Abstract
Drosophila Gcn5 is the catalytic subunit of the SAGA and ATAC histone acetylase complexes. Here, we show that mutations in Gcn5 and the ATAC component Ada2a induce a decondensation of the male X chromosome, similar to that induced by mutations in the Iswi and Nurf301 subunits of the NURF nucleosome remodelling complex. Genetic studies as well as transcript profiling analysis indicate that ATAC and NURF regulate overlapping sets of target genes during development. In addition, we find that Ada2a chromosome binding and histone H4-Lys12 acetylation are compromised in Iswi and Nurf301 mutants. Our results strongly suggest that NURF is required for ATAC to access the chromatin and to regulate global chromosome organization.
Introduction
Two classes of regulatory factor have been found to induce distinct states of gene activity by using the energy of ATP hydrolysis to physically remodel the nucleosomal arrangements (Cairns, 2005) or post-transcriptionally modify histones (Peterson & Laniel, 2004).
Drosophila nucleosome remodelling complexes can be divided into two main classes, depending on whether the ATPase catalytic subunit involved is the SWI2/SNF2 homologue Brahma or the Iswi protein (Elfring et al, 1994). Iswi was purified in three complexes, ACF, CHRAC and NURF, which all increase accessibility to chromatin templates in biochemical assays (Tsukiyama & Wu, 1995; Ito et al, 1997; Varga-Weisz et al, 1997). Accordingly, genetic analyses pointed to a role of these complexes in transcriptional regulation (Deuring et al, 2000; Xiao et al, 2001; Badenhorst et al, 2002, 2005).
The yeast Gcn5 protein was the first transcriptional coactivator identified with histone acetyltransferase (HAT) activity (Brownell et al, 1996). From yeast to human, Gcn5 orthologues operate as a catalytic subunit in various multiprotein complexes that contain Ada2- and Ada3-related coactivators, Spt proteins and TATA-binding protein-associated factors. Drosophila Gcn5 was purified biochemically in two complexes, SAGA and ATAC, which preferentially acetylate nucleosomal histones H3 and H4, respectively (Guelman et al, 2006). The complexes include distinct Ada2 relatives: SAGA contains the Ada2b protein, whereas ATAC contains the Ada2a protein (Kusch et al, 2003; Muratoglu et al, 2003). Gcn5 was found to be essential for in vivo acetylation of larval polytene chromosomes at positions lysine 9 (K9)/K14 of histone H3 and K5/K12 of histone H4 (Carre et al, 2005; Ciurciu et al, 2006). In addition, mutations in the Ada2b gene result in a loss of acetylation of residues H3-K9/K14, whereas mutations in the Ada2a gene only affect acetylation of residues H4-K5/K12 (Qi et al, 2004; Pankotai et al, 2005; Ciurciu et al, 2006). These data indicate that SAGA and ATAC have distinct substrate specificity for histone residues, which in turn could determine distinct functions or downstream regulatory events.
Numerous studies established that HAT and nucleosome remodelling complexes act synergistically to regulate chromatin structure and gene expression in yeast and human (Featherstone, 2002). In Drosophila, H4-K16 acetylation by Mof was shown to antagonize Iswi function in vivo (Corona et al, 2002) and to negatively regulate interactions between Iswi and its nucleosomal substrate in vitro (Clapier et al, 2002). Interactions between HAT and remodelling complexes remain otherwise poorly characterized in this organism. Here, we provide evidence for functional interactions between the ATAC and NURF complexes. In addition, our results show that NURF is required for proper histone acetylation of chromosomes by ATAC and indicate that this interplay is involved in the maintenance of higher order chromosome structure.
Results
Genetic interactions between ATAC and Iswi
To test whether Gcn5 and Iswi act in the same gene regulatory pathway during development, we analysed genetic interactions between Iswi and Gcn5 mutant alleles. We noticed first that homozygous Iswi2 or Gcn5E333st animals die at the end of the third larval instar, whereas double homozygous Iswi2 Gcn5E333st animals die during the first larval instar, indicating that the combination of zygotic Iswi and Gcn5 loss of function impairs early development more severely than either mutation alone.
