4, 35). In the SFV/Dj RNA-transfected cells, these changes were similar to those reported previously (4). The ER of these cells presented areas of convoluted membranes in which protein self-assembly resulted in the formation of hemispherical structures (Fig. 2A; see also Fig. 4A). Higher magnification of these structures revealed the budding of virus-like particles towards the ER lumen (Fig. 2B). This phenomenon was not detected in cells transfected with the SFV/Dj-C173 RNA (Fig. 2C) or control cells transfected with the SFV/β-Gal RNA (not shown), in which the ER was homogeneously distributed throughout the cytoplasm. In contrast, specific ultrastructural changes were observed in BHK-21 cells transfected with the SFV/Dj-C191 RNA (Fig. 3). In these cells, the convoluted ER membrane displayed a high proportion of linear, electron-dense material (Fig. 3A, arrows). Careful examination of these electron-dense structures suggested that they formed by adhesion to the cytosolic side of these convoluted ER membranes. In some electron micrographs, a dense midline was observed in these structures (Fig. 3B) that resembled those found in intercellular complexes, such as desmosomes (17, 19). Large numbers of virus-like particles budding from these ER convoluted membranes towards the ER lumen were observed in these cells transfected with the SFV/Dj-C191 RNA (Fig. 3C to E). This suggests that HCV budding may be initiated by the core protein, as previously described for retroviruses and their core (Gag) polyprotein (16). Indeed, the envelope (Env) proteins are not required for the budding of retroviruses, and expression of the gag gene alone results in the production of membrane-enveloped “Gag” particles similar to immature virions (16). The incorporation of retrovirus Env glycoproteins into virions is likely to require a direct interaction with viral Gag proteins during assembly, although direct evidence supporting this mechanism has proved to be difficult to obtain (16, 33, 44). In the HCV model, the core, E1, and E2 proteins are cleaved from a single polyprotein inserted into the ER membrane. Thus, budding of the core protein is thought to occur in membrane domains containing the E1 and E2 proteins, and virions are thought to be formed by the passive incorporation of envelope proteins. However, as in the retrovirus model (15, 33), a specific interaction between the HCV core and Env proteins may help to guarantee the efficient incorporation of E1-E2 heterodimers at the virion surface (25). Our results may account for the release of nonenveloped nucleocapsids in the sera of chronic HCV carriers (28). These envelope-free nucleocapsids have been well characterized, but it remains unclear whether they are released into the bloodstream by the secretory pathway or are due to damage to the infected hepatocytes (28). Our model, showing that the core protein initiates HCV budding towards the ER lumen, suggests that these particles may be secreted from infected hepatocytes into the bloodstream. This possibility is also consistent with the observation that HCV core protein produced in bacteria self-assembles into nucleocapsid-like particles in cell-free experiments (21). However, we observed no nucleocapsid-like particles in BHK-21 cells producing the HCV core protein without the E1 signal sequence. These results contrast with those obtained in cell-free assays, in which the first 124 N-terminal residues of the core protein are sufficient for self-assembly into nucleocapsid-like particles. This may reflect differences between cell-free and cellular assays, in which the targeting of the HCV core to the ER and its association with that compartment appear to be required to initiate the formation of a virus-like particle. The assembly of the C191 core protein into virus-like particles at the ER membrane could also explain its higher intracellular level as observed by Western blotting and enzyme immunoassay compared with that of the C173 core protein. In such a multimerized state, the C191 core protein could be less degraded by the proteasome pathway than the cytosolic, C173 core protein.
FIG. 2.
Electron micrographs of ultrathin sections of BHK-21 cells electroporated with SFV/Dj (A and B) or SFV/Dj-C173 (C) RNA. The large arrow in panel A indicates ER, revealing a dilated lumen and convoluted membranes, the arrowheads in panel B indicate virus-like particles budding from these convoluted membranes towards the ER lumen, and the arrows in panel C indicate normal, homogeneously distributed ER structures throughout the cytoplasm. Bars, 1 μm (A), 100 nm (B), and 200 nm (C).
FIG. 4.
Electron micrographs of ultrathin sections of BHK-21 cells electroporated with SFV/Dj (A) or SFV/Dj-D111A (B) RNA. (A) A typical area shows convoluted ER membranes in which the budding of virus-like particles occurs. (B) In the presence of the D111A mutation, the formation of these convoluted ER membranes is retained but no virus particles are detected in these membranes or in the ER lumen. Bars, 1 μm (A) and 500 nm (B).
