Abstract
DNA replication is an extremely accurate process and cells have evolved intricate control mechanisms to ensure that each region of their genome is replicated only once during S phase. Here, we compare what is known about the processes that prevent re-replication in prokaryotic and eukaryotic cells by using the model organisms Escherichia coli and Schizosaccharomyces pombe as examples. Although the underlying molecular details are different, the logic behind the control mechanisms is similar. For example, after initiation, crucial molecules required for the loading of replicative helicases in both prokaryotes and eukaryotes are inactivated until the next cell cycle. Furthermore, in both systems the β-clamp of the replicative polymerase associates with enzymatic activities that contribute to the inactivation of the helicase loaders. Finally, recent studies suggest that the control mechanism that prevents re-replication in both systems also increases the synthesis of DNA building blocks.
Keywords: initiation of DNA replication, helicase loading, Escherichia coli, Schizosaccharomyces pombe, DNA building blocks
Introduction
Genomic DNA is organized differently in prokaryotic and eukaryotic cells. The bacterium Escherichia coli contains a single circular chromosome and replication is initiated bi-directionally from a fixed origin (oriC; Fig 1). Consequently, a single initiation event will ensure replication of the entire 4.6 Mbp genome in a process that is completed when the two divergent replication forks collide at the opposite side of the circular chromosome.
Figure 1.
Initiation of replication at multiple origins in Escherichia coli and Schizosaccharomyces pombe. Origins that have not yet initiated are shown in green, whereas those initiated or passively replicated are shown in red. The E. coli cell is fast-growing with S + G2 phases spanning more than two generation times. Consequently, initiations occur in synchrony at four cellular origins. For simplicity, only one chromosome with six autonomously replicating sequences is shown in the S. pombe cell. Four of these are firing, whereas two are being passively replicated.
In eukaryotic cells, the genomic DNA is distributed between multiple chromosomes that are contained within the nucleus. At S phase, replication is simultaneously initiated from many different origins that are scattered throughout the genome (Fig 1), and replication is completed when all replication forks have either met a convergent fork from an adjacent origin or reached the telomeres at the end of the chromosomes.
In the fission yeast Schizosaccharomyces pombe, many potential replication origins are found in intragenic regions of the 12.5 Mbp genome distributed across three chromosomes. However, only a limited subset of these is used in a given S phase. Furthermore, the specific origins that actually fire vary from one S phase to another, suggesting that origin selection occurs by a stochastic mechanism.
Replicating once, and only once
Despite these differences in organization, both cell types are faced with the challenge of ensuring that the entire genome is replicated once, and only once, in any given S phase. At face value, this problem seems to be different depending on whether cells have a single origin or many scattered ones. However, under optimal nutritional conditions, E. coli cells are able to grow with a doubling time that is much shorter than the time required for replication and segregation of the chromosome (S + G2 phases). Consequently, initiation of replication occurs one, two or even three generations before cell birth, depending on the growth rate (Cooper & Helmstetter, 1968). Fast-growing cells are therefore born with chromosomes containing several active origins of replication, and such cells are also able to coordinate initiation at multiple—but identical—origins (Fig 1).
In both E. coli and fission yeast, initiation of replication is coupled to cell growth and is triggered by a specific signal that is generated when the cell has obtained a critical mass. Once activated, each replication origin is inhibited from re-firing until the next S phase (see below). Furthermore, in eukaryotic cells, passive replication by an incoming fork also prevents an origin that has not yet fired from firing until the next S phase. Together, these mechanisms ensure that the entire genome is replicated only once in each cell cycle.
Mechanisms of initiation
Initiation of replication in both E. coli and S. pombe occurs by a series of discrete steps. First, the origin recognition complex (ORC) is formed by the recruitment of replication factors to origin sequences. Subsequently, loading of the replicative helicase converts the ORC into a pre-replicative complex (pre-RC)—a process often referred to as ‘licensing'. This paves the way for loading of the polymerase itself; in both organisms, the crucial step seems to be loading of the helicase.
