Abstract
XRCC4-like factor (XLF)—also known as Cernunnos—has recently been shown to be involved in non-homologous end-joining (NHEJ), which is the main pathway for the repair of DNA double-strand breaks (DSBs) in mammalian cells. XLF is likely to enhance NHEJ by stimulating XRCC4–ligase IV-mediated joining of DSBs. Here, we report mechanistic details of XLF recruitment to DSBs. Live cell imaging combined with laser micro-irradiation showed that XLF is an early responder to DSBs and that Ku is essential for XLF recruitment to DSBs. Biochemical analysis showed that Ku–XLF interaction occurs on DNA and that Ku stimulates XLF binding to DNA. Unexpectedly, XRCC4 is dispensable for XLF recruitment to DSBs, although photobleaching analysis showed that XRCC4 stabilizes the binding of XLF to DSBs. Our observations showed the direct involvement of XLF in the dynamic assembly of the NHEJ machinery and provide mechanistic insights into DSB recognition.
Keywords: Cernunnos, Ku, non-homologous end-joining, XLF, XRCC4
Introduction
DNA double-strand breaks (DSBs) are among the most cytotoxic of DNA lesions, and failure to repair them results in genomic instability and subsequent malignant transformation. Non-homologous end-joining (NHEJ) is the main repair mechanism of DSBs in mammalian cells (Weterings & van Gent, 2004). The initial step of NHEJ is the binding of Ku to both ends of the broken DNA molecules. Ku consists of two subunits, Ku70 and Ku80, and has an extremely high affinity for free ends of double-stranded DNA (Smith & Jackson, 1999). On binding to DNA, Ku recruits a serine/threonine kinase called DNA-dependent protein kinase catalytic subunit (DNA-PKCS) to the DSBs (Smith & Jackson, 1999; Lees-Miller & Meek, 2003). The DNA-PKCS kinase activity is essential for the efficient progression of NHEJ reactions (Kienker et al, 2000; Kurimasa et al, 1999). Recently, the early response of both Ku and DNA-PKCS to the onset of DSBs has been shown using live cell imaging, which indicated that Ku (Mari et al, 2006) and DNA-PKCS (Uematsu et al, 2007) are recruited to laser-induced DSBs in a matter of seconds. The complex of Ku and DNA-PKCS is important in bringing two DNA ends into close proximity (Cary et al, 1997). The joining of synapsed DNA ends is carried out by XRCC4–DNA ligase IV (LigIV), which form a tight complex and stabilize each other (Bryans et al, 1999).
The XRCC4-like factor (XLF)/Cernunnos was recently identified as a gene responsible for a human genetic disorder characterized by defective increased radiosensitivity and severe combined immunodeficiency (Dai et al, 2003). Ahnesorg et al (2006) isolated a new XRCC4-interacting factor by yeast two-hybrid screening and named it XLF. Buck et al (2006) identified the responsible gene by complementation screening of human complementary DNA library and referred to it as Cernunnos. XLF and Cernunnos were found to be identical (here, we refer to XLF/Cernunnos as XLF for simplicity). XLF binds to the XRCC4–LigIV complex in vivo and in vitro (Ahnesorg et al, 2006; Callebaut et al, 2006) and stimulates ligase activity of XRCC4–LigIV in vitro (Hentges et al, 2006; Tsai et al, 2007). Purified XLF associates with DNA (Hentges et al, 2006) in a DNA-length-dependent manner (Lu et al, 2007). XLF stably associates with 83 bp or longer DNA, but not with shorter DNAs (Lu et al, 2007). These observations indicate that XLF is a member of the NHEJ pathway and modulates the ligase activity of XRCC4–LigIV through a direct interaction with XRCC4. A recent study showed that the knockdown of XRCC4, LigIV or DNA-PKCS decreases the amount of XLF in detergent-resistant insoluble chromatin fractions of DSB-induced cells (Wu et al, 2007). NHEJ factors have been thought to be recruited sequentially from Ku to XRCC4–LigIV, and XLF has been presumed to act in a final ligation step of NHEJ mainly through its interaction with XRCC4 (Sekiguchi & Ferguson, 2006).