FIG. 3.
Electron micrographs of ultrathin sections of BHK-21 cells electroporated with SFV/Dj-C191 RNA. (A and B) Areas of convoluted ER membranes displaying adhesion via their cytosolic sides (arrows). In panel B, an electron-dense midline similar to those found in intercellular adhesion complexes such as the desmosome can be observed. (C) A typical area of convoluted ER, revealing double-layered membranes and the budding of virus-like particles from these membranes into the ER lumen. (D and E) High-magnification images of some virus-like particles (arrowheads) budding into the ER. In panel D, note the virus-like budding (arrowhead) occurring close to a region in which ER membranes are adhering via their cytosolic sides (arrow). Bars, 100 nm (A, B, D, and E) and 200 nm (C).
The virus-like particles obtained following transfection with the core C191 construct were similar in terms of ultrastructure to those obtained following transfection with the Dj construct encoding the core-E1-E2 polyprotein. However, in BHK-21 cells transfected with the Dj-C191 construct, electron-dense ER membranes were frequently associated as a double layer, resembling an intercellular adhesion complex. These layered membranes may result from molecular interactions between the HCV core proteins present on the cytosolic side of convoluted ER membrane domains. This phenomenon was repeatedly observed in BHK-21 cells transfected with the Dj-C191 construct but was never seen in BHK-21 cells transfected with the Dj construct. The reasons for this difference are unclear, but there may be an interaction between the HCV core and envelope proteins during or after polyprotein processing (25, 27). Such an interaction may change the transmembrane topology of the core protein in the context of polyprotein production (25, 27).
The amino acid sequence of the core is highly conserved among the six genotypes of HCV (7). Indeed, the core sequence of our Dj clone is fully conserved in various infectious HCV clones (2, 18, 20, 46). We found that the introduction of a single D111A mutation into the core sequence completely altered the formation of HCV-like particles. In BHK-21 cells transfected with the Dj-D111A construct, numerous convoluted ER membranes that resembled those observed in cells transfected with SFV/Dj RNA were observed by electron microscopy (Fig. 4B). However, despite intensive searching of these convoluted membranes and the ER lumen, we could detect no virus-like particle in cells transfected with SFV/Dj-D111A RNA. This situation is very different from that for cells transfected with the Dj construct, despite similar levels of core protein detection by Western blotting or enzyme-linked immunosorbent assay. This suggests that the D111A mutation hampers HCV core particle formation. A similar observation was made with another clone of our original Dj construct, bearing the same D111A mutation (data not shown). Interestingly, a D-to-A mutation at position 183 of the Gag protein has been found to abolish assembly of the HIV-1 core protein (40, 42). It should also be noted that D111 in the HCV core forms part of a DPR domain fully conserved in HCV sequences and in the sequence of the closely related GB virus B (6), reflecting the importance of this domain in HCV and GB virus B biology. It would be of interest to investigate the consequences of such a mutation for the assembly of HCV core protein in cell-free assays developed by other groups (21).
In conclusion, our HCV virus-like particle model demonstrates that the HCV core protein constitutes the budding apparatus and that the aspartic acid at position 111 in this protein plays a critical role in budding. This model may be useful for studies of the mechanisms of HCV morphogenesis and for mapping of other amino acids playing crucial roles in virus assembly. As the recommended treatment for HCV infection is of only limited efficacy (14, 48), this model could also be useful in screening HCV assembly inhibitors, which may provide the basis for new antiviral strategies.
Acknowledgments
We thank Mario Mondelli and Jean Dubuisson for providing the monoclonal human anti-core antibody and the monoclonal murine anti-E2 antibody, respectively. We are indebted to Jean-Alain Jarricot, Ortho Clinical Diagnostics, for providing the HCV core antigen assays.
This work was supported by a grant from the Réseau National Hépatite (Ministère de la Recherche), by grant “Hépatite C” from the Agence Nationale pour la Recherche sur le sida (ANRS), and by grant “pseudoviruses” (IFR 82 Transposons and Virus) from the Région Centre, France. Emmanuelle Blanchard and Malika Ait-Goughoulte are supported by fellowships provided by the Région Centre and the Ministère de la Recherche, respectively.