The S. pombe ORC is a six-subunit complex that consists of the three proteins Orc1, Orc2 and Orc4, each of which contain an AAA+ ATP-binding domain (Kong & DePamphilis, 2002). Orc proteins specifically associate with origins of replication (autonomously replicating sequences (ARSs); Fig 2). One of the ORC subunits, Orc4, contains several AT-hook motifs in its amino-terminal domain, which interact with the AT-rich origin sequences. The ORC is bound to chromatin throughout the cell cycle and therefore its binding is unlikely to regulate initiation (Lygerou & Nurse, 1999). In E. coli, the ORC is formed by DnaA, which is the only protein that is specific to replication initiation at oriC. The DnaA protein associated with either ATP or ADP binds to three 9-bp binding sites within the origin of replication called R1, R2 and R4 (reviewed by Kaguni, 2006; Mott & Berger, 2007). Similar to the situation in yeast, the ORC remains bound to the origin of replication throughout most of the cell cycle (Samitt et al, 1989).
Figure 2.
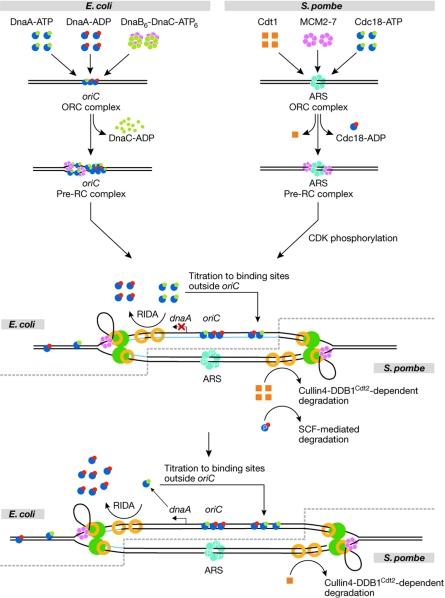
Mechanisms of replication initiation, and the prevention of re-replication in Escherichia coli and Schizosaccharomyces pombe. The symbols for the different replication proteins are indicated in the figure with the exception of the replicative DNA polymerase (green) and the DNA-loaded β-clamp–proliferating cell nuclear antigen (PCNA; yellow rings). The replicative DNA polymerase is indicated as a dimer, although recent evidence suggests, at least in E. coli, that the replicase might be a trimer (McInerney et al, 2007). For simplicity, DnaA and Cdc18 have been given the same symbol as these proteins share homology in important functional domains. In the steps after pre-replicative complex formation, the E. coli process is shown on the upper strand/leftward fork, whereas the fission yeast system is illustrated by the lower strand/rightward fork. In E. coli, the newly synthesized DNA is unmethylated at GATC sites (blue, middle panel)—that is, DNA is hemi-methylated immediately after passage of the replication fork. As the fork progresses further away, both strands become methylated (lower panel). In fission yeast, Cdk phosphorylation of ORC and Cdc18 (indicated by ‘P') prevent binding of the MCM2–7 complex to the ORC and causes SCF-mediated destruction of Cdc18. ARS, autonomously replicating sequence; CDK, cyclin-dependent kinase; MCM, minichromosome maintenance; ORC, origin recognition complex; Pre-RC, pre-replicative complex; RIDA, regulatory inactivation of DnaA; SCF, an E3 ubiquitin ligase.
The transition from the ORC-like stage to a pre-RC stage represents the next step in the initiation process. In fission yeast, the pre-RC is formed in late M and G1 phases, when the two initiation factors Cdt1 and Cdc18 facilitate loading of the replicative helicase. Both proteins are cell-cycle regulated, being absent from late S phase until cells exit mitosis (see below). Cdc18 is yet another AAA+ ATPase, whereas the biochemical function of Cdt1 is unknown. Our understanding of pre-RC formation is largely based on studies of Cdc6, the budding yeast orthologue of Cdc18, but we anticipate that the mechanism in fission yeast is similar. The presence of Cdc18 and Cdt1 at the ORC enables the recruitment of several complexes of minichromosome maintenance (MCM) 2–7 hexamers, in a cyclic process that requires hydrolysis of ATP on both Cdc18 and on ORC subunits (Randell et al, 2006). The MCM2–7 complex is believed to act as the replicative helicase, although biochemical evidence for this is still circumstantial.
