Abstract
Advances in genomic technology have improved our understanding of the genetic events that parallel breast cancer development. Because almost all mammary carcinomas develop in the terminal duct lobular units of the breast, understanding the events involved in mammary gland development make it possible to recognize those events that, when altered, contribute to breast neoplasia. In this review we focus on lobular carcinomas, discussing the pathology, development, and progression of premalignant lobular lesions from a genomic point of view. We highlight studies utilizing genomic approaches and describe how these investigations have furthered our understanding of the complexity of premalignant breast lesions.
Introduction
Breast cancer has been hypothesized to develop through a linear histological progression from hyperplasia and in situ carcinoma to invasive cancer, a progression model that is possibly similar to that described for colon cancer [1,2]. It has been suggested that this process is accompanied by increasing genomic instability, among other hallmarks of cancer [3]. Recent advances in genomic technology have improved our understanding of the accumulation of genetic events that parallel breast cancer development; they have also revealed the complexity of premalignant lesions. Two major breast cancer subtypes are ductal and lobular, and the genetics of premalignant lobular breast lesions has only recently been explored. Findings suggest that the genomic events identified in hyperplasias and in situ carcinomas may be causative for the development of premalignant lesions, thus triggering or disrupting the downstream events that lead to disease progression [4]. In this review we focus on the development, pathology, and progression of lobular breast lesions, and describe our current understanding of premalignant lesions from a genomic point of view.
An overview of mammary gland development
General structure of the mammary gland
The mammary gland is composed of an organized ductal network. Embedded within the stroma, the branching duct system leads from the collecting ducts via the segmental and subsegmental ducts to the terminal duct lobular units (TDLUs) [5]. Two cell types compose the epithelium of the duct and lobule system, namely luminal (secretory) cells and myoepithelial cells. The myoepithelial cell layer is found between the luminal epithelial cell layer and the basement membrane [5,6].
Mammary development and cancer
Almost all mammary carcinomas develop within the TDLU or the terminal ducts that enter the lobular units. An understanding of the development of the TDLU is fundamental to discerning the events that are involved in neoplastic growth. From studies of mouse mammary development and human mammary epithelial cells grown in three-dimensional culture, acini structures have been recapitulated and manipulated in order to elucidate organization and development [5,7,8].
The acini that compose the TDLU are spheroid structures with a central lumen surrounded by a layer of polarized epithelial cells [7]. The epithelial cells that line the acini have polarity markers, which include (but are not limited to) epithelial (E)-cadherin and αβ-integrins [7,9]. Blocking E-cadherin causes selective disorganization of the luminal cells; this relationship between tissue structure and developmental morphogenesis has been corroborated in mouse models (for review [10]). The bilayered configuration of the ducts and the acinar structure are established during TDLU development. Following epithelial cell polarization, apoptotic signals counter the proliferative signals produced by the cells residing in the luminal space, allowing for hollowing of the acini in a process known as luminal morphogenesis. Subsequent regulation of epithelial cell size, number, and survival maintains this structure [11].
Molecular factors important to the development of premalignant breast lesions
Pathways implicated in luminal morphogenesis
Specific pathways have been implicated in the development of the structures that comprise the mammary gland. The above mentioned process of luminal morphogenesis [7] is one example. In vitro studies using three-dimensional cultures have identified that proper formation of the hollow bilayered acinar structure at the end of a terminal duct involves the initiation of proliferative and apoptotic signals, leading to the formation of the lumen. In particular, the cells located in the center of the luminal space have been reported to lack AKT pathway signals [11,12]. A lack of AKT survival signals would allow for selective cell death, which, in the case of luminal morphogenesis, contributes to the formation of the hollow lumen.
It is clear that the proliferative and apoptotic signals fundamental to luminal morphogenesis exist in a delicate balance. Further evidence for this involves colony-stimulating factor (CSF)1 receptor, a protein tyrosine kinase transmembrane receptor that, when activated by its ligand CSF1, signals through Ras, allowing for cell growth and differentiation. Studies using three-dimensional mammary culture models have reported that constitutive CSF1 receptor signaling causes uncontrolled proliferation and altered E-cadherin mediated adhesion, thereby disrupting normal mammary structure and development [13]. Interestingly, loss of E-cadherin expression is also a significant feature of lobular carcinomas [14,15].
In general, these studies provide additional evidence that altering the balance of proliferative and apoptotic signals can lead to the accumulation of cells within the luminal space of the acini [12], which is a characteristic of neoplastic breast lesions. However, not all of the players that are involved in luminal morphogenesis have yet been identified. Continued research in this area will further expand our understanding of this essential developmental process in the mammary gland.
Mammary gland microenvironment
The microenvironment and/or adjacent cells may also contribute to the development of premalignant breast lesions. The stroma within the mammary gland is composed of adipocytes, fibroblasts, inflammatory cells, blood vessels, and extracellular matrix. These components contribute instructive and permissive signals during development. Many stromal factors that are essential to mammary gland development have also been found to be associated with cancer. For example, the interaction between the epithelium and the extracellular matrix promotes branching morphogenesis during normal mammary development [16]. Transforming growth factor-β, a key player in tumorigenesis, is a stromal factor that functions to prevent inappropriate branching. If transforming growth factor-β signaling is inhibited, excess ductal branching results [17]. Thus, as this example suggests, although the epithelium is responsible for proliferation, invasion, and tumorigenic potential, the microenvironment within which premalignant lesions develop can play an important role.
