Abstract
The maintenance of correct mitochondrial shape requires numerous proteins that act on the surface or inside of the organelle. Although the soluble F-box protein Mfb1 was recently found to associate peripherally with mitochondria and to regulate organelle connectivity in budding yeast, how it localizes to mitochondria is unknown. Here, we show that two tetratricopeptide repeat proteins—the general preprotein import receptor Tom70 (a component of translocase of the outer membrane) and its paralogue Tom71—are required for Mfb1 mitochondrial localization. Mitochondria in cells lacking Tom70 and Tom71 form short tubules and aggregates, aberrant morphologies similar to those observed in the mfb1-null mutant. In addition, Mfb1 interacts with Tom71 in vivo, and binds to mitochondria through Tom70 in vitro. Our data indicate an unexpected role for Tom70 in recruitment of soluble proteins to the mitochondrial surface, and indicate that Tom71 has a specialized role in Mfb1-mediated mitochondrial morphogenesis.
Keywords: TPR proteins, mitochondrial morphology, protein targeting, F-box proteins, Saccharomyces cerevisiae
Introduction
Mitochondria form elaborate tubular networks that constantly remodel their shape and distribution in response to intra- and extracellular cues (Chan, 2006a; Hoppins et al, 2007). Defects in mitochondrial shape and distribution are linked to human neurodegenerative disorders, highlighting the physiological relevance of organelle dynamics (Chan, 2006b). Studies in yeast and multicellular eukaryotes continue to identify new proteins required for mitochondrial morphogenesis, indicating that establishment and maintenance of mitochondrial architecture is a complex process (Okamoto & Shaw, 2005; Frazier et al, 2006).
A large number of soluble and membrane-integrated proteins act on the surface or inside of mitochondria to regulate organelle shape. Most of these morphology proteins are synthesized as precursors (preproteins) in the cytoplasm, targeted to mitochondria, sorted to the correct mitochondrial subcompartments and refolded/assembled into functional complexes through the general protein transport pathways (Neupert & Herrmann, 2007). An emerging group of soluble morphology proteins mature in the cytoplasm and transiently or stably associate with the surface of mitochondria. Examples of the latter include Mdm30 and Mfb1, two peripherally associated F-box proteins that form distinct complexes on the cytoplasmic face of the outer membrane (Fritz et al, 2003; Dürr et al, 2006; Escobar-Henriques et al, 2006; Kondo-Okamoto et al, 2006). F-box proteins can function as components of Skp1–Cul1–F-box-protein (SCF) ubiquitin–ligase complexes, in which they bind to substrates for ubiquitin-mediated proteolysis; however, they are also found in non-SCF protein complexes that carry out various cellular functions. Although Mdm30 has been shown to regulate degradation of a known mitochondrial fusion protein, Mfb1 regulates the length and connectivity of mitochondrial tubules by an unknown mechanism. Mitochondrial targeting of Mfb1—and presumably Mdm30—is likely to be essential for function; however, factors that recruit these F-box proteins to the mitochondrial surface have not been identified.
Here, we report that Mfb1 interacts with Tom71 (a component of translocase of the outer membrane; Bömer et al, 1996; Schlossmann et al, 1996), a tetratricopeptide repeat (TPR) protein that is a paralogue of the mitochondrial preprotein import receptor Tom70 (Hines et al, 1990; Steger et al, 1990). Moreover, we show that binding of Mfb1 to mitochondria requires Tom70 in vitro. Our data establish that Tom70 and Tom71 have overlapping functions in Mfb1 mitochondrial localization that are essential for the maintenance of mitochondrial morphology.
Results
Mfb1 interacts with Tom71
To identify proteins that recruit Mfb1 to the surface of mitochondria, we carried out biochemical tandem affinity purification and mass spectrometry for Mfb1-specific interacting proteins (data not shown). This approach identified Tom71, a mitochondrial protein anchored to the cytoplasmic face of the outer membrane (Bömer et al, 1996; Schlossmann et al, 1996). Tom71 is a paralogue of the mitochondrial preprotein import receptor Tom70 (Hines et al, 1990; Steger et al, 1990).