Assembly of the replisome at the pre-RC and activation of the replication process involve several additional factors, including Cdc45 and the GINS complex (reviewed by Legouras et al, 2006). In addition, the activity of two protein kinases is required at this stage: cyclin-dependent protein kinase (Cdk)—the main driver of the cell cycle in fission yeast—and the conserved Hsk1–Dfb1 kinase (also known as Ddk). The phosphorylation targets for Cdk were recently identified as Sdl2 and Sdl3 in budding yeast (Zegerman & Diffley, 2007; Tanaka et al, 2007).
In E. coli, pre-RC formation is initiated by further binding of DnaA to the weaker recognition sites within the origin—that is, R3 and R5, which are indifferent to the nucleotide-bound status of DnaA—and to three I-boxes (McGarry et al, 2004; Kawakami et al, 2005) that are specific for DnaA-ATP. With the help of the accessory proteins IHF, HU and DiaA (Ryan et al, 2002; Keyamura et al, 2007), this induces formation of a DnaA–DNA nucleoprotein complex on oriC, where the DNA is remodelled to a right-handed DNA wrapped around a right-handed DnaA-ATP filament (Erzberger et al, 2006). The DnaA–DNA complex promotes duplex opening in an adjacent AT-rich region. This open complex is stabilized by the binding of DnaA-ATP to specific 6-bp sequences found in the single-stranded region (Speck & Messer, 2001). The requirement for DnaA-ATP in origin remodelling explains why this configuration of the protein is limiting for initiation in vivo (Nishida et al, 2002; Riber et al, 2006). Subsequently, the DnaA protein recruits the hexameric DnaB helicase associated with ATP-bound DnaC as a B6C6 complex to the single-stranded region of the open complex. DnaC loads the DnaB helicase on the open complex to promote further duplex opening to form the pre-RC stage. During this process, ATP is hydrolysed and DnaC is released. In E. coli, there does not seem to be any control on the pre-RC stage and the transition to replication proceeds immediately by the loading of two or three DNA polymerase III holoenzymes on the origin (McInerney et al, 2007). When dNTPs are present, replication can then commence (Herrick & Sclavi, 2007).
The spatial arrangement of DnaA protein domains involved in nucleotide binding, DNA binding and oligomerization is similar to the fission yeast initiation factor Cdc18, and it has been suggested that the helical DNA-binding domain could direct similar functioning AAA+ domains to their respective origins (Erzberger et al, 2002). It is worth noting that both DnaA and Cdc18 can switch between active and inactive configurations depending on the nature of the bound nucleotide, and that this molecular switch is one of the determinants for initiation control.
Cascades of initiation
As discussed above, rapidly growing E. coli cells contain many origins of replication that all fire simultaneously (Fig 1), and synchronous initiation presumably results from the release of the DnaA protein from the first initiated origin in a cell. This will momentarily increase the DnaA:oriC ratio for remaining ‘old' origins and their initiation will follow in a cascade-like manner—known as the initiation cascade (Lobner-Olesen et al, 1994). Eukaryotic cells are faced with a similar problem: origins are selected by a stochastic mechanism, and therefore there is a risk that large chromosomal regions will occasionally remain unreplicated during any given S phase. Analogous to the initiation cascade model, it has been proposed that a crucial replication factor is rate limiting for initiation. As replication proceeds, this factor is released and can be redistributed to other origins, thereby increasing their probability of firing (Lucas et al, 2000). The nature of this factor has not been established, but it would have to be a protein that is not degraded in the initiation processthat is, not Cdt1 or Cdc18 (see below).
Mechanisms to prevent re-initiation
In both E. coli and S. pombe, inactivation of the helicase loader proteins has a crucial role in preventing immediate re-firing of a recently activated origin. In E. coli, the DnaA protein is the target for this regulation, whereas fission yeast cells regulate both Cdt1 and Cdc18. In both systems, inhibition of re-replication is accomplished both by physically preventing pre-RC assembly and by a reduction in the activity of AAA+ ATPase proteins. This is mediated by post-translational inactivation of the proteins and by the modulation of gene expression.