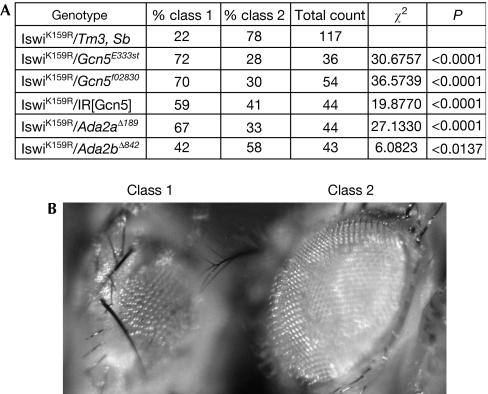
We then took advantage of a transgenic construct that expresses a dominant-negative form of Iswi. Expression of IswiK159R in eye-antennal imaginal discs leads to small, rough eyes in about 20% of adults (Deuring et al, 2000; Fig 1). The occurrence of this phenotype was significantly increased in heterozygous Gcn5/+ mutant backgrounds as well as after Gcn5 RNA interference (RNAi) knockdown (P<0.0001; Fig 1). A similar enhancement of the IswiK159R eye phenotype was observed in an Ada2a/+ mutant background, whereas an Ada2b mutant allele had only a modest effect (P<0.0001 and P=0.0137, respectively). These results point to a functional interaction between ATAC and Iswi during development.
Figure 1.
ATAC and Iswi interact functionally during Drosophila development. Decreased Gcn5 or Ada2a expression enhances the Iswi dominant-negative phenotype induced by expression of IswiK159R in the eye. (A) Enhancement of IswiK159R dominant-negative phenotype by Gcn5 and Ada2a mutant alleles. eye-Gal4, UAS-IswiK159R/Tm3, Ser females were crossed with RNAi trigger UAS-IR[Gcn5] transgenic males or Tm3, Sb balanced Gcn5E333st, Gcn5f02830, Ada2aΔ189 or Ada2bΔ842 heterozygous males. Progenies of the indicated genotype were scored for eye defects as class 1 (>50% reduction in size) or class 2 (<50% reduction in size). The eye-Gal4, UAS-IswiK159R chromosome is indicated in the table as IswiK159R. Results are expressed as percentages relative to the total count of siblings of the indicated genotype. Control IswiK159R/Tm3, Sb siblings were counted from the progeny of the eye-Gal4, UAS-IswiK159R/Tm3, Ser × Gcn5E333st/Tm3, Sb cross. Equivalent counts were obtained for IswiK159R/Tm3, Sb siblings from the other crosses. Statistical significance was first assessed by a χ2 homogeneity test with 5 degrees of freedom for error, which indicated that class-1 and class-2 frequencies vary with the tested genotypes (P<0.0001). Pairwise χ2 tests were then performed for each mutant genotype as indicated, taking the IswiK159R/Tm3, Sb as reference. χ2 and P-values indicate that class-1 and class-2 frequencies differ significantly from the control in all mutant combinations except in Ada2bΔ842 (P=0.0137). (B) A representative class-1 eye-Gal4, UAS-IswiK159R/Gcn5E333st adult fly is shown together with a representative class-2 eye-Gal4, UAS-IswiK159R/Tm3, Sb adult fly.
Gcn5, Ada2a and Nurf301 regulate common target genes
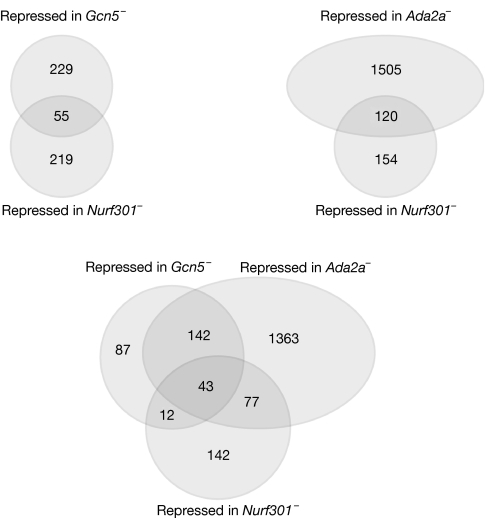
In an attempt to define ATAC target genes, we compared whole-genome transcript profiles of homozygous Gcn5 and Ada2a mutant larvae with those of corresponding heterozygous larvae at the end of the third larval instar (supplementary Table 1 online). Among the genes significantly affected in mutants as compared with control samples (P<0.05), a total of 284 and 1,625 genes were decreased by at least a factor of three in Gcn5 and Ada2a mutants, respectively. Previous whole-genome expression profile analyses have shown that loss of function of Nurf301, a specific subunit of the Iswi-containing NURF complex, results in a threefold downregulation of 274 genes at the end of the third larval instar (Badenhorst et al, 2005). Strikingly, among these 274 genes, 55 (P=2.6 × 10−6) and 120 (P=8.0 × 10−7) genes are also downregulated in Gcn5 and Ada2a mutants, respectively (Fig 2). Moreover, 43 genes are repressed in all three mutants (supplementary Table 2 online).