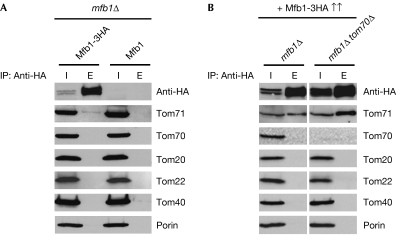
Specificity of the Mfb1–Tom71 interaction was confirmed by co-precipitation assays (Fig 1A). This co-immunoprecipitation was inefficient but specific, as no significant precipitation was detected in control experiments using wild-type Mfb1. In addition, overexpression of Mfb1 strongly increased Tom71 co-precipitation (Fig 1B). Under these conditions, we did not detect co-precipitation of Mfb1 with Tom70, Tom20, Tom22 and Tom40, the main components of the TOM complex, and Porin, an abundant outer membrane protein. In addition, interactions of Mfb1 with endogenous Tom20 and Tom40 were not detected in co-precipitation assays using Tom20 and Tom40 antibodies (data not shown). The fact that Tom71 co-precipitation was enhanced in the absence of Tom70 indicates that Tom70 is not required for the Mfb1–Tom71 interaction.
Figure 1.
Tom71 interacts with Mfb1. Mitochondria isolated from (A) mfb1Δ cells expressing Mfb1-3HA or wild-type Mfb1 and (B) mfb1Δ or mfb1Δ tom70Δ cells overexpressing Mfb1-3HA from the TPI promoter (Mfb1-3HA↑↑) were solubilized and subjected to immunoprecipitation using HA antibody-conjugated agarose. The mitochondrial extract inputs (I) and eluted immunoprecipitates (E) are shown. TOM, translocase of the outer membrane.
Mfb1 localization is disrupted in tom70Δ tom71Δ cells
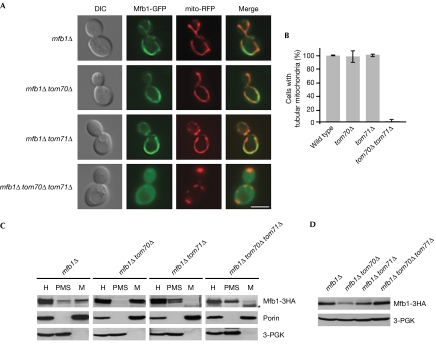
We evaluated the localization patterns of coexpressed Mfb1-green fluorescent protein (GFP) and mitochondria-targeted red fluorescent protein (mito-RFP; Fig 2A). Mfb1-GFP colocalized with mito-RFP and restored normal mitochondrial morphology in mfb1Δ cells (Kondo-Okamoto et al, 2006). Mitochondrial localization of Mfb1-GFP was not altered in mfb1Δ tom71Δ or mfb1Δ tom70Δ cells. Strikingly, Mfb1-GFP remained in the cytoplasm, and localized poorly to mitochondria in the absence of both Tom70 and Tom71. In addition, mitochondria lacking Tom70 and Tom71 were discontinuous and aggregated, aberrant morphologies reported previously in mfb1Δ cells (Kondo-Okamoto et al, 2006). Nearly 100% of tom70Δ tom71Δ cells showed mitochondrial morphology defects (Fig 2B). These observations indicate that Tom70 and Tom71 have overlapping functions in Mfb1 mitochondrial localization.
Figure 2.
Mfb1 mitochondrial targeting is disrupted in cells lacking Tom70 and Tom71. (A) Mfb1-GFP localization and mitochondria visualized by mito-RFP in mfb1Δ, mfb1Δ tom70Δ, mfb1Δ tom71Δ and mfb1Δ tom70Δ tom71Δ cells. Scale bar: 5 μm. (B) Mitochondrial morphology in wild-type, tom70Δ, tom71Δ and tom70Δ tom71Δ cells expressing mito-GFP. (C) Subcellular fractionation of strains used in (A) expressing Mfb1-3HA. The cell homogenate (H) was separated into post-mitochondrial supernatant (PMS) and mitochondrial pellet (M). Porin and 3-PGK were monitored as mitochondria and cytoplasm markers, respectively. The asterisk indicates nonspecific bands. (D) Steady-state levels of Mfb1-3HA in total cell extracts of strains used in (C). 3-PGK was monitored as a loading control. DIC, differential interference contrast; GFP, green fluorescent protein; mito-RFP, mitochondria-targetted red fluorescent protein; PGK, phosphoglycerate kinase; TOM, translocase of the outer membrane.