Prevention of pre-RC assembly
The E. coli origin of replication is rich in GATC sites, which is the substrate for the Dam methyltransferase. As methylation is a post-replicational process, newly replicated origins are methylated on only one strand (Fig 2). These hemi-methylated origins are bound (sequestered) by SeqA, a protein with high affinity for hemi-methylated GATC sites (Lu et al, 1994). Sequestration renders the origin inaccessible to DnaA for approximately one-third of the generation time to prevent immediate re-initiation (Campbell & Kleckner, 1990; von Freiesleben et al, 2000).
Origin sequestration is instrumental not only in preventing the immediate re-initiation at an origin, but also in preparing the origin for the next round of initiation. During sequestration, the DnaA protein is only able to bind the high affinity sites R1, R2 and R4, to re-set the origin to the ORC stage (Nievera et al, 2006). Although sequestration lasts less than one generation, it ensures that successive initiations at the same origin are separated by a doubling time, because it provides a time window during which the origin cannot be initiated and the amount of DnaA-ATP is reduced to a level that cannot sustain initiation (see below). Consequently, a period of growth is necessary before origins of replication are released from sequestration and can re-initiate.
In eukaryotic cells, the increase in Cdk activity that initiates S phase has an additional function in preventing re-replication during S, G2 and M phases. The importance of this mechanism follows from the observation that G2 cells can be manipulated to erroneously enter another round of S phase by temporarily inactivating a temperature-sensitive Cdk allele (Hayles et al, 1994). Cdk seems to inhibit re-initiation of DNA replication partly by phosphorylating subunits in the ORC (Fig 2), thereby preventing de novo assembly of pre-RCs until Cdk activity becomes low again as cells exit mitosis (Nguyen et al, 2001; Vas et al, 2001). Phosphorylation of the ORC has not been reported to involve a sequestration mechanism as in E. coli; presumably it simply prevents recruitment of Cdt1 and Cdc18.
Post-translational inactivation of AAA+ proteins
The activity of the DnaA protein is reduced during S phase by a process known as the ‘regulatory inactivation of DnaA' (RIDA; Fig 2), in which the active ATP-bound DnaA protein is converted to the inactive ADP-bound form by ATP hydrolysis (Katayama et al, 1998). RIDA activity involves two proteins: the DnaA-related protein Hda (Kato & Katayama, 2001) and the β-clamp of the DNA polymerase (Pol) III holoenzyme (encoded by the dnaN gene; Katayama et al, 1998). These proteins form a complex even before the clamp is loaded onto the DNA (Kawakami et al, 2006). However, only the DNA-loaded β-subunit of Pol III in complex with the Hda protein stimulates the ATPase activity of DnaA to promote conversion of DnaA-ATP to the inactive DnaA-ADP (Su'etsugu et al, 2004). At the end of the initiation process, hydrolysis of DnaA-ATP by RIDA is accelerated because new replication forks are formed, and more β-clamps are loaded onto the DNA.
In fission yeast, the two helicase-loader proteins Cdc18 and Cdt1 also become inactivated after initiation of replication, but here this is accomplished by physical degradation rather than biochemical inactivation. The increase in Cdk activity that brings about S phase also causes phosphorylation of Cdc18, which targets the protein for SCF-mediated ubiquitination and subsequent degradation by proteolysis (Fig 2; Jallepalli et al, 1997). The importance of this regulation is clear from the fact that ectopic over-production of Cdc18 causes massive re-initiation of DNA replication (Nishitani & Nurse, 1995).
The Cdt1 protein also becomes degraded after successful initiation of DNA replication, but this process does not require Cdk. Instead, Cdt1 is targeted for degradation by a different E3 ubiquitin ligase, the Cullin4–Ddb1–Roc1 complex (Ralph et al, 2006). Interestingly, Cdt1 ubiquitination is tightly coupled to its function in initiation by means of two different mechanisms. First, substrate recognition requires a specific adaptor protein, the WD40-repeat protein Cdt2, which becomes transcriptionally induced when cells enter S phase (Liu et al, 2005). Second, Cdt1 only becomes ubiquitinated when it is associated with the proliferating cell nuclear antigen (PCNA) processivity clamp of the polymerase (Fig 2; Arias & Walter, 2006, Jin et al, 2006; Nishitani et al, 2006; Senga et al, 2006). Presumably, Cdt1 molecules are consumed when they have been actively engaged in initiation. Therefore, enzymatic activities that negatively regulate helicase-loader proteins seem to associate with the processivity clamp in both E. coli and S. pombe.