Figure 2.
Gcn5, Ada2a and Nurf301 regulate a common set of genes at the end of the larval phase. Venn diagrams describing the overlap of genes downregulated by at least a factor of three in Gcn5, Ada2a and Nurf301 mutants at the end of the third larval instar.
To validate our data set, we further analysed Ultrabithorax (Ubx), engrailed (en) and heat-shock protein 70 (hsp70) candidate gene expression in Gcn5 mutants. All three genes were shown to have reduced expression in Nurf301 mutants (Badenhorst et al, 2002). We also found that they are downregulated in Gcn5 mutant larvae. In addition, heat-shock induction of hsp70 was reduced to 50% and 35% of the wild-type induction level in Gcn5E333st and Gcn5f02830 mutant larvae, respectively—an effect similar to that observed in Nurf301 mutants (supplementary Fig S1 online).
These data indicate that ATAC and NURF complexes are involved in the transcriptional regulation of a common set of target genes. In agreement, HAT-containing and nucleosome remodelling complexes were suggested to act synergistically at promoters to regulate target gene transcription (Featherstone, 2002). Although we cannot rule out that our genetic and transcript profiling data reflect indirect interactions, a similar interplay between NURF and ATAC might exist at Drosophila target promoters.
A role of ATAC and NURF in X chromosome structure
Genetic loss of function of Iswi as well as of the NURF subunit Nurf301 leads to a marked decondensation of the male X chromosome, indicating a role of this remodelling complex in the higher order chromosome structure (Deuring et al, 2000; Badenhorst et al, 2002; Corona et al, 2002). These observations prompted us to search for similar chromosome defects in Gcn5 and Ada2a mutants.
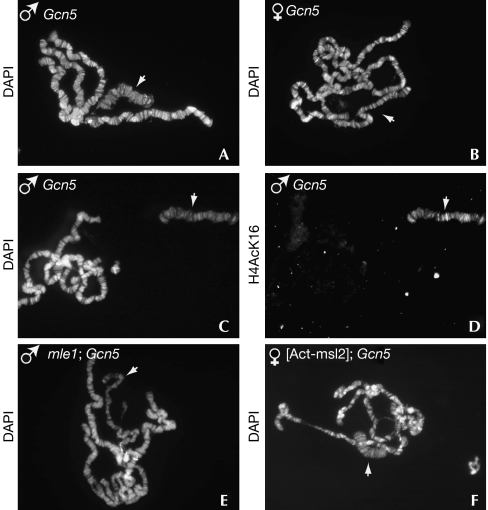
We observed that Gcn5 mutant males display a specific and highly penetrant decondensation of the X chromosome that is never observed in Gcn5 mutant females (Fig 3A,B). In addition, previous studies had shown that the parallel alignment of interbands of polytene chromosomes is often disrupted in Gcn5 mutants, leading to an altered banding pattern (Ciurciu et al, 2006).
Figure 3.
Gcn5 mutations induce male-specific X-chromosome decondensation. Polytene chromosomes from salivary glands were prepared from males and females homozygous for the (A,B) Gcn5f02830 or (C,D) Gcn5E333st mutations and stained with 4,6-diamidino-2-phenylindole (DAPI) or an H4-AcK16 antibody, as indicated. (E) DAPI staining of polytene chromosomes from mle1/mle1; Gcn5E333st/Gcn5f02830 mutant male larvae. (F) DAPI staining of polytene chromosomes from Act-msl2/+; Gcn5E333st/Gcn5f02830 mutant female larvae. White arrows indicate X chromosomes.