Biochemical fractionation assays were used to determine whether Tom70 and Tom71 are required for Mfb1 mitochondrial association (Fig 2C). In the presence of Tom70 and Tom71, Mfb1-3HA was found in the mitochondria-enriched pellet (M) and post-mitochondrial supernatant (PMS; Kondo-Okamoto et al, 2006). In the absence of Tom70, almost all Mfb1-3HA cofractionated in M, indicating that Tom70 is not required for Mfb1 mitochondrial association. By contrast, most of the Mfb1-3HA was found in the PMS from mfb1Δ tom71Δ and mfb1Δ tom70Δ tom71Δ cells. These different fractionation patterns were not due to changes in Mfb1-3HA protein levels in the double and triple mutant strains, although the Mfb1-3HA levels were lower in mfb1Δ tom70Δ cells (Fig 2D). These results are consistent with our findings that Mfb1 interacts with Tom71 in the absence of Tom70, and that Mfb1 does not form a stable complex with Tom70 in co-precipitation assays.
Finally, the preprotein receptors Tom20 and Tom22, and Tom5 that transfers preproteins from surface receptors to the TOM channel (Pfanner et al, 2004), are not required for Mfb1 mitochondrial localization (supplementary Figs S1,S2 online). Together, our data support the idea that Tom70 and Tom71 mediate Mfb1 mitochondrial targeting independently of other preprotein-interacting receptors.
Membrane-anchored Mfb1 in tom70Δ tom71Δ cells
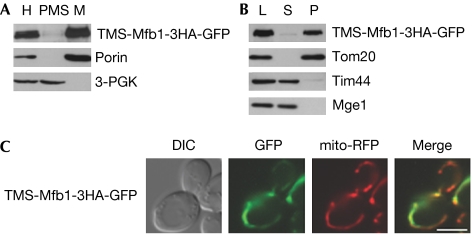
Aberrant mitochondrial morphology in cells lacking Tom70 and Tom71 might result from defects in mitochondrial localization of other morphology proteins. To test this possibility, we expressed TMS-Mfb1-3HA-GFP in mfb1Δ tom70Δ tom71Δ cells. This membrane-anchored Mfb1 variant contains the amino-terminal transmembrane segment of Tom20 and restores normal mitochondrial morphology in mfb1Δ cells (Kondo-Okamoto et al, 2006). Subcellular fractionation and carbonate extraction assays indicated that TMS-Mfb1-3HA-GFP was stably associated with mitochondria and integrated into membranes in the absence of Tom70 and Tom71 (Fig 3A,B).
Figure 3.
An outer membrane-anchored Mfb1 variant can rescue mitochondrial morphology defects in cells lacking Tom70 and Tom71. (A) Subcellular fractionation of mfb1Δ tom70Δ tom71Δ cells expressing TMS-Mfb1-3HA-GFP was carried out as described in Fig 2C. Porin and 3-PGK were monitored as mitochondria and cytoplasm markers, respectively. (B) Carbonate extraction assays for mitochondria isolated from cells used in (A). Mitochondria were treated with Na2CO3 and separated into soluble supernatant (S) and membrane pellet (P) fractions. Tom20 (integral membrane protein), Tim44 (peripheral membrane protein) and Mge1 (soluble protein) were monitored as controls. (C) TMS-Mfb1-3HA-GFP localization and mitochondria visualized by mito-RFP in mfb1Δ tom70Δ tom71Δ cells. Scale bar: 5 μm. GFP, green fluorescent protein; L, loaded mitochondria; M, mitochondrial pellet; mito-RFP, mitochondria-targeted red fluorescent protein; PMS, post-mitochondrial supernatant; TOM, translocase of the outer membrane.