REFERENCES
- 1.Acosta-Rivero, N., J. C. Aguilar, A. Musacchio, V. Falcon, A. Vina, M. C. de la Rosa, and J. Morales. 2001. Characterization of the HCV core virus-like particles produced in the methylotrophic yeast Pichia pastoris. Biochem. Biophys. Res. Commun. 287:122-125. [DOI] [PubMed] [Google Scholar]
- 2.Beard, M. R., G. Abell, M. Honda, A. Carroll, M. Gartland, B. Clarke, K. Suzuki, R. Lanford, D. V. Sangar, and S. M. Lemon. 1999. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology 30:316-324. [DOI] [PubMed] [Google Scholar]
- 3.Blanchard, E., D. Brand, and P. Roingeard. 2003. Endogenous virus and hepatitis C virus-like particle budding in BHK-21 cells. J. Virol. 77:3888-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchard, E., D. Brand, S. Trassard, A. Goudeau, and P. Roingeard. 2002. Hepatitis C virus-like particle morphogenesis. J. Virol. 76:4073-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blight, K.-J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 6.Bukh, J., C. L. Apgar, and M. Yanagi. 1999. Toward a surrogate model for hepatitis C virus: an infectious molecular clone of the GB virus-B hepatitis agent. Virology 262:470-478. [DOI] [PubMed] [Google Scholar]
- 7.Bukh, J., R. H. Purcell, and R. H. Miller. 1994. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc. Natl. Acad. Sci. USA 91:8239-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 9.Clayton, R. F., A. Owsianka, J. Aitken, S. Graham, D. Bhella, and A. H. Patel. 2002. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J. Virol. 76:7672-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esposito, G., E. Scarselli, A. Cerino, M. U. Mondelli, N. La Monica, and C. Traboni. 1995. A human antibody specific for hepatitis C virus core protein: synthesis in a bacterial system and characterization. Gene 164:203-209. [DOI] [PubMed] [Google Scholar]
- 12.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heathcote, E. J., M. L. Shiffman, W. G. Cooksley, G. M. Dusheiko, S. S. Lee, L. Balart, R. Reindollar, R. K. Reddy, T. L. Wright, A. Lin, J. Hoffman, and J. De Pamphilis. 2000. Peginterferon alfa-2a in patients with chronic hepatitis C and cirrhosis. N. Engl. J. Med. 343:1673-1680. [DOI] [PubMed] [Google Scholar]
- 15.Hourioux, C., D. Brand, P.-Y. Sizaret, F. Lemiale, S. Lebigot, F. Barin, and P. Roingeard. 2000. Identification of the glycoprotein 41TM cytoplasmic tail domains of human immunodeficiency virus type 1 that interact with Pr 55 gag particles. AIDS Res. Hum. Retrovir. 16:1141-1147. [DOI] [PubMed] [Google Scholar]
- 16.Hunter, E. 1994. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin. Virol. 5:71-83. [Google Scholar]
- 17.Jamora, C., and E. Fuchs. 2002. Intercellular adhesion, signalling and the cytoskeleton. Nat. Cell Biol. 4:101-108. [DOI] [PubMed] [Google Scholar]
- 18.Kato, N., M. Hijikata, Y. Ootsuyama, M. Nakagawa, S. Ohkoshi, T. Sugimura, and K. Shimotohno. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 87:9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitajima, Y. 2002. Mechanisms of desmosome assembly and disassembly. Clin. Exp. Dermatol. 27:684-690. [DOI] [PubMed] [Google Scholar]
- 20.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 21.Kunkel, M., M. Lorinczi, R. Rijnbrand, S. M. Lemon, and S. J. Watowich. 2001. Self-assembly of nucleocapsid-like particles from recombinant hepatitis C virus core protein. J. Virol. 75:2119-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lidenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 23. Liu, Q., C. Tackney, R. A. Bhat, A. M. Prince, and P. Zhang. 1997. Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J. Virol. 71:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo, S. Y., F. Masiarz, S. B. Hwang, M. M. Lai, and J. H. Ou. 1995. Differential subcellular localization of hepatitis C virus core gene products. Virology 213:455-461. [DOI] [PubMed] [Google Scholar]
- 25.Lo, S. Y., M. J. Selby, and J. H. Ou. 1996. Interaction between hepatitis C virus core protein and E1 envelope protein. J. Virol. 70:5177-5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 27.Ma, H. C., C. H. Ke, T. Y. Hsieh, and S. Y. Lo. 2002. The first hydrophobic domain of the hepatitis C virus E1 protein is important for interaction with the capsid protein. J. Gen. Virol. 83:3085-3092. [DOI] [PubMed] [Google Scholar]