Although several functions of XLF have been described, limited information is available about the precise pathway of XLF recruitment to DSBs in the functional assembly of the NHEJ machinery. Here, we focused our efforts on the spatial and temporal dynamics of XLF at DSBs. Our data show that Ku is essential for the in vivo recruitment of XLF to DSBs. XRCC4 is dispensable for in vivo XLF recruitment to DSBs but stabilizes the recruited XLF at DSBs. We show biochemical evidence of a unique interaction between Ku and XLF on DNA, which provides a new mechanistic insight into DNA damage recognition by repair proteins. Together, our data provide a new model of protein assembly in the NHEJ pathway.
Results
XLF rapidly accumulates at laser-induced DSBs
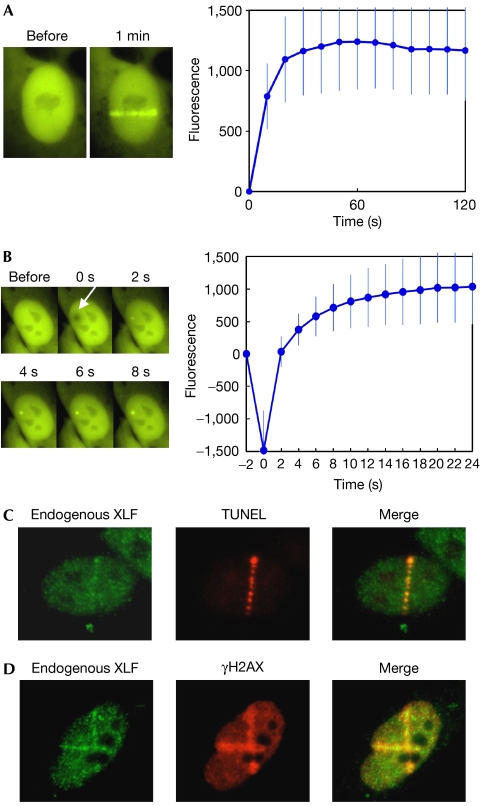
To study the molecular dynamics of XLF at DSBs in living cells, we used a live cell imaging system that was coupled to a 365-nm laser micro-irradiation device (Uematsu et al, 2007). This system allowed us to generate DSBs in small defined regions in the cell nucleus and to monitor the spatio–temporal characteristics of protein recruitment to those DSBs. We expressed yellow fluorescent protein (YFP)-XLF in human 1BR3 cells. We confirmed that the protein level of exogenously expressed YFP-XLF was comparable to the endogenous XLF and that YFP tagging did not interfere with the protein interaction of XLF with XRCC4 (supplementary Fig S1 online). DSBs were introduced into the nucleus of these cells using 365-nm laser micro-irradiation and resulted in the immediate accumulation of YFP-XLF at the micro-irradiated region (Fig 1A). A clear and defined focus of YFP-XLF was visible in a few seconds after micro-irradiation (Fig 1B), indicating that XLF, similar to Ku (Mari et al, 2006) and DNA-PKCS (Uematsu et al, 2007), is rapidly recruited to DSBs in vivo. We measured the accumulation of YFP-XRCC4 under identical experimental conditions. The accumulation of YFP-XRCC4 at DSBs was weaker than XLF (supplementary Fig S2B online), but the kinetics of XLF and XRCC4 accumulation were similar (supplementary Fig S2C online).
Figure 1.
Accumulation of XLF at laser-induced DNA double-strand breaks. (A) Accumulation of YFP-XLF at DSBs. Human 1BR3 cells expressing YFP-XLF were micro-irradiated across the nuclei. A typical example of the situation before and after (1 min) irradiation is shown (left). For kinetics analysis, the cell nucleus was exposed to a single burst of the laser, and images were obtained at 10 s intervals for 2 min. XLF accumulation at DSB sites was quantified and depicted in a graph (right). Mean values of the fluorescence intensities at each time point were calculated from ten independent measurements. (B) Rapid accumulation of YFP-XLF at DSBs. Images were taken at 2 s intervals. A typical example (left) and mean fluorescence intensities of ten independent measurements (right) are shown as in (A). The arrow indicates the site of micro-irradiation. (C) Colocalization of endogenous XLF protein and DSBs. Human 1BR3 cells were micro-irradiated and fixed. DSBs were labelled using TUNEL staining (red). Subsequently, the cells were stained with an XLF antibody (green). (D) Colocalization of XLF and γH2AX. 1BR3 cells were micro-irradiated, fixed and co-immunostained with XLF (green) and γH2AX (red) antibodies. DSB, double-strand break; TUNEL, TdT-mediated dUTP nick end labelling; XLF, XRCC4-like factor; YFP, yellow fluorescent protein; γH2AX, phosphorylated form of histone H2AX.