Fluorescence microscopy indicated that TMS-Mfb1-3HA-GFP colocalized with mito-RFP (89% of cells) and restored mitochondrial tubular networks in mfb1Δ tom70Δ tom71Δ cells (51% of cells; Fig 3C). These observations indicate that morphology defects in cells lacking Tom70 and Tom71 are caused largely by the dissociation of Mfb1 from mitochondria.
Tom70 domain required for Mfb1 recruitment
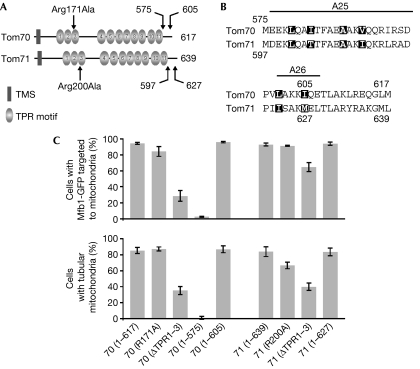
Tom70 and Tom71 share a similar domain structure: an N-terminal transmembrane segment, 11 TPR motifs and a C-terminal extension (Fig 4A). The crystal structure of yeast Tom70 was recently determined, indicating two distinct TPR modules (Wu & Sha, 2006). The TPR motifs 1–3 and 4–11 form a peptide-binding groove for Hsp70 (heat shock protein 70) and a putative binding pocket for mitochondrial preproteins, respectively (Brix et al, 2000; Young et al, 2003; Chan et al, 2006; Wu & Sha, 2006; Fig 4A). The C-terminal extension consists of two α-helices, which are predicted to form an interface for Tom70 dimerization (Wu & Sha, 2006; Fig 4B).
Figure 4.
Tom70 and Tom71 domain analyses for Mfb1 mitochondrial targeting and morphology maintenance. (A) Schematic representation of Tom70 and Tom71 domain structures. Amino-acid substitutions generated in the TPR3 motifs and sites for the carboxy-terminal deletions are indicated by arrows. (B) Amino-acid sequence alignment of the Tom70 and Tom71 C-terminal extensions that are predicted to form two α-helices, A25 and A26. Residues involved in possible homo-dimer formation are highlighted. (C) Mfb1-GFP localization and mitochondrial morphology (visualized by MitoTracker Red staining) in mfb1Δ tom70Δ tom71Δ cells expressing Tom70 and Tom71 variants. GFP, green fluorescent protein; TOM, translocase of the outer membrane; TPR, tetratricopeptide repeat.
To define the Tom70 and Tom71 regions required for Mfb1 mitochondrial localization and morphology maintenance, Mfb1-GFP localization and mitochondrial morphology were quantified in mfb1Δ tom70Δ tom71Δ cells expressing mutant Tom70 or Tom71 proteins (Fig 4C). Although Tom71 (1–597) was not stably expressed, other mutant proteins were expressed at levels similar to, or higher than wild-type proteins (supplementary Fig S3 online). In cells with either the Tom70 Arg171Ala mutant, which does not bind to Hsp70 in vitro (Young et al, 2003), or the corresponding Tom71 Arg200Ala mutant, Mfb1-GFP mitochondrial localization (84% and 91%, respectively) was similar to cells expressing wild-type Tom70 and Tom71 (93–94%). Deletion of the Tom70 and Tom71 regions, including the TPR1–3 motifs (ΔTPR1–3), did not completely disrupt Mfb1-GFP mitochondrial targeting (29% and 65%, respectively). These results indicate that the putative Hsp70-binding sites of Tom70 and Tom71 are not essential for Mfb1 mitochondrial localization.
Tom70 (1–605) and Tom71 (1–627), mutant proteins that lack the last 12 amino acids but might retain the A25 and A26 α-helices (Fig 4B), restored Mfb1-GFP mitochondrial targeting in mfb1Δ tom70Δ tom71Δ cells (96% and 94%, respectively). By contrast, Mfb1-GFP was poorly targeted to mitochondria in cells expressing Tom70 (1–575), which lacks the entire C-terminal extension (3%). These targeting phenotypes correlated well with mitochondrial morphology profiles. Together, our data indicate that the last two α-helices of Tom70 are important for Mfb1 mitochondrial localization and morphology maintenance.