Pathology of lobular breast lesions
Histological description
Lobular neoplasia represents a broad range of noninvasive (in situ) proliferations of 'lobular cells' in the breast. These lesions share the distinctive cytology of invasive lobular cells and an almost exclusive distribution within the acini of a TDLU; such characteristics give these lesions a signature histologic appearance.
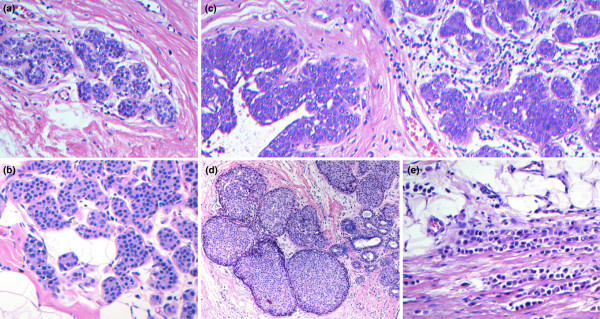
The classic cells of lobular neoplasia are small and round, with uniform bland nuclei [18]. The nuclei are often central but can be paracentral with an occasional cytoplasmic vacuole, representing an invagination of the cell membrane as evidenced by electron microscopy [19]. The loss of E-cadherin in lobular cells imparts the discohesive quality apparent on microscopic sections and reflects a characteristic pattern of pagetoid spread and diffuse invasion [20,21]. Although morphologically distinct, lobular neoplasia is quite varied in terms of the extent of disease both within the TDLUs and within the breast in general, and as such is classified into significant clinical entities (as verified by follow up). These classifications include minimal atypical lobular hyperplasia (MALH), atypical lobular hyperplasia (ALH), with or without ductal involvement by cells of ALH (DIALH), lobular carcinoma in situ (LCIS), and invasive lobular carcinoma (ILC; Figure 1)
Figure 1.
Lobular neoplasia. The classification of lobular neoplasia is determined by the extent of disease both within the terminal duct lobular units and within the breast in general. The clinical entities that fall into the classification of lobular neoplasia include (a) minimal atypical lobular hyperplasia (magnification: 200×), (b) atypical lobular hyperplasia (ALH; 200×), (c) ALH with ductal involvement by cells of ALH (200×), (d) lobular carcinoma in situ (100×), and (e) invasive lobular carcinoma (200×). All lesions were stained with hematoxylin and eosin.
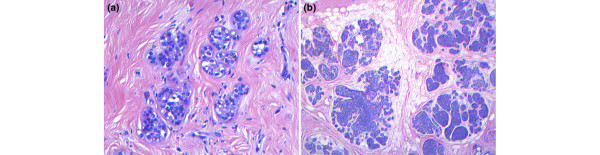
Most lobular lesions are classified as ALH, which is defined as the expansion of a lobular unit with a uniform population of cells. In a TDLU, evident expansion of at least one acinar space, with a minimum of eight cells across, is mandatory for the diagnosis of ALH [22]. The designation MALH is appropriate in cases in which the lobular unit is populated by cells of ALH with minimal to no expansion of the acini (four to five cells across an acinar diameter). The distinction between ALH and MALH is important because MALH represents a significantly lower risk for subsequent development of invasive carcinoma than does classic ALH. Regressed ALH is a pattern of ALH that may be identified in postmenopausal women in which the ALH lesion is less developed; has a more varied and scant population of lobular cells with degenerating changes, as compared with classical examples; and may lack the minimum criteria for diagnosis (Page DL, personal communication; Figure 2a). These morphologic characteristics may prove to be related to the lower associated cancer risk reported in postmenopausal women diagnosed with ALH.
Figure 2.
Suggested new entities of lobular neoplasia. (a) Regressed atypical lobular hyperplasia (ALH; magnification 200×) is a pattern of ALH present in postmenopausal women, in which the ALH lesion is less developed; has a more varied population of lobular cells than classical ALH, with the presence of apoptotic bodies; and meets the minimum criteria for diagnosis. (b) Tumor-forming lobular carcinoma in situ (LCIS; 20×) is defined by clustering of LCIS in at least two adjacent lobular units, and may have specific implications for cancer risk and required excision. Lesions were stained with hematoxylin and eosin.
DIALH describes the pattern of pagetoid spread that cells of ALH exhibit when traveling down ducts, undermining the normal ductal epithelium. This lesion is usually observed in association with ALH or LCIS [23]. Moreover, LCIS represents the higher spectrum of ALH, with marked distention and distortion of more than 50% of acini within a lobular unit, and may be designated tumor forming when clustered (Figure 2b).