Modulation of gene expression
E. coli does not seem to regulate DnaA activity by degrading the protein; however, in addition to RIDA, a second mechanism for reducing DnaA activity in the post-initiation period exists. This method uses the sequestration mechanism to reduce expression of the dnaA gene. On replication, the dnaA gene promoter region, which is rich in GATC sequences, is hemi-methylated and sequestered for the same time period as the origin of replication. Sequestration of the dnaA promoter completely blocks transcription of the dnaA gene (Campbell & Kleckner, 1990). As the dnaA gene is close to the origin, sequestration of dnaA is virtually coincident with sequestration of oriC, and de novo DnaA synthesis is prevented during the origin sequestration period (Fig 2). In cells in which origin and dnaA gene sequestration no longer coincide, DnaA synthesis continues during origin sequestration. In such cells, re-initiations occasionally occur at some origins within the same cell cycle (Riber & Lobner-Olesen, 2005).
Transcription of the genes encoding the helicase-loader proteins Cdc18 and Cdt1 also oscillates in fission yeast and is high in late mitosis and G1 (Hofmann & Beach, 1994; Kelly et al, 1993). However, this is actively controlled by the cell-cycle-regulated MBF transcription factor complex rather than by an intricate system that monitors ongoing replication.
Titrating DnaA to reservoir sites
During origin sequestration, replication generates new DnaA protein-binding sites outside oriC. These titrate DnaA protein away from the origin and, in the absence of de novo DnaA synthesis (Campbell & Kleckner, 1990), efficiently reduce the intracellular concentration of DnaA protein available for initiation (Fig 2). The E. coli chromosome contains a hierarchy of 308 evenly distributed R-type DnaA boxes with different affinities for the DnaA protein. The datA locus, which contains five R-type DnaA boxes, seems to have the highest DnaA-binding capacity, and might bind to several hundred molecules of DnaA protein associated with either ATP or ADP. The datA locus is located approximately 470 Kbp away from oriC and is replicated within the period of origin sequestration during which no new DnaA protein is synthesized. This generates a sink for free DnaA protein (Kitagawa et al, 1998).
Coupling nucleotide synthesis to chromosome replication
In most cells, the intracellular concentration of DNA precursors (dNTPs) is low and can only sustain limited chromosome replication unless they are continuously synthesized to match the demand of ongoing replication forks. Upregulation of dNTP synthesis in S phase is carefully controlled because imbalances between the four individual nucleotide pools, as well as balanced deviation from the normal level, are mutagenic (reviewed by Mathews, 2006). In both E. coli and fission yeast, dNTPs are synthesized from their corresponding NTPs exclusively by the ribonucleotide reductase (RNR) complex. RNR is a heterodimeric tetramer consisting of two large and two small subunits. RNR activity is the rate-limiting step in dNTP synthesis.
The RNR subunits of E. coli are encoded by the nrdAB operon, and nrdAB expression is adjusted to DNA synthesis (reviewed by Herrick & Sclavi, 2007). Transcription of nrdAB is induced at the time of initiation by a DnaA-independent mechanism (Jacobson & Fuchs, 1998). Superimposed on this cell-cycle regulation is modulation of transcription by the DnaA protein (Augustin et al, 1994). DnaA was initially reported to stimulate nrdAB transcription (Jacobson & Fuchs, 1998) although a recent study indicates that DnaA-ATP—but not DnaA-ADP—is an efficient repressor of its transcription (Gon et al, 2006). The nrdAB expression level is therefore determined by the DnaA-ATP:DnaA-ADP ratio.
The RIDA-imposed variation in DnaA-ATP:DnaA-ADP ratio throughout the cell cycle (Kurokawa et al, 1999) could therefore couple dNTP synthesis to the elongation step of chromosome replication. Before initiation, when the cellular DnaA-ATP:DnaA-ADP ratio is high (Kurokawa et al, 1999), DnaA regulation would favour nrdAB repression. After initiation, RIDA is accelerated, resulting in a reduced DnaA-ATP:DnaA-ADP ratio and consequently an increase in nrdAB transcription. Therefore, the RNR level is increased in S phase, resulting in an increased synthesis of dNTPs to match the demand from the ongoing replication forks.
Precursor synthesis in eukaryotic cells is also adjusted to ongoing DNA replication by the regulation of RNR activity; however, the molecular basis is different. In S. pombe, transcription of the gene encoding the large subunit (Cdc22) is cell-cycle regulated (Fernandez Sarabia et al, 1993). In addition, assembly of RNR is actively prevented outside S phase by the presence of the RNR inhibitor protein Spd1 (Liu et al, 2003). When cells enter S phase, Spd1 is degraded by the same pathway that downregulates the Cdt1 helicase loader—that is, the Cullin4–Ddb1–Roc1 E3 ubiquitin ligase and the adaptor protein Cdt2 (Holmberg et al, 2005; Liu et al, 2005). It is unclear whether the degradation of Spd1—similar to the degradation of Cdt1—is coupled to PCNA.
Perspectives
The development of the eukaryotic type of genome organization—with multiple chromosomes and many scattered origins of replication—was probably important for the expansion of genome size that allowed the development of complex organisms. Taken at face value, control of replication seems to be organized differently in prokaryotic and eukaryotic cells; however, the control mechanisms found in the two systems seem to regulate the same steps in the process. First, in both prokaryotes and eukaryotes the crucial step in the establishment of a replication origin is loading of the replicative helicase. This process is mediated when the concentration of the helicase-loading AAA+ ATPases builds up to a certain threshold in the cell. In E. coli, this seems to be the rate-limiting step; loading of the replicative polymerase and initiation immediately follows. In S. pombe, further progress requires the action of S-phase-activating kinases. Second, once an origin of replication has fired, re-firing is prevented for a period of time. In both systems, this is accomplished by a combination of physical modification of the origin and/or associated protein factors (by sequestration or by phosphorylation), such that the helicase loader cannot access it, and by removing the helicase-loader activity. Eukaryotic cells literally get rid of the protein by switching on ubiquitin-mediated degradation. The prokaryotic cell does not have this option and therefore it is dependent on several other methods of reducing the active concentration of the helicase loader, such as through hydrolysis of its bound ATP, binding of the loader to unproductive sites or downregulation of its expression. The development of ubiquitin-mediated protein degradation made these mechanisms redundant.
In this review, we have attempted to draw parallels between the basic mechanisms that prevent re-replication in two simple uni-cellular model organisms. Failure to restrict replication to once per cell cycle leads to DNA damage through the generation of double-stranded breaks and can result in development of tumours (reviewed by Arias & Walter, 2007). It is therefore not surprising that metazoans have evolved additional mechanisms—such as inactivation of Cdt1 by Geminin binding—to minimize the likelihood of untimely replication initiations.

Olaf Nielsen

Anders Løbner-Olesen
Acknowledgments
We thank R. Egel, S. MacNeill and O. Fleck for their comments on the manuscript. Work in our laboratories is supported by The Danish Cancer Society, the Lundbeck Foundation, The Novo-Nordisk Foundation, the Danish Medical Research Council and the Danish Natural Science Research Council.
References
- Arias EE, Walter JC (2006) PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol 8: 84–90 [DOI] [PubMed] [Google Scholar]
- Arias EE, Walter JC (2007) Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21: 497–518 [DOI] [PubMed] [Google Scholar]
- Augustin LB, Jacobson BA, Fuchs JA (1994) Escherichia coli Fis and DnaA proteins bind specifically to the nrd promoter region and affect expression of an nrd–lac fusion. J Bacteriol 176: 378–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JL, Kleckner N (1990) E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62: 967–979 [DOI] [PubMed] [Google Scholar]
- Cooper S, Helmstetter CE (1968) Chromosome replication and the division cycle of Escherichia coli B/r. J Mol Biol 31: 519–540 [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Pirruccello MM, Berger JM (2002) The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J 21: 4763–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Mott ML, Berger JM (2006) Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol 13: 676–683 [DOI] [PubMed] [Google Scholar]
- Fernandez Sarabia MJ, McInerny C, Harris P, Gordon C, Fantes P (1993) The cell cycle genes cdc22+ and suc22+ of the fission yeast Schizosaccharomyces pombe encode the large and small subunits of ribonucleotide reductase. Mol Gen Genet 238: 241–251 [DOI] [PubMed] [Google Scholar]
- Gon S, Camara JE, Klungsoyr HK, Crooke E, Skarstad K, Beckwith J (2006) A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J 25: 1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J, Fisher D, Woollard A, Nurse P (1994) Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2–mitotic B cyclin complex. Cell 78: 813–822 [DOI] [PubMed] [Google Scholar]
- Herrick J, Sclavi B (2007) Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol Microbiol 63: 22–34 [DOI] [PubMed] [Google Scholar]
- Hofmann JF, Beach D (1994) cdt1 is an essential target of the Cdc10/Sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. EMBO J 13: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg C, Fleck O, Hansen HA, Liu C, Slaaby R, Carr AM, Nielsen O (2005) Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev 19: 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson BA, Fuchs JA (1998) A 45 bp inverted repeat is required for cell cycle regulation of the Escherichia coli nrd operon. Mol Microbiol 28: 1307–1314 [DOI] [PubMed] [Google Scholar]
- Jallepalli PV, Brown GW, Muzi-Falconi M, Tien D, Kelly TJ (1997) Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev 11: 2767–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC (2006) A family of diverse Cul4–Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 23: 709–721 [DOI] [PubMed] [Google Scholar]
- Kaguni JM (2006) DnaA: controlling the initiation of bacterial DNA replication and more. Annu Rev Microbiol 60: 351–375 [DOI] [PubMed] [Google Scholar]
- Katayama T, Kubota T, Kurokawa K, Crooke E, Sekimizu K (1998) The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 94: 61–71 [DOI] [PubMed] [Google Scholar]
- Kato J, Katayama T (2001) Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J 20: 4253–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami H, Keyamura K, Katayama T (2005) Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J Biol Chem 280: 27420–27430 [DOI] [PubMed] [Google Scholar]
- Kawakami H, Su'etsugu M, Katayama T (2006) An isolated Hda–clamp complex is functional in the regulatory inactivation of DnaA and DNA replication. J Struct Biol 156: 220–229 [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P (1993) The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell 74: 371–382 [DOI] [PubMed] [Google Scholar]
- Keyamura K, Fujikawa N, Ishida T, Ozaki S, Su'etsugu M, Fujimitsu K, Kagawa W, Yokoyama S, Kurumizaka H, Katayama T (2007) The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes Dev 21: 2083–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa R, Ozaki T, Moriya S, Ogawa T (1998) Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev 12: 3032–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong D, DePamphilis ML (2002) Site-specific ORC binding, pre-replication complex assembly and DNA synthesis at Schizosaccharomyces pombe replication origins. EMBO J 21: 5567–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Nishida S, Emoto A, Sekimizu K, Katayama T (1999) Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J 18: 6642–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legouras I, Xouri G, Dimopoulos S, Lygeros J, Lygerou Z (2006) DNA replication in the fission yeast: robustness in the face of uncertainty. Yeast 23: 951–962 [DOI] [PubMed] [Google Scholar]