Tom70 is necessary for Mfb1 binding to mitochondria
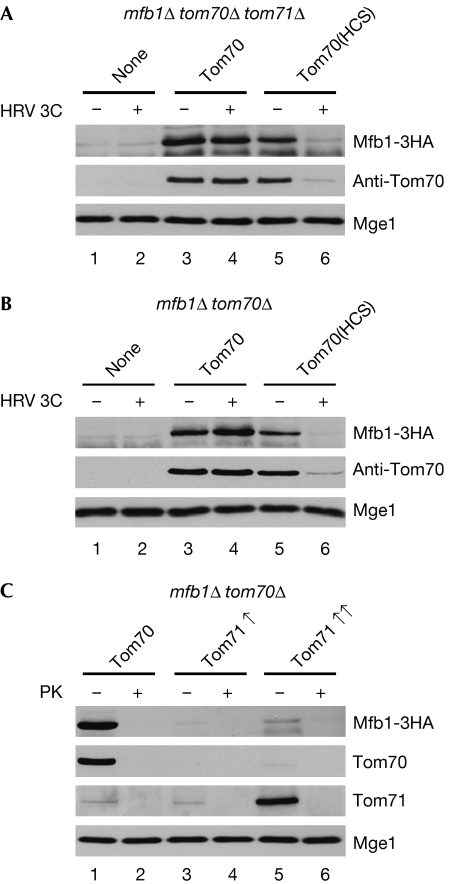
Although Mfb1 localizes to tom71Δ mitochondria in vivo (Fig 2A), it dissociates from tom71Δ mitochondria during subcellular fractionation (Fig 2B). In addition, Tom70 does not form a stable complex with Mfb1 in co-precipitation assays (Fig 1A,B). On the basis of these results, we hypothesized that Tom70 transiently interacts with Mfb1 during mitochondrial targeting. To test this possibility, we carried out in vitro binding assays. Previous studies using similar assays (Hines et al, 1990; Steger et al, 1990) have successfully shown transient receptor–preprotein interactions that cannot be detected by co-precipitation and subcellular fractionation. Consistent with the in vivo localization (Fig 2A), Mfb1-3HA did not bind to mitochondria in the absence of both Tom70 and Tom71 (Fig 5A, lane 1). By contrast, Mfb1 co-fractionated with mitochondria isolated from mfb1Δ tom70Δ tom71Δ cells expressing Tom70 (Fig 5A, lane 3). These results indicate that Tom70 is required for Mfb1 mitochondrial targeting, raising the possibility that this receptor protein recruits Mfb1.
Figure 5.
Mfb1 binds to mitochondria through Tom70 and Tom71 in vitro. Mitochondria isolated from (A) mfb1Δ tom70Δ tom71Δ cells expressing Tom70 or Tom70(HCS), a variant with an HRV 3C processing site, (B) mfb1Δ tom70Δ cells expressing Tom70 or Tom70(HCS) or (C) mfb1Δ tom70Δ cells expressing Tom70 or Tom71 at 2- to 3-fold (Tom71↑, from a low-copy plasmid with the TOM71 promoter) and 10- to 15-fold (Tom71↑↑, from a multi-copy plasmid with the TOM71 promoter) higher levels were pretreated with (+) or without (−) HRV 3C protease (80 U/ml) or proteinase K (PK; 50 mg/ml) at 4°C for 30 min and incubated with post-mitochondrial supernatant prepared from mfb1Δ tom70Δ tom71Δ cells overexpressing Mfb1-3HA. The samples were subjected to sedimentation using a 25% sucrose cushion and the separated mitochondrial fractions were analysed. Mge1 was monitored as a mitochondrial loading control. TOM, translocase of the outer membrane.
As Tom70 is the general import receptor for mitochondrial preproteins, it is possible that an unknown Mfb1-interacting factor localizes to mitochondria through Tom70. In this case, loss of Tom70 would block the biogenesis of this unknown factor and secondarily disrupt Mfb1 mitochondrial targeting. To test this possibility, we generated a functional variant of Tom70, Tom70 (HCS), which contains a cleavage site specific for HRV 3C protease. The cleavage site was inserted between the N-terminal transmembrane segment and the cytoplasmic domain of Tom70. On treatment of mitochondria with HRV 3C protease, Tom70(HCS) is released from the mitochondrial surface without affecting other mitochondrial proteins. Mfb1-3HA efficiently bound mock-treated mitochondria isolated from mfb1Δ tom70Δ tom71Δ cells expressing Tom70(HCS) (Fig 5A, lane 5). By contrast, binding of Mfb1-3HA to HRV 3C-pretreated mitochondria was significantly reduced (Fig 5A, lane 6). HRV 3C treatment did not affect binding of Mfb1-3HA to mfb1Δ tom70Δ tom71Δ and mfb1Δ tom71Δ mitochondria (Fig 5A, lanes 2 and 4, respectively). These results support the idea that Tom70 recruits Mfb1 to mitochondria through direct or indirect interaction.
Our live cell imaging, subcellular fractionation and co-precipitation assays indicate that Mfb1 can localize to mitochondria through interaction with Tom71 (Figs 1A,2A,B). Surprisingly, Mfb1-3HA did not bind to mfb1Δ tom70Δ mitochondria (Fig 5B, lane 1), although binding was restored by expressing wild-type Tom70 or Tom70(HCS) in this strain (Fig 5B, lanes 3 and 5, respectively). HRV 3C treatment specifically disrupted Mfb1-3HA binding to mitochondria containing Tom70(HCS) (Fig 5B, lane 6) without affecting binding to mfb1Δ tom70Δ and mfb1Δ mitochondria (Fig 5B, lanes 2 and 4, respectively). When introduced into mfb1Δ tom70Δ cells, an additional copy of TOM71 did not restore Mfb1-3HA mitochondrial binding (Fig 5C, lane 3), whereas Mfb1-3HA efficiently bound mitochondria on expression of TOM70 (Fig 5C, lane 1). Interestingly, overexpression of Tom71 resulted in weak but significant Mfb1-3HA mitochondrial binding (Fig 5C, lane 5). Proteinase K pretreatment eliminated co-fractionation of Mfb1-3HA with mitochondria, which verified that binding occurred through a proteinacious factor (Fig 5C, lanes 2,4 and 6). Together, these results are consistent with the idea that Tom71 is able to recruit Mfb1 to mitochondria, although this targeting activity is reduced in vitro.
Discussion
Here, we have shown that Tom71 is required for Mfb1 mitochondrial localization in the absence of Tom70. Conversely, Tom70 is indispensable for localizing Mfb1 to mitochondria in cells lacking Tom71. These homologous proteins probably have overlapping functions in Mfb1 localization. Concomitant with the accumulation of Mfb1 in the cytoplasm, mitochondria show aberrant shapes in cells lacking Tom70 and Tom71. A functional membrane-anchored Mfb1 variant can significantly rescue mitochondrial morphology defects in the tom70Δ tom71Δ double mutant, indicating that aberrant mitochondrial morphologies in the double mutant are primarily due to dissociation of Mfb1 from the organelle. However, we do not exclude the possibilities that Tom70 and Tom71 are required for localization of other morphology proteins, and that the latter proteins have direct roles in mitochondrial morphogenesis. Consistent with these ideas, mitochondrial morphology defects are associated with the loss of Tom70 function in filamentous fungi, which do not have Mfb1 homologues (Jamet-Vierny et al, 1997; Grad et al, 1999).
In the absence of Tom71, Mfb1 dissociates from mitochondria during subcellular fractionation, even in the presence of Tom70. By contrast, Mfb1 binds to mitochondria lacking Tom71 in vitro, in a Tom70-dependent manner. How can these results be reconciled? One interpretation is that Tom70 might act as a receptor that only transiently interacts with Mfb1. Conversely, Tom71 might act as an assembly partner rather than a transient receptor for Mfb1. Alternatively, Tom70 and Tom71 might function in both Mfb1 mitochondrial targeting and association, which are not mutually exclusive. It is also unclear whether Mfb1 interacts with Tom70 and Tom71 directly or indirectly. Further studies are needed to clarify these issues.