Next, the accumulation of endogenous XLF protein at laser-induced DSBs was verified. 1BR3 cells were micro-irradiated, fixed and stained with an XLF antibody. As shown in Fig 1C, endogenous XLF was enriched at DSB sites in the micro-irradiated region. The presence of DSBs was confirmed by TdT-mediated dUTP nick end labelling staining. XLF colocalized with a phosphorylated form of histone H2AX (γH2AX), an established marker for DSBs (Thiriet & Hayes, 2005; Fig 1D). Specificity of the XLF antibody was verified using XLF RNA interference, followed by immunostaining (supplementary Fig S3 online).
XRCC4 stabilizes XLF at DSB sites
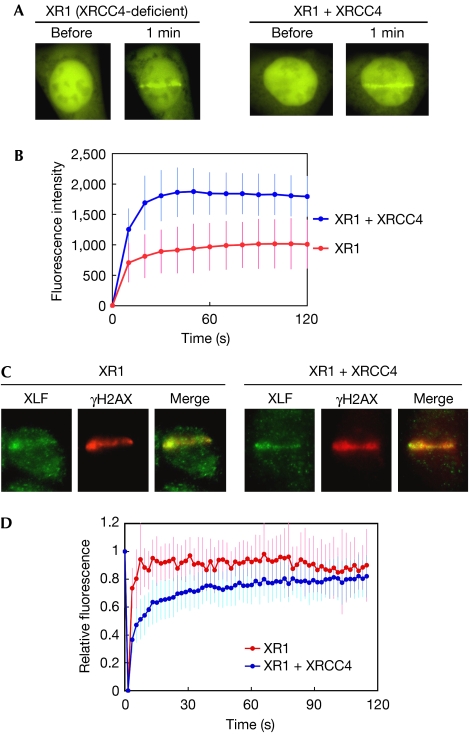
As XLF was originally identified as an XRCC4-interacting protein (Ahnesorg et al, 2006), we tested whether XRCC4 is required for XLF recruitment to DSBs. YFP-XLF was expressed in an XRCC4-deficient cell (XR1), in which XRCC4 alleles were completely deleted (Li et al, 1995), and in XRCC4-complemented XR1 cells (supplementary Fig S4 online). Recruitment of YFP-XLF to DSBs was monitored by using live cell imaging. Unexpectedly, YFP-XLF accumulated at the damaged sites in both XR1 and XRCC4-complemented XR1 cells (Fig 2A). Quantification of the fluorescence intensities showed that the signal intensity at DSBs in XR1 cells was significantly lower than that of the complemented XR1 (Fig 2B), although YFP-XLF was expressed at equal levels in both cells. These observations indicate that XRCC4 is dispensable for the recruitment of XLF to DSBs, but that the presence of XRCC4 increases the overall number of XLF molecules recruited to DSBs. We carried out immunostaining of the endogenous XLF in both micro-irradiated and complemented XR1 cells, and confirmed that the accumulation of the endogenous XLF occurs irrespective of the presence or absence of XRCC4 (Fig 2C).
Figure 2.