- Liu C, Powell KA, Mundt K, Wu L, Carr AM, Caspari T (2003) Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev 17: 1130–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Poitelea M, Watson A, Yoshida SH, Shimoda C, Holmberg C, Nielsen O, Carr AM (2005) Transactivation of Schizosaccharomyces pombe cdt2+ stimulates a Pcu4–Ddb1–CSN ubiquitin ligase. EMBO J 24: 3940–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobner-Olesen A, Hansen FG, Rasmussen KV, Martin B, Kuempel PL (1994) The initiation cascade for chromosome replication in wild-type and Dam methyltransferase deficient Escherichia coli cells. EMBO J 13: 1856–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Campbell JL, Boye E, Kleckner N (1994) SeqA: a negative modulator of replication initiation in E. coli. Cell 77: 413–426 [DOI] [PubMed] [Google Scholar]
- Lucas I, Chevrier-Miller M, Sogo JM, Hyrien O (2000) Mechanisms ensuring rapid and complete DNA replication despite random initiation in Xenopus early embryos. J Mol Biol 296: 769–786 [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Nurse P (1999) The fission yeast origin recognition complex is constitutively associated with chromatin and is differentially modified through the cell cycle. J Cell Sci 112: 3703–3712 [DOI] [PubMed] [Google Scholar]
- Mathews CK (2006) DNA precursor metabolism and genomic stability. FASEB J 20: 1300–1314 [DOI] [PubMed] [Google Scholar]
- McGarry KC, Ryan VT, Grimwade JE, Leonard AC (2004) Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc Natl Acad Sci USA 101: 2811–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney P, Johnson A, Katz F, O'Donnell M (2007) Characterization of a triple DNA polymerase replisome. Mol Cell 27: 527–538 [DOI] [PubMed] [Google Scholar]
- Mott ML, Berger JM (2007) DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol 5: 343–354 [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Li JJ (2001) Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411: 1068–1073 [DOI] [PubMed] [Google Scholar]
- Nievera C, Torgue JJ, Grimwade JE, Leonard AC (2006) SeqA blocking of DnaA–oriC interactions ensures staged assembly of the E. coli pre-RC. Mol Cell 24: 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida S, Fujimitsu K, Sekimizu K, Ohmura T, Ueda T, Katayama T (2002) A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication: evidence from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J Biol Chem 277: 14986–14995 [DOI] [PubMed] [Google Scholar]
- Nishitani H, Nurse P (1995) p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell 83: 397–405 [DOI] [PubMed] [Google Scholar]
- Nishitani H et al. (2006) Two E3 ubiquitin ligases, SCF–Skp2 and DDB1–Cul4, target human Cdt1 for proteolysis. EMBO J 25: 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph E, Boye E, Kearsey SE (2006) DNA damage induces Cdt1 proteolysis in fission yeast through a pathway dependent on Cdt2 and Ddb1. EMBO Rep 7: 1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell JC, Bowers JL, Rodriguez HK, Bell SP (2006) Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2–7 helicase. Mol Cell 21: 29–39 [DOI] [PubMed] [Google Scholar]
- Riber L, Lobner-Olesen A (2005) Coordinated replication and sequestration of oriC and dnaA are required for maintaining controlled once-per-cell-cycle initiation in Escherichia coli. J Bacteriol 187: 5605–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riber L, Olsson JA, Jensen RB, Skovgaard O, Dasgupta S, Marinus MG, Lobner-Olesen A (2006) Hda-mediated inactivation of the DnaA protein and dnaA gene autoregulation act in concert to ensure homeostatic maintenance of the Escherichia coli chromosome. Genes Dev 20: 2121–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan VT, Grimwade JE, Nievera CJ, Leonard AC (2002) IHF and HU stimulate assembly of pre-replication complexes at Escherichia coli oriC by two different mechanisms. Mol Microbiol 46: 113–124 [DOI] [PubMed] [Google Scholar]
- Samitt CE, Hansen FG, Miller JF, Schaechter M (1989) In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J 8: 989–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, Dutta A (2006) PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J Biol Chem 281: 6246–6252 [DOI] [PubMed] [Google Scholar]
- Speck C, Messer W (2001) Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J 20: 1469–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su'etsugu M, Takata M, Kubota T, Matsuda Y, Katayama T (2004) Molecular mechanism of DNA replication-coupled inactivation of the initiator protein in Escherichia coli: interaction of DnaA with the sliding clamp-loaded DNA and the sliding clamp–Hda complex. Genes Cells 9: 509–522 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H (2007) CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332 [DOI] [PubMed] [Google Scholar]
- Vas A, Mok W, Leatherwood J (2001) Control of DNA rereplication via Cdc2 phosphorylation sites in the origin recognition complex. Mol Cell Biol 21: 5767–5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Freiesleben U, Krekling MA, Hansen FG, Lobner-Olesen A (2000) The eclipse period of Escherichia coli. EMBO J 19: 6240–6248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF (2007) Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285 [DOI] [PubMed] [Google Scholar]