Although Tom70 is a general receptor that mediates import of preproteins, including those destined for the outer membrane, inner membrane and matrix (Chan et al, 2006), it has never been implicated in targeting of soluble proteins to the mitochondrial surface. Our study indicates that Tom70 also recruits Mfb1, a soluble protein, to the surface of mitochondria. In mammalian cells, Tom70 facilitates targeting of Mcl-1, a Bcl-2 family member that inserts loosely into the mitochondrial outer membrane and regulates programmed cell death (Chou et al, 2006). Thus, the role of Tom70 in protein recruitment to the mitochondrial surface has been conserved during evolution.
Despite the fact that Tom71 is a paralogue sharing high similarity with Tom70, Tom71 has only a minor role in mitochondrial protein transport (Schlossmann et al, 1996; Koh et al, 2001), and its primary function has not been described. Here, we show that Tom71 forms a complex with Mfb1 and maintains Mfb1 mitochondrial association. Tom71 might have arisen from a genome duplication event and co-evolved with Mfb1 to gain a specialized role in mitochondrial morphogenesis.
Methods
Yeast strains and growth conditions. Yeast strains were generated in the W303 backgrounds. Strains and plasmids used in this study are listed in the supplementary information online. Standard genetic and molecular biology methods were used for yeast (Burke et al, 2000) and bacterial (Sambrook & Russell, 2001) strains. Yeast cells were grown as described previously (Kondo-Okamoto et al, 2003).
Quantification of mitochondrial morphology and targeting. Mitochondrial morphology and Mfb1 targeting were scored in cells expressing mito-GFP, or mito-RFP and Mfb1-GFP grown to log phase (OD600=0.4–1.7) at 30°C in dextrose media, and stained with MitoTracker Red. Phenotypes were quantified in 100 cells in three experiments. Data are the average of all experiments, with bars indicating standard deviations.
Biochemical studies. Mitochondrial isolation, subcellular fractionation, carbonate extraction, immunoprecipitation, in vitro binding assays and immunoblot analysis were carried out as described previously (Frederick et al, 2004; Kondo-Okamoto et al, 2006) with some modifications (see the supplementary information online).
Microscopy. Images of yeast cells were acquired and processed as described previously (Kondo-Okamoto et al, 2006).
Supplementary information is available at EMBO Reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We appreciate discussions with the lab members of J.M.S. We thank P. Krishna for mass spectrometry at the University of Utah Core Facility. N.K.-O. and K.O. are grateful to Y. Ohsumi and Ohsumi lab members for their generous support during the final stage of this study. This work was supported by grants from the National Institutes of Health (GM53466) (J.M.S.), the University of Utah Seed Grant Program (J.M.S.), the United Mitochondrial Disease Foundation (K.O.) and the American Heart Association (K.O.). This paper is dedicated to the memory of Ronald A. Butow.
References
- Bömer U, Pfanner N, Dietmeier K (1996) Identification of a third yeast mitochondrial Tom protein with tetratrico peptide repeats. FEBS Lett 382: 153–158 [DOI] [PubMed] [Google Scholar]
- Brix J, Ziegler GA, Dietmeier K, Schneider-Mergener J, Schulz GE, Pfanner N (2000) The mitochondrial import receptor Tom70: identification of a 25 kDa core domain with a specific binding site for preproteins. J Mol Biol 303: 479–488 [DOI] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T (2000) Methods in Yeast Genetics. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory [Google Scholar]
- Chan DC (2006a) Mitochondrial fusion and fission in mammals. Annu Rev Cell Dev Biol 22: 79–99 [DOI] [PubMed] [Google Scholar]
- Chan DC (2006b) Mitochondria: dynamic organelles in disease, aging, and development. Cell 125: 1241–1252 [DOI] [PubMed] [Google Scholar]
- Chan NC, Likic VA, Waller RF, Mulhern TD, Lithgow T (2006) The C-terminal TPR domain of Tom70 defines a family of mitochondrial protein import receptors found only in animals and fungi. J Mol Biol 358: 1010–1022 [DOI] [PubMed] [Google Scholar]
- Chou CH, Lee RS, Yang-Yen HF (2006) An internal EELD domain facilitates mitochondrial targeting of Mcl-1 via a Tom70-dependent pathway. Mol Biol Cell 17: 3952–3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr M, Escobar-Henriques M, Merz S, Geimer S, Langer T, Westermann B (2006) Nonredundant roles of mitochondria-associated F-box proteins Mfb1 and Mdm30 in maintenance of mitochondrial morphology in yeast. Mol Biol Cell 17: 3745–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Henriques M, Westermann B, Langer T (2006) Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1. J Cell Biol 173: 645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier AE, Kiu C, Stojanovski D, Hoogenraad NJ, Ryan MT (2006) Mitochondrial morphology and distribution in mammalian cells. Biol Chem 387: 1551–1558 [DOI] [PubMed] [Google Scholar]
- Frederick RL, McCaffery JM, Cunningham KW, Okamoto K, Shaw JM (2004) Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway. J Cell Biol 167: 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz S, Weinbach N, Westermann B (2003) Mdm30 is an F-box protein required for maintenance of fusion-competent mitochondria in yeast. Mol Biol Cell 14: 2303–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad LI, Descheneau AT, Neupert W, Lill R, Nargang FE (1999) Inactivation of the Neurospora crassa mitochondrial outer membrane protein TOM70 by repeat-induced point mutation (RIP) causes defects in mitochondrial protein import and morphology. Curr Genet 36: 137–146 [DOI] [PubMed] [Google Scholar]
- Hines V, Brandt A, Griffiths G, Horstmann H, Brutsch H, Schatz G (1990) Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J 9: 3191–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Lackner L, Nunnari J (2007) The machines that divide and fuse mitochondria. Annu Rev Biochem 76: 751–780 [DOI] [PubMed] [Google Scholar]
- Jamet-Vierny C, Contamine V, Boulay J, Zickler D, Picard M (1997) Mutations in genes encoding the mitochondrial outer membrane proteins Tom70 and Mdm10 of Podospora anserina modify the spectrum of mitochondrial DNA rearrangements associated with cellular death. Mol Cell Biol 17: 6359–6366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh JY, Hajek P, Bedwell DM (2001) Overproduction of PDR3 suppresses mitochondrial import defects associated with a TOM70 null mutation by increasing the expression of TOM72 in Saccharomyces cerevisiae. Mol Cell Biol 21: 7576–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo-Okamoto N, Shaw JM, Okamoto K (2003) Mmm1p spans both the outer and inner mitochondrial membranes and contains distinct domains for targeting and foci formation. J Biol Chem 278: 48997–49005 [DOI] [PubMed] [Google Scholar]
- Kondo-Okamoto N, Ohkuni K, Kitagawa K, McCaffery JM, Shaw JM, Okamoto K (2006) The novel F-box protein Mfb1p regulates mitochondrial connectivity and exhibits asymmetric localization in yeast. Mol Biol Cell 17: 3756–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM (2007) Translocation of proteins into mitochondria. Annu Rev Biochem 76: 723–749 [DOI] [PubMed] [Google Scholar]
- Okamoto K, Shaw JM (2005) Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet 39: 503–536 [DOI] [PubMed] [Google Scholar]
- Pfanner N, Wiedemann N, Meisinger C, Lithgow T (2004) Assembling the mitochondrial outer membrane. Nat Struct Mol Biol 11: 1044–1048 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory [Google Scholar]
- Schlossmann J, Lill R, Neupert W, Court DA (1996) Tom71, a novel homologue of the mitochondrial preprotein receptor Tom70. J Biol Chem 271: 17890–17895 [DOI] [PubMed] [Google Scholar]
- Steger HF, Söllner T, Kiebler M, Dietmeier KA, Pfaller R, Trülzsch KS, Tropschug M, Neupert W, Pfanner N (1990) Import of ADP/ATP carrier into mitochondria: two receptors act in parallel. J Cell Biol 111: 2353–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sha B (2006) Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat Struct Mol Biol 13: 589–593 [DOI] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU (2003) Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112: 41–50 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information