- 28. Maillard, P., K. Krawczynski, J. Nitkiewicz, C. Bronnert, M. Sidorkiewicz, P. Gounon, J. Dubuisson, G. Faure, R. Crainic, and A. Budkowska. 2001. Nonenveloped nucleocapsids of hepatitis C virus in the serum of infected patients. J. Virol. 75:8240-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 30. McLauchlan, J. 2000. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J. Viral Hepat. 7:2-14. [DOI] [PubMed] [Google Scholar]
- 31.McLauchlan, J., M. K. Lemberg, G. Hope, and B. Martoglio. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukhopadhyay, S., P. R. Chipman, E. M. Hong, R. J. Kuhn, and M. G. Rossmann. 2002. In vitro-assembled alphavirus core-like particles maintain a structure similar to that of nucleocapsid cores in mature virus. J. Virol. 76:11128-11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murakami, T., and E. O. Freed. 2000. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J. Virol. 74:3548-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roingeard, P., S. Lu, C. Sureau, M. Freschlin, B. Arbeille, M. Essex, and J.-L. Romet-Lemonne. 1990. Immunocytochemical and electron microscopic study of hepatitis B virus antigen and complete particle production in hepatitis B virus DNA transfected HepG2 cells. Hepatology 11:277-285. [DOI] [PubMed] [Google Scholar]
- 36.Santolini, E., G. Migliaccio, and N. La Monica. 1994. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol. 68:3631-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki, R., Y. Matsuura, T. Suzuki, A. Ando, J. Chiba, S. Harada, I. Saito, and T. Miyamura. 1995. Nuclear localization of the truncated hepatitis C virus core protein with its hydrophobic C terminus deleted. J. Gen. Virol. 76:53-61. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki, R., K. Tamura, J. Li, K. Ishii, Y. Matsuura, T. Miyamura, and T. Suzuki. 2001. Ubiquitin-mediated degradation of hepatitis C virus core protein is regulated by processing at its carboxyl terminus. Virology 280:301-309. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka, Y., K. Hanada, M. Mizokami, A. E. T. Yeo, J. W.-K. Shih, T. Gojobori, and H. J. Alter. 2002. A comparison of the molecular clock of hepatitis C virus in the United States and Japan predicts that hepatocellular carcinoma incidence in the United States will increase over the next two decades. Proc. Natl. Acad. Sci. USA 99:15584-15589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang, C., Y. Ndassa, and M. F. Summers. 2002. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 9:537-543. [DOI] [PubMed] [Google Scholar]
- 41.Tellinghuisen, T. L., and C. M. Rice. 2002. Interaction between hepatitis C virus proteins and host cell factors. Curr. Opin. Microbiol. 5:419-427. [DOI] [PubMed] [Google Scholar]
- 42.Von Schwedler, U. K., T. L., Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weihofen, A., K. Binns, M. K. Lemberg, K. Ashman, and B. Martoglio. 2002. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 296:2215-2218. [DOI] [PubMed] [Google Scholar]
- 44.West, J. T., S. K. Weldon, S. Wyss, X. Lin, Q. Yu, M. Thali, and E. Hunter. 2002. Mutation of the dominant endocytosis motif in human immunodeficiency virus type 1 gp41 can complement matrix mutations without increasing Env incorporation. J. Virol. 76:3338-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamanaka, T., M. Uchida, and T. Doi. 2002. Innate form of HCV core protein plays an important role in the localization and the function of HCV core protein. Biochem. Biophys. Res. Commun. 294:521-527. [DOI] [PubMed] [Google Scholar]
- 46.Yanagi, M., M. St. Claire, M. Shapiro, S. U. Emerson, R. H. Purcell, and J. Bukh. 1998. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology 244:161-172. [DOI] [PubMed] [Google Scholar]
- 47.Yasui, K., T. Wakita, K. Tsukiyama-Kohara, S. I. Funahashi, M. Ichikawa, T. Kajita, D. Moradpour, J. R. Wands, and M. Kohara. 1998. The native form and maturation process of hepatitis C virus core protein. J. Virol. 72:6048-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeuzem, S., S. V. Feinman, J. Rasenack, E. J. Heathcote, M. Y. Lai, E. Gane, J. O'Grady, J. Reichen, M. Diago, A. Lin, J. Hoffman, and M. J. Brunda. 2000. Peginterferon alfa-2a in patients with chronic hepatitis C. N. Engl. J. Med. 343:1666-1672. [DOI] [PubMed] [Google Scholar]