Breast cancer risk
The clinical implications for cancer vary between the different subtypes of lobular neoplasia. Diagnostic ALH has been shown to be a general indicator for bilateral cancer risk, with a fourfold to fivefold increase in relative risk compared with the general population [22,24-26]. It should be noted that although ALH may be linked to increased risk in both breasts, a recent study has shown that the ipsilateral breast (compared with the contralateral breast) is somewhat more likely to develop subsequent invasive disease [25,27]. Cancer risk in ALH also appears to vary with patient age; an initial diagnosis of ALH in women older than 55 years (postmenopausal) has been linked to a lower associated cancer risk, which is not true of lesions in younger premenopausal women [25,27]. Interestingly, it is in the lesions found in postmenopausal women that we observe regressed ALH (see above). Taken together, these factors suggest that regressed ALH may have morphologic characteristics, and possibly a genomic signature, that distinguish it from classic ALH.
Lesions that exhibit sufficient distention and distortion for the diagnosis of LCIS have been found to carry an associated breast cancer risk twice that of ALH [28,29]. Although DIALH was initially believed to increase risk similarly to LCIS [23], a more recent study involving a longer period of follow up reported that this increased risk was not statistically significant [27].
Lobular verses ductal lesions
Compared with lobular lesions, atypical ductal hyperplasia and ductal carcinoma in situ (DCIS) exhibit varied patterns, including solid, cribriform, and micropapillary. In terms of risk, atypical ductal hyperplasia appears to carry a slightly lower generalized increased cancer risk than does ALH, with a relative risk of about three to four times that in the general population [24,25]. DCIS is a localized premalignant lesion and, if not excised, is highly likely to progress locally to invasive carcinoma.
Pathologic indications for progression
Lobular neoplasia is a definitive risk indicator for the development of breast cancer. This association was first demonstrated by Foote and Stewart in 1941 [30], with a report that described the co-occurrence of LCIS and ILC in 60% of cases in their study. These seminal findings began the investigations into the evolution of lobular neoplasia and were recently reproduced by Bratthauer and Tavassoli [31], who reported a 23% coincident incidence of invasive cancer in association with LCIS, with 86% of the invasive tumors being ILC. This evidence is quite compelling but the association remains morphologic and the capacity to predict progression is limited. However, underlying genomic alterations, as discussed in the following sections, may significantly enhance our ability to predict neoplastic behavior.
The genomic point of view
In the past, loss of heterozygosity (LOH) [32], fluorescence in situ hybridization [33], and chromosomal comparative genomic hybridization (CGH) [34] were techniques used to identify the genetic alterations underlying solid human neoplasms. More recently, however, there has been an increasing use of array-CGH [35], a high-throughput technique for the high-resolution detection of copy number alterations at a genome-wide level.
Whereas chromosomal-CGH allows for the identification of regions of DNA gain/loss of more than 20 megabases throughout the genome by hybridization of a sample to metaphase chromosomes immobilized on a glass slide [34], array-CGH has the same theoretical end-point but with a refined process and resolution. Most recently, using a genomic DNA template and tiling bacterial artificial chromosome (BAC) microarray, copy number alterations can be identified at the submegabase level [36]. Array-CGH can be manipulated to evaluate the genome of any cell and has been applied to the identification and differentiation of breast cancers [37,38].
Array-CGH data has been proven to complement gene expression data obtained from cDNA microarrays [37,39,40]. Coupling the gain/loss of chromosomal regions with the increase/decrease of gene expression has identified significantly altered genes that are putative mediators of tumor formation or progression.
Genomics and premalignant lobular breast lesions
Genomic alterations in lobular neoplasia
Molecular studies of ALH are sparse, limited by the small size and rarity of these lesions. To date, only two studies have investigated ALH for genomic alterations. Lu and colleagues [41] examined ALH (and LCIS) lesions, as well as the associated invasive breast cancers, using chromosomal-CGH and identified alterations at chromosomes 6, 16, 17, and 22. These alterations were found at similar frequencies in ALH and LCIS. Based on the CGH profiles and the average number of genomic alterations found in both types of lesion (2.9 in both), those investigators suggested that ALH and LCIS represent the same genetic stage of development.
More recently, Mastracci and coworkers [4] profiled solitary ALH lesions using a whole-genome tiling BAC-array. This study represents the most in-depth investigation of the ALH genome. The reported genomic signature identified and further refined the alterations reported by Lu and colleagues [41]. Novel regions of copy number gain and loss, including gain at 2p11.2 and loss at 7p11-p11.1 and 22q11.1, were determined to be common to ALH. In this study, the authors also investigated solitary LCIS [4]. Novel regions of copy number alteration were found to be common to LCIS, including gain at 20q13.13 and loss at 19q13.2-q13.31. In both ALH and LCIS the region of loss at 16q21-q23.1 was identified to be altered. Furthermore, genes within regions of alteration common to both ALH and LCIS were identified to be involved in the process of luminal morphogenesis.
To date, no studies have investigated genomic alterations in MALH or DIALH. Three studies have analyzed LCIS with adjacent invasive carcinoma using array-CGH platforms. Shelley Hwang and coworkers [42] evaluated 24 synchronous LCIS and ILC lesions and found alterations of 16q loss, 1q gain, 11q14-11qter loss, and 11q11-q13 loss. Moreover, in their case report, Nyante and colleagues [43] evaluated three synchronous lesions, including DCIS, LCIS, and ILC. They detected two different genomic signatures. The LCIS and ILC were found to harbor loss at 1p, 16q, and 17p/q, whereas the DCIS exhibited loss at 6q and 11q and gain at 10p, 11q, and 15q. The third study, that reported by Morandi and coworkers [44], investigated the clonality of synchronous lobular neoplasia and ILC. They identified common regions of alteration, including gain at 19p, 6p, 1p, 3p, 16p, 21q and 2q, and loss at 16q and 19q. From this study lobular neoplastic lesions were reported to be genetically related to ILC and consequently categorized as precursors.
Alterations found in invasive lobular carcinomas
Loss of 16q is one of the most frequent events found in breast cancer and appears to be the most consistent alteration found in ILC by both chromosomal-CGH and array-CGH. Chromosomal-CGH studies identified a myriad of alterations, but these reports only agree that lobular carcinomas have a recurrent loss of 16q (where the E-cadherin gene [CDH1] is located), a frequent gain of 1q, and are different from invasive ductal carcinomas (IDCs) [45-49]. These particular genomic alterations were again recently identified in a study of classic lobular carcinomas, which employed both high-resolution CGH and array-CGH [50]. Moreover, an array-CGH study conducted by Roylance and colleagues [51] identified that ILC and low-grade IDC exhibit loss of the whole arm of 16q, whereas high-grade IDC shows regional gain and loss on 16q but infrequent whole-arm loss. However, despite the studies distinguishing lobular from ductal carcinomas, and notwithstanding the advancements in high-throughput technology, there is currently no accepted genomic signature specific for ILC.
A comprehensive list of the alterations identified to date in lobular carcinomas is presented in Table 1.
Table 1.
Published reports of genomic alterations identified by comparative genomic hybridization in lobular carcinoma
| Cases studied | Noted lobular-specific alterations | ||||||
| Reference | ALH/LCIS | Invasive | Other | Type of CGH | Gain | Loss | Observations |
| Lu and coworkers [41] | ALH and LCIS | IBC (adjacent in 6 cases) | Chromosomal | 6q | 16p, 16q, 17p, and 22q | Alterations were found at a similar high frequency in LCIS and ALH | |
| Buerger and coworkers [45] | LCIS | ILC (adjacent LCIS in 4 cases) | DCIS | Chromosomal | 1q | 16q | LCIS characterized by low average rate of copy number changes; no evidence of amplification in LCIS |
| Gunther and coworkers [46] | ILC and IDC | Chromosomal | 16q, 17q, and 22 | Lower frequency of gain at 8q in ILC compared with IDC; changes of equal frequency include gain at 1q, and loss at 19p and parts of 1p and 11q | |||
| Weber-Mangal and coworkers [47] | ILC and IDC | IBC | Chromosomal | 1q, 8q, and 11q | 16q, 17p, and 22q | Lobular alterations were identified in a table of alterations; however, the study investigated alterations in breast cancer in general | |
| Nishizaki and coworkers [48] | ILC and IDC | Chromosomal | 1q | 16q | Compared ILC with IDC; IDC had higher frequency of gain at 8q and 20q | ||
| Etzell and coworkers [49] | LCIS | Chromosomal | 1q | 8p, 12q24, 16q, and 17p | Correlated 16q loss with loss of expression of E-cadherin by immunohistochemistry | ||
| Mastracci and coworkers [4] | ALH and LCIS | SMRT BAC-array | 2p11.2 and 20q13.13 | 7p11.2, 16q21-q23.1, 19q13.2, and 22q11.1 | Alterations found in common between ALH and LCIS; also identified changes that were specific to either ALH or LCIS | ||
| Loo and coworkers [38] | ILC | IDC | BAC-array | 1q32, 8p23, 11q13, and 11q14 | 16q23 and 16q24 | Found differences between ILC and IDC, stratified by histologic type and estrogen receptor status | |
| Hwang and coworkers [42] | LCIS | ILC (synchronous) | BAC-array | 1q | 11q11-q13, 11q14-qter, and 16q | Clonality was suggested for the genetic relationship between LCIS and ILC | |
| Nyante and coworkers [43] | LCIS | ILC | DCIS | BAC-array | 1p, 16q, and 17p/q | A different profile was identified for DCIS | |
| Morandi and coworkers [44] | ILC | Oligo-array | 1p, 2q, 3p, 6p, 16p, 19p, and 21q | 16q and 19q | Lobular neoplastic lesions are genetically related to ILC and can be categorized as precursors | ||
| Reis-Filho and coworkers [50] | ILC | High-resolution CGH and array-CGH | 1q, 5p, 7q, 11p, 11q, 12q, 14q, 16p, 18p, 19p+q, and 20p+q | 11q, 13q, 16q, 18q, and Xq | Lobular carcinomas have greater genetic complexity and a higher number of recurrent genomic changes than previously reported with other techniques | ||
| Roylance and coworkers [51] | ILC and IDC | 16q BAC-array | 16q (whole chromosome arm) | Alterations on 16q are common to both IDC and ILC; higher grade IDCs have more complex changes on 16q | |||
The 'cases studied' column notes the breast lesion investigated in each study, i.e. ALH/LCIS, Invasive carcinoma or other breast lesions. The 'noted lobular-specific alterations' column notes the chromosomal gains and losses identified in each study. ALH, atypical lobular hyperplasia; BAC, bacterial artificial chromosome; DCIS, ductal carcinoma in situ; IBC, invasive breast cancer; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; LCIS, lobular carcinoma in situ ; LN, lobular neoplasia; SMRT, submegabase resolution tiling array.
Are we closer to understanding the development of premalignant lobular breast lesions?
Cases containing ALH and/or LCIS without adjacent invasive carcinomas are known to occur in only 0.5% to 3.8% of breast cases that are otherwise benign. Despite their rarity, these cases spark intrigue with respect to progression. Epidemiologic studies have not provided concrete evidence for progression, but rather have deemed these lesions to be risk indicators. From a molecular perspective, the cells that characterize premalignant lesions may not have acquired all of the 'hallmarks' of cancer cells. Those properties relating to the development of the normal mammary gland are potential candidates for malfunction in the cells that define premalignant breast lesions.
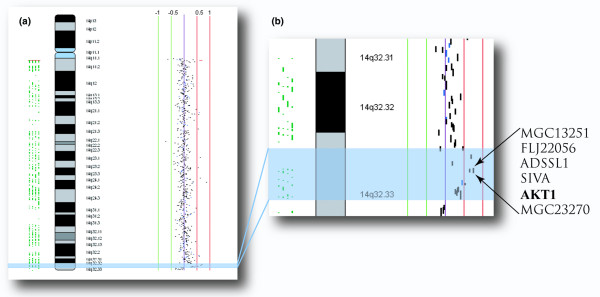
Specific clues to the development of lobular neoplastic lesions have only recently been described. Unbalanced proliferative and apoptotic signals that contribute to altered luminal morphogenesis may provide biologic cues that initiate lobular neoplastic proliferation. Genomic alterations, described by array-CGH, in regions that contain genes involved in luminal morphogenesis provided the first evidence for the link between these clinical entities and the developmental process [4]. One example is AKT1, a gene identified to be altered at the 14q32.33 locus in both ALH and LCIS (Figure 3). A lack of AKT pathway survival signals is characteristic of the cells located in the lumen of the acinar structure during the process of luminal morphogenesis. An increase in the amount of AKT1 would provide the pro-proliferative signals necessary for the survival and proliferation of these cells.
Figure 3.
Genomic techniques. Genomic techniques can identify alterations that are fundamental and possibly functionally significant to development of premalignant breast lesions. (a) An example of a gain on chromosome 14 is illustrated in this ideogram (data from Mastracci and coworkers [4]). The specific region of gain at 14q32.33 (highlighted in blue) was identified in atypical lobular hyperplasia and lobular carcinoma in situ. (b) The region of gain at 14q32.33 magnified and showing the genes encompassed by the amplified bacterial artificial chromosomes. AKT1 has been implicated in the process of luminal morphogenesis, and a copy number gain of the region containing AKT1 could produce the reported effect on this developmental process.
Do certain genomic alterations give clues to progression?
Synchronous lesions of LCIS and ILC appear to have a high degree of similarity. Molecular studies investigating these lesions for LOH [52], protein expression of the E-cadherin adhesion complex [15], and mutations in CDH1 and LOH at 16q [14] suggest a relationship between adjacent lesions. As outlined in the preceding section, recent array-CGH studies have also reported similarities in the genomic alterations identified in adjacent lobular lesions.
From a molecular point of view it has been suggested that ALH and LCIS are nonobligate precursor lesions. Thus, for progression to ensue, these solitary lobular neoplastic lesions would require additional molecular or genetic events to take place. Interestingly, a recent study conducted in a mouse mammary tumor model investigated the somatic loss of E-cadherin [53]. This study provided evidence that loss of E-cadherin in a p53-deficient background plays a role in the initiation, progression, and metastasis of mammary carcinomas. In particular, these mice developed tumors that appeared strikingly similar to human ILC, with evidence of anoikis resistance, angiogenesis, and acquired potential for stromal invasion. Thus, in premalignant lobular lesions the detection of loss of 16q (the locus that harbors the E-cadherin gene) might point to possible progression toward an invasive phenotype, given the frequency of this alteration in invasive breast cancer and the functional effect of loss of CDH1.
Additional chromosomal alterations, including loss at 1p, 11q and 17p/q, and gain at 1q, identified in synchronous lobular lesions may also contribute to progression [41-43]. The presence of these alterations in both the invasive and adjacent premalignant lesions suggests that genes within these altered regions may alter the proliferation program or affect cell death, thereby contributing to and/or maintaining the invasive properties of these cells. In general, alterations identified in adjacent in situ and invasive lobular lesions could provide a selective advantage during tumor initiation, thereby encouraging tumor development and progression.
Is there genomic evidence to support lobular breast cancer progression?
Compelling evidence for progression has yet to be reported. Advances in the resolution of array-CGH technology allow specific identification of genomic breakpoints. As such, studies identifying identical genomic breakpoints in solitary lobular neoplastic lesions and solitary ILC would provide strong support for progression from premalignant to invasive disease. Loss at 16q and gain at 1q have been suggested to be frequent events linked to progression, but identical breakpoints within these, or novel loci, would provide direct evidence linking genomic alterations to breast cancer progression.
Conclusion
Recent advances in genomic technology have provided new tools for in-depth, fine-scale mapping of genomes, allowing successful study of small premalignant breast lesions at the genome level. An understanding of the events that are involved in mammary gland development makes it possible to recognize those events that, when altered, contribute to breast neoplasia. However, recent genomic studies have only begun to uncover clues to neoplastic development in the breast. Definitive proof of the progression to invasive disease will require the discovery of identical breakpoints in adjacent lesions using array-CGH technology. Continued evolution of the technology and further investigation will contribute to a better understanding of the pathways and processes that are involved in the development and progression of lobular breast cancer.
Abbreviations
ALH = atypical lobular hyperplasia; BAC = bacterial artificial chromosome; CGH = comparative genomic hybridization; CSF = colony-stimulating factor; DCIS = ductal carcinoma in situ; DIALH = ductal involvement by cells of ALH; IDC = invasive ductal carcinoma; ILC = invasive lobular carcinoma; LCIS = lobular carcinoma in situ; LOH = loss of heterozygosity; MALH = minimal atypical lobular hyperplasia; TDLU = terminal duct lobular unit.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
Acknowledgements
The authors would like to thank Dr David L Page for insightful discussion and invaluable assistance in the drafting of this manuscript. This work was supported by a Canadian Breast Cancer Research Alliance IDEA Grant (#018531) to TLM and WLL, the United States Army Medical Research and Materiel Command predoctoral fellowship to TLM (DAMD17-02-1-0498), and the National Cancer Institute of Canada with funds provided by the Terry Fox Run to ILA. The authors regret the omission of any citations to this manuscript.
Contributor Information
Teresa L Mastracci, Email: tmastracci@bccrc.ca.
Fouad I Boulos, Email: fouad.boulos@Vanderbilt.Edu.
Irene L Andrulis, Email: andrulis@mshri.on.ca.
Wan L Lam, Email: wanlam@bccrc.ca.
References
- Arpino G, Laucirica R, Elledge RM. Premalignant and in situ breast disease: biology and clinical implications. Ann Intern Med. 2005;143:446–457. doi: 10.7326/0003-4819-143-6-200509200-00009. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Mastracci TL, Shadeo A, Colby SM, Tuck AB, O'Malley FP, Bull SB, Lam WL, Andrulis IL. Genomic alterations in lobular neoplasia: a microarray comparative genomic hybridization signature for early neoplastic proliferation in the breast. Genes Chromosomes Cancer. 2006;45:1007–1017. doi: 10.1002/gcc.20368. [DOI] [PubMed] [Google Scholar]
- Stingl J, Raouf A, Emerman JT, Eaves CJ. Epithelial progenitors in the normal human mammary gland. J Mammary Gland Biol Neoplasia. 2005;10:49–59. doi: 10.1007/s10911-005-2540-7. [DOI] [PubMed] [Google Scholar]
- Nelson CM, Bissell MJ. Modeling dynamic reciprocity: engineering three-dimensional culture models of breast architecture, function, and neoplastic transformation. Semin Cancer Biol. 2005;15:342–352. doi: 10.1016/j.semcancer.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci USA. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/S1046-2023(03)00032-X. [DOI] [PubMed] [Google Scholar]
- Silberstein GB. Postnatal mammary gland morphogenesis. Microsc Res Tech. 2001;52:155–162. doi: 10.1002/1097-0029(20010115)52:2<155::AID-JEMT1001>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Debnath J, Walker SJ, Brugge JS. Akt activation disrupts mammary acinar architecture and enhances proliferation in an mTOR-dependent manner. J Cell Biol. 2003;163:315–326. doi: 10.1083/jcb.200304159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/S0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- Wrobel CN, Debnath J, Lin E, Beausoleil S, Roussel MF, Brugge JS. Autocrine CSF-1R activation promotes Src-dependent disruption of mammary epithelial architecture. J Cell Biol. 2004;165:263–273. doi: 10.1083/jcb.200309102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berx G, Cleton-Jansen AM, Strumane K, de Leeuw WJ, Nollet F, van Roy F, Cornelisse C. E-cadherin is inactivated in a majority of invasive human lobular breast cancers by truncation mutations throughout its extracellular domain. Oncogene. 1996;13:1919–1925. [PubMed] [Google Scholar]
- De Leeuw WJ, Berx G, Vos CB, Peterse JL, Van de Vijver MJ, Litvinov S, Van Roy F, Cornelisse CJ, Cleton-Jansen AM. Simultaneous loss of E-cadherin and catenins in invasive lobular breast cancer and lobular carcinoma in situ. J Pathol. 1997;183:404–411. doi: 10.1002/(SICI)1096-9896(199712)183:4<404::AID-PATH1148>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Fata JE, Werb Z, Bissell MJ. Regulation of mammary gland branching morphogenesis by the extracellular matrix and its remodeling enzymes. Breast Cancer Res. 2004;6:1–11. doi: 10.1186/bcr634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman BS, Werb Z. Stromal effects on mammary gland development and breast cancer. Science. 2002;296:1046–1049. doi: 10.1126/science.1067431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagensen CD, Lane N, Lattes R, Bodian C. Lobular neoplasia (so-called lobular carcinoma in situ) of the breast. Cancer. 1978;42:737–769. doi: 10.1002/1097-0142(197808)42:2<737::AID-CNCR2820420247>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Tobon H, Price HM. Lobular carcinoma in situ. Some ultrastructural observations. Cancer. 1972;30:1082–1091. doi: 10.1002/1097-0142(197210)30:4<1082::AID-CNCR2820300430>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Mastracci TL, Tjan S, Bane AL, O'Malley FP, Andrulis IL. E-cadherin alterations in atypical lobular hyperplasia and lobular carcinoma in situ of the breast. Mod Pathol. 2005;18:741–751. doi: 10.1038/modpathol.3800362. [DOI] [PubMed] [Google Scholar]
- Gonzalez MA, Pinder SE, Wencyk PM, Bell JA, Elston CW, Nicholson RI, Robertson JF, Blamey RW, Ellis IO. An immunohistochemical examination of the expression of E-cadherin, alpha- and beta/gamma-catenins, and alpha2- and beta1-integrins in invasive breast cancer. J Pathol. 1999;187:523–529. doi: 10.1002/(SICI)1096-9896(199904)187:5<523::AID-PATH296>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Page DL, Dupont WD, Rogers LW, Rados MS. Atypical hyperplastic lesions of the female breast. A long-term follow-up study. Cancer. 1985;55:2698–2708. doi: 10.1002/1097-0142(19850601)55:11<2698::AID-CNCR2820551127>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Page DL, Dupont WD, Rogers LW. Ductal involvement by cells of atypical lobular hyperplasia in the breast: a long-term follow-up study of cancer risk. Hum Pathol. 1988;19:201–207. doi: 10.1016/S0046-8177(88)80350-2. [DOI] [PubMed] [Google Scholar]
- Fitzgibbons PL, Henson DE, Hutter RV. Benign breast changes and the risk for subsequent breast cancer: an update of the 1985 consensus statement. Cancer Committee of the College of American Pathologists. Arch Pathol Lab Med. 1998;122:1053–1055. [PubMed] [Google Scholar]
- Collins LC, Baer HJ, Tamimi RM, Connolly JL, Colditz GA, Schnitt SJ. Magnitude and laterality of breast cancer risk according to histologic type of atypical hyperplasia: results from the Nurses' Health Study. Cancer. 2007;109:180–187. doi: 10.1002/cncr.22408. [DOI] [PubMed] [Google Scholar]
- Dupont WD, Page DL. Breast cancer risk associated with proliferative disease, age at first birth, and a family history of breast cancer. Am J Epidemiol. 1987;125:769–779. doi: 10.1093/oxfordjournals.aje.a114594. [DOI] [PubMed] [Google Scholar]
- Page DL, Schuyler PA, Dupont WD, Jensen RA, Plummer WD, Jr, Simpson JF. Atypical lobular hyperplasia as a unilateral predictor of breast cancer risk: a retrospective cohort study. Lancet. 2003;361:125–129. doi: 10.1016/S0140-6736(03)12230-1. [DOI] [PubMed] [Google Scholar]
- Page DL, Kidd TE, Jr, Dupont WD, Simpson JF, Rogers LW. Lobular neoplasia of the breast: higher risk for subsequent invasive cancer predicted by more extensive disease. Hum Pathol. 1991;22:1232–1239. doi: 10.1016/0046-8177(91)90105-X. [DOI] [PubMed] [Google Scholar]
- Fisher ER, Land SR, Fisher B, Mamounas E, Gilarski L, Wolmark N. Pathologic findings from the National Surgical Adjuvant Breast and Bowel Project: twelve-year observations concerning lobular carcinoma in situ. Cancer. 2004;100:238–244. doi: 10.1002/cncr.11883. [DOI] [PubMed] [Google Scholar]
- Foote F, Stewart F. Lobular carcinoma in situ: a rare form of mammary carcinoma. Am J Pathol. 1941;17:491–496. doi: 10.3322/canjclin.32.4.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratthauer GL, Tavassoli FA. Lobular intraepithelial neoplasia: previously unexplored aspects assessed in 775 cases and their clinical implications. Virchows Arch. 2002;440:134–138. doi: 10.1007/s00428-001-0541-5. [DOI] [PubMed] [Google Scholar]
- Cavenee WK, Dryja TP, Phillips RA, Benedict WF, Godbout R, Gallie BL, Murphree AL, Strong LC, White RL. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature. 1983;305:779–784. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- Devilee P, Thierry RF, Kievits T, Kolluri R, Hopman AH, Willard HF, Pearson PL, Cornelisse CJ. Detection of chromosome aneuploidy in interphase nuclei from human primary breast tumors using chromosome-specific repetitive DNA probes. Cancer Res. 1988;48:5825–5830. [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Dohner H, Cremer T, Lichter P. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. doi: 10.1002/(SICI)1098-2264(199712)20:4<399::AID-GCC12>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP, Snijders A, Albertson DG, Pinkel D, Marra MA, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo LW, Grove DI, Williams EM, Neal CL, Cousens LA, Schubert EL, Holcomb IN, Massa HF, Glogovac J, Li CI, et al. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res. 2004;64:8541–8549. doi: 10.1158/0008-5472.CAN-04-1992. [DOI] [PubMed] [Google Scholar]
- Forozan F, Mahlamaki EH, Monni O, Chen Y, Veldman R, Jiang Y, Gooden GC, Ethier SP, Kallioniemi A, Kallioniemi OP. Comparative genomic hybridization analysis of 38 breast cancer cell lines: a basis for interpreting complementary DNA microarray data. Cancer Res. 2000;60:4519–4525. [PubMed] [Google Scholar]
- Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, Ringner M, Sauter G, Monni O, Elkahloun A, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–6245. [PubMed] [Google Scholar]
- Lu YJ, Osin P, Lakhani SR, Di Palma S, Gusterson BA, Shipley JM. Comparative genomic hybridization analysis of lobular carcinoma in situ and atypical lobular hyperplasia and potential roles for gains and losses of genetic material in breast neoplasia. Cancer Res. 1998;58:4721–4727. [PubMed] [Google Scholar]
- Shelley Hwang E, Nyante SJ, Yi Chen Y, Moore D, DeVries S, Korkola JE, Esserman LJ, Waldman FM. Clonality of lobular carcinoma in situ and synchronous invasive lobular carcinoma. Cancer. 2004;100:2562–2572. doi: 10.1002/cncr.20273. [DOI] [PubMed] [Google Scholar]
- Nyante SJ, Devries S, Chen YY, Hwang ES. Array-based comparative genomic hybridization of ductal carcinoma in situ and synchronous invasive lobular cancer. Hum Pathol. 2004;35:759–763. doi: 10.1016/j.humpath.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Morandi L, Marucci G, Foschini MP, Cattani MG, Pession A, Riva C, Eusebi V. Genetic similarities and differences between lobular in situ neoplasia (LN) and invasive lobular carcinoma of the breast. Virchows Arch. 2006;449:14–23. doi: 10.1007/s00428-006-0192-7. [DOI] [PubMed] [Google Scholar]
- Buerger H, Simon R, Schafer KL, Diallo R, Littmann R, Poremba C, van Diest PJ, Dockhorn-Dworniczak B, Bocker W. Genetic relation of lobular carcinoma in situ, ductal carcinoma in situ, and associated invasive carcinoma of the breast. Mol Pathol. 2000;53:118–121. doi: 10.1136/mp.53.3.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther K, Merkelbach-Bruse S, Amo-Takyi BK, Handt S, Schroder W, Tietze L. Differences in genetic alterations between primary lobular and ductal breast cancers detected by comparative genomic hybridization. J Pathol. 2001;193:40–47. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH745>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Weber-Mangal S, Sinn HP, Popp S, Klaes R, Emig R, Bentz M, Mansmann U, Bastert G, Bartram CR, Jauch A. Breast cancer in young women (< or = 35 years): Genomic aberrations detected by comparative genomic hybridization. Int J Cancer. 2003;107:583–592. doi: 10.1002/ijc.11460. [DOI] [PubMed] [Google Scholar]
- Nishizaki T, Chew K, Chu L, Isola J, Kallioniemi A, Weidner N, Waldman FM. Genetic alterations in lobular breast cancer by comparative genomic hybridization. Int J Cancer. 1997;74:513–517. doi: 10.1002/(SICI)1097-0215(19971021)74:5<513::AID-IJC6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Etzell JE, Devries S, Chew K, Florendo C, Molinaro A, Ljung BM, Waldman FM. Loss of chromosome 16q in lobular carcinoma in situ. Hum Pathol. 2001;32:292–296. doi: 10.1053/hupa.2001.22759. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Simpson PT, Turner NC, Lambros MB, Jones C, Mackay A, Grigoriadis A, Sarrio D, Savage K, Dexter T, et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res. 2006;12:6652–6662. doi: 10.1158/1078-0432.CCR-06-1164. [DOI] [PubMed] [Google Scholar]
- Roylance R, Gorman P, Papior T, Wan YL, Ives M, Watson JE, Collins C, Wortham N, Langford C, Fiegler H, et al. A comprehensive study of chromosome 16q in invasive ductal and lobular breast carcinoma using array CGH. Oncogene. 2006;25:6544–6553. doi: 10.1038/sj.onc.1209659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayar R, Zhuang Z, Merino MJ, Silverberg SG. Loss of heterozygosity on chromosome 11q13 in lobular lesions of the breast using tissue microdissection and polymerase chain reaction. Hum Pathol. 1997;28:277–282. doi: 10.1016/S0046-8177(97)90124-6. [DOI] [PubMed] [Google Scholar]
- Derksen PW, Liu X, Saridin F, van der Gulden H, Zevenhoven J, Evers B, van Beijnum JR, Griffioen AW, Vink J, Krimpenfort P, et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell. 2006;10:437–449. doi: 10.1016/j.ccr.2006.09.013. [DOI] [PubMed] [Google Scholar]