Dynamics of XLF in XRCC4-deficient and XRCC4-complemented cells. (A) Accumulation of YFP-XLF at DSBs. YFP-XLF was expressed in XRCC4-deficient XR1 (left) and XRCC4-complemented XR1 (right) cells. DSBs were generated by micro-irradiation and XLF accumulation was monitored before and 1 min after irradiation. (B) Kinetics of YFP-XLF accumulation at DSBs. Accumulation of YFP-XLF was measured as in Fig 1B. Mean values of the fluorescence intensities at each time point were calculated from ten independent measurements. (C) Accumulation of the endogenous XLF at DSBs in XR1 (left) and XRCC4-complemented (right) cells. Cells were micro-irradiated in their nucleus, fixed and stained with XLF and γH2AX antibodies. (D) FRAP analysis of YFP-XLF in XR1 and XRCC4-complemented XR1 cells. After maximum XLF accumulation, the whole DSB region was photobleached. Images were acquired before bleaching and at 1.5 s intervals after bleaching. Mean values of the fluorescence intensities at each time point were calculated from ten independent measurements. DSB, double-strand break; FRAP, fluorescence recovery after photobleaching; XLF, XRCC4-like factor; YFP, yellow fluorescent protein; γH2AX, phosphorylated form of histone H2AX.
We subsequently studied the dynamics of DSB-recruited XLF by using fluorescence recovery after photobleaching (FRAP) (Lippincott-Schwartz et al, 2001). We micro-irradiated the nucleus of XR1 and complemented XR1 cells and incubated the irradiated cells for 10 min to allow XLF accumulation at DSBs to reach a steady state. We then photobleached the XLF accumulation area and monitored recovery of the fluorescence signals. Recovery of the fluorescence in both cells was rapid (Fig 2D), indicating a highly dynamic exchange between bound and free XLF molecules at DSBs. The recovery dynamics, however, differed significantly between XR1 and XRCC4-complemented cells. During the first 30 s, XLF exchange was markedly slower in XRCC4-complemented cells than in XR1 cells (Fig 2D); only after approximately 100 s, did the recovery in both cell lines reach similar levels. These data show that XRCC4 decreases the exchange rate of XLF at DSBs. We conclude that XRCC4 is not required for XLF recruitment but has a role in the stabilization of recruited XLF at DSBs, and that such XRCC4-mediated stabilization of XLF at DSBs could account for the higher level of overall XLF accumulation observed in XRCC4-proficient cells.
Ku recruits XLF to DSBs
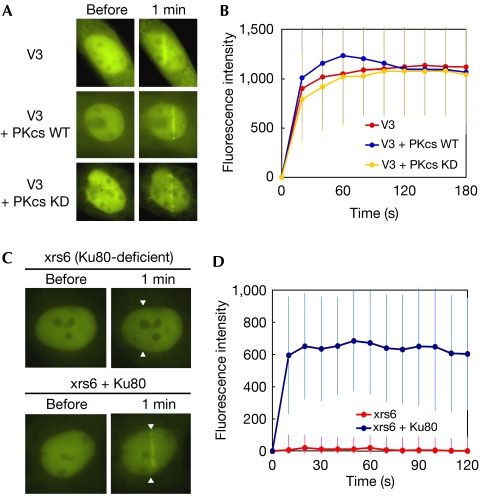
Next, we examined whether DNA-PK components are required for XLF recruitment to DSBs. First, we micro-irradiated the nuclei of DNA-PKCS-deficient (V3) cells and V3 cells complemented with wild-type or kinase-dead DNA-PKCS. In these cell lines, wild-type and kinase-dead DNA-PKCS were expressed at comparable levels (Uematsu et al, 2007). We observed clear and rapid YFP-XLF accumulation at the laser-induced damage site, with no distinguishable differences in the accumulation kinetics of the three cell lines (Fig 3A,B). This result indicates that DNA-PKCS is not involved in the recruitment process of XLF.
Figure 3.
XLF accumulation in DNA-PKCS-deficient and Ku80-deficient cells. (A) Accumulation of YFP-XLF at DSBs in DNA-PKCS-deficient (V3) cells (top), V3 cells complemented wild-type DNA-PKCS (middle) and V3 cells complemented with kinase-dead DNA-PKCS (bottom). Live cell imaging was carried out as described in Fig 1A. (B) Quantitative analysis of YFP-XLF accumulation at DSBs in DNA-PKCS-deficient and DNA-PKCS-complemented cells. Signal intensities were quantified and a mean value for each time point was calculated from ten independent measurements. (C) Accumulation of YFP-XLF at DSBs in Ku80-deficient xrs6 cells (top) and the Ku80-complemented xrs6 cells (bottom). Arrowheads indicate the sites of micro-irradiation. (D) Quantitative analysis of YFP-XLF accumulation at DSBs in xrs6 and Ku80-complemented xrs6. Images were obtained at 10 s intervals for 2 min. Signal intensities were quantified and a mean value with standard deviation at each time point was calculated from ten independent measurements. DNA-PKCS, DNA-dependent protein kinase catalytic subunit; DSB, double-strand break; KD, kinase dead; WT, wild type; XLF, XRCC4-like factor; YFP, yellow fluorescent protein.