In Drosophila males, the dosage compensation complex (DCC) containing maleless (Mle), male-specific lethal 1 (Msl1), Msl3 and the male-specific Msl2 protein recruits the Mof HAT to the X chromosome, resulting in a specific hyperacetylation of H4-K16 residues, which ensures a twofold increase of X-linked genes (Akhtar, 2003). To test whether the decondensation of the X chromosome reflects an alteration in the DCC, we analysed H4-K16 chromosome acetylation in Gcn5 mutant males. Despite its bloated appearance, we found that the X chromosome in these males is still highly acetylated on H4-K16 residues (Fig 3C,D). We therefore explored the possibility that the particular structure conferred by the DCC to the male X chromosome makes it more susceptible to loss of function of Gcn5. The X-bloated phenotype was suppressed in mle, Gcn5 double mutant males in which dosage compensation is impaired (Fig 3E). Moreover, we could induce X-chromosome decondensation by ectopically expressing Msl2 in Gcn5 mutant females to induce DCC formation (Fig 3F). These experiments indicate that normal DCC function is necessary and sufficient to alter the structure of the X chromosome in Gcn5 mutants. It is noteworthy that male X-chromosome decondensation induced by Iswi mutations shows a similar requirement for DCC function (Corona et al, 2002).
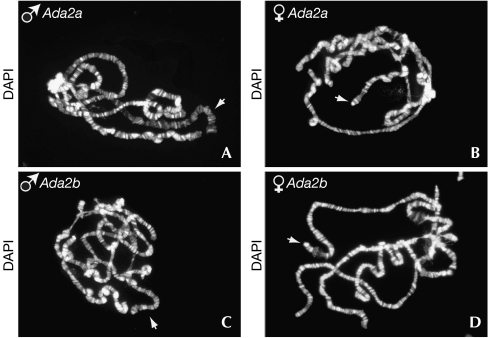
We also found a high frequency of X-bloated chromosomes in preparations from Ada2a mutant males but not females (Fig 4). Similar to the situation encountered in Gcn5 mutant males, X-bloated chromosomes from Ada2a mutants were normally hyperacetylated on H4-K16 residues (Ciurciu et al, 2006). We did not observe X-chromosome bloating in Ada2b mutants (Fig 4). These data strongly suggest that the ATAC complex is required for a correct male X-chromosome structure. It can be noted that mutations affecting several chromatin-associated proteins, including Hp1, Su(var)3-7 and Jil1, also induce male-specific X-chromosome decondensation (Deng et al, 2005; Spierer et al, 2005). As proposed for these mutations, the effect of loss of function of ATAC probably reflects a higher sensitivity of the male X-chromosome structure conferred by the activity of the dosage compensation machinery, rather than a specific role in the regulation of X-linked genes. In support of this conclusion, genome-wide profiling did not show a disproportionate set of X-linked genes among the genes affected in Gcn5 or Ada2a mutants (supplementary Table 1 online).
Figure 4.
Mutation of the ATAC subunit Ada2a induces male-specific X-chromosome bloating. Polytene chromosomes from homozygous (A,B) Ada2aΔ189 or (C,D) Ada2bΔ842 third instar mutant larvae were stained with 4,6-diamidino-2-phenylindole. White arrows indicate X chromosomes.
Iswi is required for ATAC chromosome binding
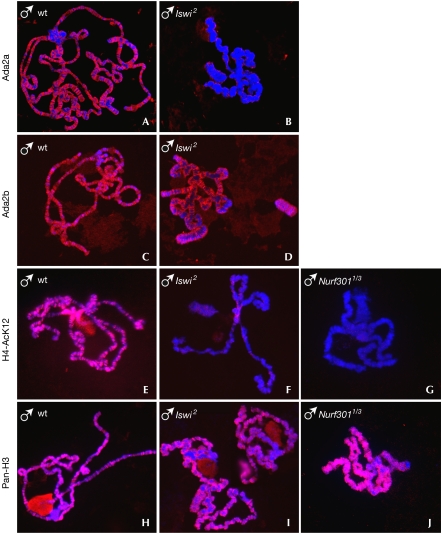
We explored further the functional relationships between NURF and ATAC. Whole-genome RNA profiling indicated that mutations in Gcn5 or Ada2a do not significantly affect the expression of Iswi in late third instar larvae. In addition, immunostaining of polytene chromosomes from either of these mutants did not show changes in the Iswi binding pattern (supplementary Fig S2A online). In turn, quantitative reverse transcription–PCR analysis indicated that the Iswi mutation does not significantly alter the level of Gcn5 and Ada2a expression (supplementary Fig S3 online). Strikingly, however, Ada2a binding was strongly reduced in the Iswi mutant compared with wild-type polytene chromosomes (Fig 5A,B; supplementary Fig S2B online). Non-specific loss of Ada2a binding could result from the general alteration of chromosomes caused by the Iswi mutation (Deuring et al, 2000). However, this does not seem to be the case, as the catalytic subunit of Pol II and, more importantly, the SAGA-specific subunit Ada2b remain associated with the Iswi mutant polytene chromosome under the same experimental conditions (Fig 5C,D; supplementary Fig S2A,B online).
Figure 5.
Binding of Ada2a to chromosomes and H4-AcK12 acetylation are impaired by mutations in components of the Iswi-containing NURF remodelling complex. Polytene chromosomes from wild-type (wt), homozygous Iswi2 and heteroallelic Nurf3011/Nurf3013 mutant males were co-stained with 4,6-diamidino-2-phenylindole (blue) and Ada2a, Ada2b, H4-AcK12 or H3-AcK9/14 (red) antibodies, as indicated.
We reasoned that a specific impairment of Ada2a binding to chromosomes should have an effect on their acetylation by ATAC. Indeed, acetylation of H4-K12 residues is markedly reduced in Iswi mutants (Fig 5E,F; supplementary Fig S4 online), whereas acetylation of H3-K9 and H3-K14 residues generated by SAGA does not seem to be affected (Fig 5H,I). A similar and specific decrease of H4-K12 acetylation of polytene chromosomes was observed in nuclei from the salivary glands of Nurf301 mutants (Fig 5G–J; supplementary Fig S4B online). In agreement with these data, western blot analysis showed a significant decrease of H4-K12 acetylation in salivary glands from mutant Iswi and Nurf301 late third instar larvae (supplementary Fig S5 online). These results strongly indicate that a functional NURF complex is required for the binding of ATAC to chromatin and for subsequent acetylation of H4-K12 residues.
Conclusions
Here we show that loss of function of the ATAC complex markedly affects the morphology of the Drosophila male X chromosome. This suggests that HATs, as previously shown for nucleosome remodelling complexes (Varga-Weisz & Becker, 2006), might also fulfil an architectural function to regulate higher order chromosome structures.
Interestingly, mutations of Hp1 and Su(var)3-7 also induce male X-chromosome bloating, suggesting that defects in heterochromatin formation lead to perturbation of higher order chromosome structure (Spierer et al, 2005). However, loss of function of ATAC probably perturbs chromosome structure through a distinct pathway: Gcn5 mutations do not suppress the variegation of heterochromatic markers and do not affect the methylation of H3-K9 residues as well as the recruitment of Hp1 to chromosomes (Carre et al, 2005; data not shown).
By contrast, we observed genetic interactions between Iswi, Gcn5 and Ada2a, and transcripts downregulated in Nurf301, Gcn5 and Ada2a mutants significantly overlap. These data indicate an interplay between ATAC and NURF. Strikingly, Iswi mutation impairs the binding of Ada2a to chromosomes, and both Iswi and Nurf301 mutations strongly reduce acetylation of H4-K12 residues. This suggests that NURF is required for the recruitment of ATAC to chromatin and for subsequent acetylation of H4-K12 residues by this complex.
Speculation
A mutation of the histone variant H2Av was shown to impair the acetylation of H4-K12 residues on polytene chromosomes (Labrador & Corces, 2003). However, this mutation, in contrast to the Gcn5 and Ada2a mutations, does not induce male X-chromosome decondensation (C.C., unpublished data). It is therefore unlikely that loss of histone H4 acetylation at K12 residues is responsible per se for perturbations of chromosome structure. Gcn5 also acetylates non-histone substrates (Sterner & Berger, 2000). Indeed, Gcn5 was recently shown to acetylate the Rsc4 subunit of the RSC remodelling complex in yeast, potentially modulating RSC binding to its nucleosomal substrate (Vandemark et al, 2007). Moreover, Drosophila Gcn5 efficiently acetylates Iswi in vitro, and its depletion in cultured cells reduces the amount of acetylated Iswi (Ferreira et al, 2007). We speculate that NURF provides ATAC with access to chromatin and that acetylation of Iswi by Gcn5 might, in turn, be important for regulating its remodelling activity. Thus, the perturbation of such an interaction might result in changes in the global chromosome structure as well as in downregulation of NURF target genes.
Methods
Fly strains. The Act-[msl2] transgenic line and mle1 mutants were provided by B. Baker and the Iswi and Nurf301 mutant stocks were provided by P. Badenhorst. The Gcn5E333st null mutant stock has been previously characterized (Carre et al, 2005). Homozygous Gcn5f02830 null mutants (Excelexis collection) were fully rescued by a Gcn5 genomic construct. Homozygous Ada2aΔ189 and Ada2bΔ842 null mutants were obtained by genetic crossing as described (Pankotai et al, 2005). The enhancement of the IswiK159R dominant-negative phenotype in eyes was examined by crossing an eye-Gal4, UAS-IswiK159R/Tm3, Ser stock (Papoulas et al, 2001) with UAS-IR[Gcn5] RNAi (Roignant et al, 2003), Gcn5E333st/Tm3 Sb, Gcn5f02830/Tm3 Sb, Ada2aΔ189/Tm3 Sb or Ada2bΔ842/Tm3 Sb stocks.
Polytene chromosome staining. Immunostaining of polytene chromosomes was performed as described (Zink & Paro, 1995). Antibodies against H4-AcK16 (Turner et al, 1992), H4-AcK12 and H3-AcK9/K14 (Upstate Laboratory, Millipore, Saint-Quentin-en-Yvenlines, France) were used at 1:200 dilution. The Iswi antibody (provided by J. Tamkun) was used at 1:150 dilution. Pol II, Ada2a and Ada2b antibodies were used as described (Muratoglu et al, 2003).
Whole-genome expression analysis. RNA was isolated from homozygous and heterozygous Gcn5E333st and Ada2aΔ189 third instar larvae before pupariation and was labelled and hybridized to Affymetrix Drosophila genome arrays, as detailed in the supplementary information online. A full set of microarray data (experiment E-MEXP-1208) is available from the ArrayExpress database (http:www.ebi.ac.uk/arrayexpress/).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Table 1
supplementary Table 2
supplementary Figs S1–S5
Acknowledgments
We thank I. Bourdeix, M.A. Madillies and R. Castelo for their kind help in statistical analysis, and C. Thibault and the Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC) Affymetrix facility for analysing the expression pattern of the mutant flies. We thank H. Thomassin, F. Azorin and P. Avner for critical reading of the manuscript. This work was supported by grants from the Centre National de la Recherche Scientifique (CNRS), the Pasteur Institute, the Association pour la Recherche Contre le Cancer (ARC) (7742/4383/3202), the Réseau National des Génopoles (no. 260 to L.T. and C.A.) and the Hungarian Science Fund (OTKA T046414 to I.M.B.). C.C. is a ARC fellow and A.C. is an Research Training Networks (RTN) Marie Curie research fellow supported by grant HPRN-CT-2004-504228.
References
- Akhtar A (2003) Dosage compensation: an intertwined world of RNA and chromatin remodelling. Curr Opin Genet Dev 13: 161–169 [DOI] [PubMed] [Google Scholar]
- Badenhorst P, Voas M, Rebay I, Wu C (2002) Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev 16: 3186–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst P, Xiao H, Cherbas L, Kwon SY, Voas M, Rebay I, Cherbas P, Wu C (2005) The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev 19: 2540–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD (1996) Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84: 843–851 [DOI] [PubMed] [Google Scholar]
- Cairns BR (2005) Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr Opin Genet Dev 15: 185–190 [DOI] [PubMed] [Google Scholar]
- Carre C, Szymczak D, Pidoux J, Antoniewski C (2005) The histone H3 acetylase dGcn5 is a key player in Drosophila melanogaster metamorphosis. Mol Cell Biol 25: 8228–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciurciu A, Komonyi O, Pankotai T, Boros IM (2006) The Drosophila histone acetyltransferase Gcn5 and transcriptional adaptor Ada2a are involved in nucleosomal histone H4 acetylation. Mol Cell Biol 26: 9413–9423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Nightingale KP, Becker PB (2002) A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res 30: 649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona DF, Clapier CR, Becker PB, Tamkun JW (2002) Modulation of ISWI function by site-specific histone acetylation. EMBO Rep 3: 242–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Zhang W, Bao X, Martin JN, Girton J, Johansen J, Johansen KM (2005) The JIL-1 kinase regulates the structure of Drosophila polytene chromosomes. Chromosoma 114: 173–182 [DOI] [PubMed] [Google Scholar]
- Deuring R et al. (2000) The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell 5: 355–365 [DOI] [PubMed] [Google Scholar]
- Elfring LK, Deuring R, McCallum CM, Peterson CL, Tamkun JW (1994) Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol 14: 2225–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone M (2002) Coactivators in transcription initiation: here are your orders. Curr Opin Genet Dev 12: 149–155 [DOI] [PubMed] [Google Scholar]
- Ferreira R, Eberharter A, Bonaldi T, Chioda M, Imhof A, Becker PB (2007) Site-specific acetylation of ISWI by GCN5. BMC Mol Biol 8: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelman S, Suganuma T, Florens L, Weake V, Swanson SK, Washburn MP, Abmayr SM, Workman JL (2006) The essential gene wda encodes a WD40 repeat subunit of Drosophila SAGA required for histone H3 acetylation. Mol Cell Biol 26: 7178–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT (1997) ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90: 145–155 [DOI] [PubMed] [Google Scholar]
- Kusch T, Guelman S, Abmayr SM, Workman JL (2003) Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol Cell Biol 23: 3305–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrador M, Corces VG (2003) Phosphorylation of histone H3 during transcriptional activation depends on promoter structure. Genes Dev 17: 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratoglu S et al. (2003) Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol Cell Biol 23: 306–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankotai T, Komonyi O, Bodai L, Ujfaludi Z, Muratoglu S, Ciurciu A, Tora L, Szabad J, Boros I (2005) The homologous Drosophila transcriptional adaptors ADA2a and ADA2b are both required for normal development but have different functions. Mol Cell Biol 25: 8215–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoulas O, Daubresse G, Armstrong JA, Jin J, Scott MP, Tamkun JW (2001) The HMG-domain protein BAP111 is important for the function of the BRM chromatin-remodeling complex in vivo. Proc Natl Acad Sci USA 98: 5728–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA (2004) Histones and histone modifications. Curr Biol 14: R546–R551 [DOI] [PubMed] [Google Scholar]
- Qi D, Larsson J, Mannervik M (2004) Drosophila Ada2b is required for viability and normal histone H3 acetylation. Mol Cell Biol 24: 8080–8089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant JY, Carre C, Mugat B, Szymczak D, Lepesant JA, Antoniewski C (2003) Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA 9: 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spierer A, Seum C, Delattre M, Spierer P (2005) Loss of the modifiers of variegation Su(var)3-7 or HP1 impacts male X polytene chromosome morphology and dosage compensation. J Cell Sci 118: 5047–5057 [DOI] [PubMed] [Google Scholar]
- Sterner DE, Berger SL (2000) Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev 64: 435–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C (1995) Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell 83: 1011–1020 [DOI] [PubMed] [Google Scholar]
- Turner BM, Birley AJ, Lavender J (1992) Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell 69: 375–384 [DOI] [PubMed] [Google Scholar]
- Vandemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR (2007) Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol Cell 27: 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz PD, Becker PB (2006) Regulation of higher-order chromatin structures by nucleosome-remodelling factors. Curr Opin Genet Dev 16: 151–156 [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB (1997) Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 388: 598–602 [DOI] [PubMed] [Google Scholar]
- Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu D, Wu C (2001) Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell 8: 531–543 [DOI] [PubMed] [Google Scholar]
- Zink D, Paro R (1995) Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J 14: 5660–5671 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Table 1
supplementary Table 2
supplementary Figs S1–S5