Then we micro-irradiated the nuclei of Ku80-deficient (xrs6) and Ku80-complemented xrs6 cells. XLF did not accumulate at the micro-irradiated region in xrs6 cells, whereas rapid accumulation of XLF was observed in the Ku80-complemented xrs6 cells (Fig 3C,D). These results show that Ku is essential for the recruitment of XLF to DSBs.
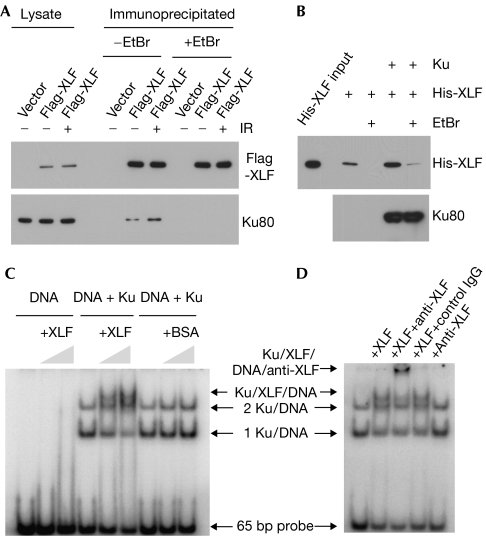
To obtain further insights into the role of Ku in XLF recruitment, we carried out protein interaction assays. First, we examined whether Ku co-precipitates with XLF from human cell lysates. We expressed Flag-tagged XLF in human embryonic kidney (HEK) 293 cells and then immunoprecipitated Flag-XLF using a Flag antibody. As shown in Fig 4A, we observed co-precipitation of Ku with Flag-XLF. Treatment of the cells with γ-rays increased co-precipitation of Ku with XLF. However, this Ku–XLF association was sensitive to ethidium bromide, which intercalates into DNA strands and interferes with DNA–protein interactions (Lai & Herr, 1992). Therefore, our results indicate that the observed co-precipitation of Ku with XLF was mediated by DNA molecules in the lysates.
Figure 4.
Complex formation of XLF and Ku on DNA. (A) Ethidium bromide-sensitive co-precipitation of Ku with Flag-XLF. 1BR3 cells were transfected with Flag-XLF and treated with or without γ-rays at 10 Gy. Immunoprecipitation was carried out with a Flag antibody in the presence or absence of ethidium bromide, and the co-precipitation of Ku80 was analysed by western blotting. (B) Ku facilitates the XLF–DNA association. DNA-cellulose was reacted with or without purified Ku. The purified XLF was then incubated with DNA-cellulose with or without Ku. Ethidium bromide was added to the reactions as indicated. After washing, proteins bound on DNA-cellulose were analysed by western blotting. (C) EMSA of XLF and Ku. A 32P-labeled 65-bp DNA probe was reacted with purified Ku, XLF and BSA as indicated. Protein–DNA complexes were resolved by polyacrylamide gel electrophoresis and subsequently visualized by autoradiography. (D) Supershift assay using an XLF antibody. Ku and DNA probe were reacted with XLF, anti-XLF and control antibodies as indicated. BSA, bovine serum albumin; EtBr, ethidium bromide; DSB, double-strand break; EMSA, electrophoretic mobility shift assay; XLF, XRCC4-like factor; YFP, yellow fluorescent protein.
The ethidium bromide-sensitive association of XLF and Ku prompted us to explore the possibility that the Ku–XLF interaction occurs on DNA. We mixed purified XLF with DNA-cellulose beads in the presence or absence of purified Ku, and then measured the amount of XLF that bound to the DNA-cellulose (Fig 4B). When XLF alone was mixed with DNA-cellulose, we observed only minor association of XLF with DNA-cellulose, and this XLF–DNA association was sensitive to ethidium bromide. These results indicate that the XLF–DNA association is relatively weak in the absence of Ku. However, when Ku was prebound to DNA-cellulose and subsequently incubated with XLF, we observed a marked increase in XLF–DNA association that showed slight resistance to ethidium bromide. These results show that Ku facilitates the XLF–DNA association.
To study further the influence of Ku on the XLF–DNA interaction, we carried out electrophoretic mobility shift assays (EMSA) to identify XLF–Ku-DNA species. XLF–DNA association has been reported to be DNA length dependent. Purified XLF binds to 83-bp or longer DNA fragments, but fails to form a stable complex with shorter DNA (Lu et al, 2007). We confirmed that our purified XLF protein binds to a 100-bp DNA probe but not to a 65-bp probe (supplementary Fig S5B online), which is consistent with the previous observations. Ku forms a tight complex with DNA as short as 14 bp in length (Yoo et al, 1999). We incubated Ku with a 65-bp DNA probe and observed two discrete Ku–DNA species using EMSA (Fig 4C). When Ku and XLF were incubated together with the DNA probe, the protein–DNA species migrated more slowly than the Ku–DNA species using EMSA. The addition of an XLF antibody caused a supershift of the XLF–Ku-DNA species (Fig 4D). These results show that the presence of Ku abolished the length dependency of the XLF–DNA association. We showed that highly purified XLF and Ku do not form a stable complex in the absence of DNA (supplementary Fig S5C online). Together, we conclude that XLF interacts with Ku on DNA.
Discussion
Our study shows that XLF is recruited to DSBs in the initial phase of the NHEJ process, as shown by the fact that XLF accumulation at DSBs occurred just as rapidly as Ku and DNA-PKCS (Mari et al, 2006; Uematsu et al, 2007). Our finding that XLF recruitment to DSBs is fully dependent on the presence of Ku gives rise to a model in which Ku has a crucial role in the assembly of the early NHEJ complex, acting as a scaffold for the recruitment of other NHEJ core factors. Studies of DNA-PKCS (Uematsu et al, 2007), XRCC4 (Mari et al, 2006) and XLF in NHEJ-deficient cell lines indicate that the recruitment of these core factors to DSBs is independent of each other. A recent study of biochemical fractionation indicates that XLF deficiency does not affect the migration of XRCC4 and LigIV into a detergent-resistant nuclear fraction upon DNA damage (Wu et al, 2007). It will be interesting to study the precise roles of XLF in the dynamics of other NHEJ factors in vivo.
XRCC4 is dispensable for XLF recruitment to DSBs but stabilizes the recruited XLF. XRCC4 is known to interact with DNA-PKCS (Hsu et al, 2002) and XLF (Ahnesorg et al, 2006; Callebaut et al, 2006). Although we have previously reported a very weak to absent XRCC4–Ku interaction, Mari et al (2006) have shown an interaction of both proteins on chemical crosslinking, indicating a transient interaction with a putative role in the NHEJ complex. The many protein interactions of XRCC4 could act to tether XLF to the NHEJ machinery, reduce the turnover of XLF and consequently increase the amount of XLF at DSBs. Our finding that XRCC4 stabilizes the association of XLF with DSBs indicates that the correct assembly of the whole repair complex stabilizes the individual components of the complex. Calsou et al (2003) showed that the stable association of XRCC4–LigIV to DNA ends requires DNA-PKCS. Recently, Wu et al (2007) showed by cellular fractionation that XLF exists in a detergent-resistant chromatin fraction after treatment of DSB-inducing agents and that knockdown of XRCC4, LigIV or DNA-PKCS decreases the amount of XLF in the insoluble fraction of DSB-induced cells. These observations support the idea that after recruitment to DSBs, NHEJ factors at DSBs stabilize each other.
XLF recruitment was exclusively Ku-dependent, but DNA-PKCS was dispensable for XLF recruitment, although DNA-PK phosphorylates XLF (Wu et al, 2007). Similar DNA-PKCS-independent recruitment to DSBs was reported for XRCC4 (Mari et al, 2006). We showed previously that fully functional DNA-PK is required for efficient NHEJ (Kurimasa et al, 1999), and that kinase activity of DNA-PKCS is dispensable for the recruitment of DNA-PKCS but essential for the efficient progress of NHEJ repair (Uematsu et al, 2007). These observations indicate that one of the main functions of DNA-PK is the promotion of NHEJ reactions after protein recruitment to DSBs. In light of this, it will be interesting to test whether DNA-PK modulates XLF functions, such as stimulation of LigIV activity, through phosphorylation.
Previous research using purified XLF protein documented that XLF preferentially binds to longer DNA molecules, indicating that the observed XLF–DNA interaction in vitro occurs on DNA strands rather than at DNA ends (Lu et al, 2007). This raised questions as to how XLF specifically recognizes DSBs in vivo. Here, we have shown that the presence of Ku abolishes the size limitation of XLF–DNA binding, indicating that Ku is directly responsible for the specific recruitment of XLF to DSBs, or at least alters the DNA-binding characteristics of XLF. Knowing the architecture of the Ku–DNA–XLF complex might be crucial to understanding the molecular mechanism of DNA damage recognition in NHEJ.
In conclusion, our data shows the direct involvement of XLF in the early phases of the assembly of the NHEJ machinery through the interaction with Ku on DNA. In view of our findings and the known ligation-stimulatory activity of XLF, it seems likely that XLF functions to link DSB recognition to ligation steps, rather than acting as a simple auxiliary XRCC4-associating factor. We suggest a new model in which the recruitment of DNA-PKCS, XRCC4 and XLF to DSBs is independent of each other and the recruited factors are assembled into a stable complex by several protein–protein interactions. Further study of the XLF functions during protein assembly and NHEJ reactions will help to determine the molecular details of the NHEJ pathway.
Methods
Live cell imaging and laser micro-irradiation. Live cell imaging combined with laser micro-irradiation was carried out as described previously (Uematsu et al, 2007). Briefly, YFP-XLF was expressed by transfection using FuGENE 6 (Roche, Mannheim, Germany). Fluorescence in a living cell was monitored by using an Axiovert 200M microscope (Carl Zeiss MicroImaging, Thornwood, NY, USA). A 365-nm pulsed nitrogen laser (Spectra-Physics, Mountain View, CA, USA) was directly coupled to the epifluorescence path of the microscope. DSBs were generated in a defined area of the nucleus by micro-irradiation with the 365-nm laser. For quantitative analyses, we used standardized irradiation conditions (80% laser output at 10 Hz for 400 ms) to generate the same amount of DSBs in each experiment. Time-lapse images were taken by an AxioCam HRm camera and fluorescence intensities of micro-irradiated or non-irradiated within the cell nucleus were determined using the Axiovison Software, version 4.5 (Carl Zeiss). All measurements were corrected for nonspecific bleaching during monitoring.
FRAP analysis. FRAP analysis was carried out on an LSM 510 Meta confocal microscope, as described previously (Uematsu et al, 2007). Briefly, cells expressing YFP-XLF were micro-irradiated with the 365-nm laser as described above. After 10 min incubation at 37°C to allow maximum accumulation of YFP-XLF at the DSB sites, the whole DSB site was photobleached with a pulse of 514-nm argon laser. Time-lapse imaging before and after photobleaching and fluorescence quantification were carried out as described above. All measurements were corrected for nonspecific monitor bleaching.
Other materials and experimental procedures are described in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary information
Acknowledgments
We thank B.P.C. Chen for providing the V3 cell lines, and S. Burma, A. Davis and L. Ortega for their critical reading of the manuscript. This work was supported by grants from the National Institutes of Health (CA50519 and PO1-CA92584) to D.J.C.
References
- Ahnesorg P, Smith P, Jackson SP (2006) XLF interacts with the XRCC4–DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 124: 301–313 [DOI] [PubMed] [Google Scholar]
- Bryans M, Valenzano MC, Stamato TD (1999) Absence of DNA ligase IV protein in XR-1 cells: evidence for stabilization by XRCC4. Mutat Res 433: 53–58 [DOI] [PubMed] [Google Scholar]
- Buck D et al. (2006) Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124: 287–299 [DOI] [PubMed] [Google Scholar]
- Callebaut I, Malivert L, Fischer A, Mornon JP, Revy P, de Villartay JP (2006) Cernunnos interacts with the XRCC4 x DNA–ligase IV complex and is homologous to the yeast nonhomologous end-joining factor Nej1. J Biol Chem 281: 13857–13860 [DOI] [PubMed] [Google Scholar]
- Calsou P, Delteil C, Frit P, Drouet J, Salles B (2003) Coordinated assembly of Ku and p460 subunits of the DNA-dependent protein kinase on DNA ends is necessary for XRCC4–ligase IV recruitment. J Mol Biol 326: 93–103 [DOI] [PubMed] [Google Scholar]
- Cary RB, Peterson SR, Wang J, Bear DG, Bradbury EM, Chen DJ (1997) DNA looping by Ku and the DNA-dependent protein kinase. Proc Natl Acad Sci USA 94: 4267–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y et al. (2003) Nonhomologous end joining and V(D)J recombination require an additional factor. Proc Natl Acad Sci USA 100: 2462–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentges P et al. (2006) Evolutionary and functional conservation of the DNA non-homologous end-joining protein, XLF/Cernunnos. J Biol Chem 281: 37517–37526 [DOI] [PubMed] [Google Scholar]
- Hsu HL, Yannone SM, Chen DJ (2002) Defining interactions between DNA-PK and ligase IV/XRCC4. DNA Repair 1: 225–235 [DOI] [PubMed] [Google Scholar]
- Kienker LJ, Shin EK, Meek K (2000) Both V(D)J recombination and radioresistance require DNA-PK kinase activity, though minimal levels suffice for V(D)J recombination. Nucleic Acids Res 28: 2752–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimasa A, Kumano S, Boubnov NV, Story MD, Tung CS, Peterson SR, Chen DJ (1999) Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol Cell Biol 19: 3877–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JS, Herr W (1992) Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA 89: 6958–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller SP, Meek K (2003) Repair of DNA double strand breaks by non-homologous end joining. Biochimie 85: 1161–1173 [DOI] [PubMed] [Google Scholar]
- Li Z et al. (1995) The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell 83: 1079–1089 [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Snapp E, Kenworthy A (2001) Studying protein dynamics in living cells. Nat Rev Mol Cell Biol 2: 444–456 [DOI] [PubMed] [Google Scholar]
- Lu H, Pannicke U, Schwarz K, Lieber MR (2007) Length-dependent binding of human XLF to DNA and stimulation of XRCC4.DNA ligase IV activity. J Biol Chem 282: 11155–11162 [DOI] [PubMed] [Google Scholar]
- Mari PO et al. (2006) Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci USA 103: 18597–18602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi JM, Ferguson DO (2006) DNA double-strand break repair: a relentless hunt uncovers new prey. Cell 124: 260–262 [DOI] [PubMed] [Google Scholar]
- Smith GC, Jackson SP (1999) The DNA-dependent protein kinase. Genes Dev 13: 916–934 [DOI] [PubMed] [Google Scholar]
- Thiriet C, Hayes JJ (2005) Chromatin in need of a fix: phosphorylation of H2AX connects chromatin to DNA repair. Mol Cell 18: 617–622 [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Kim SA, Chu G (2007) Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc Natl Acad Sci USA 104: 7851–7856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu N et al. (2007) Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol 177: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings E, van Gent DC (2004) The mechanism of non-homologous end-joining: a synopsis of synapsis. DNA Repair 3: 1425–1435 [DOI] [PubMed] [Google Scholar]
- Wu PY, Frit P, Malivert L, Revy P, Biard D, Salles B, Calsou P (2007) Interplay between Cernunnos-XLF and nonhomologous end-joining proteins at DNA ends in the cell. J Biol Chem 282: 31937–31943 [DOI] [PubMed] [Google Scholar]
- Yoo S, Kimzey A, Dynan WS (1999) Photocross-linking of an oriented DNA repair complex. Ku bound at a single DNA end. J Biol Chem 274: 20034–20039 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary information