Abstract
The Groucho (Gro)/transducin-like enhancer of split family of transcriptional corepressors are implicated in many signalling pathways that are important in development and disease, including those mediated by Notch, Wnt and Hedgehog. Here, we describe a genetic screen in Drosophila that yielded 50 new gro alleles, including the first protein-null allele, and has two mutations in the conserved Q oligomerization domain that have been proposed to have an essential role in corepressor activity. One of these latter mutations, encoding an amino-terminal protein truncation that lacks part of the Q domain, abolishes oligomerization in vitro and renders the protein unstable in vivo. Nevertheless, the mutation is not a null: maternal mutant embryos have intermediate segmentation phenotypes and relatively normal terminal patterning suggesting that the mutant protein retains partial corepressor activity. Our results show that homo-oligomerization of Gro is not obligatory for its action in vivo, and that Gro represses transcription through more than one molecular mechanism.
Keywords: Drosophila, Groucho, repression, segmentation, transcription
Introduction
Groucho (Gro)/transducin-like enhancer of split (TLE) proteins are recruited to act as corepressors for many different families of transcription factors, including RUNX, Nkx, Hes and LEF1/TCF, and are crucial in many developmental signalling pathways, including Notch and Wnt (Chen & Courey, 2000; Gasperowicz & Otto, 2005). Gro is required in Drosophila development for many processes, including terminal patterning, segmentation, sex determination, dorsal–ventral patterning and neurogenesis (reviewed by Chen & Courey, 2000). In vertebrates, Gro/TLE proteins participate in a diverse range of processes, including neurogenesis, osteogenesis and haematopoiesis (reviewed by Gasperowicz & Otto, 2005), and have been implicated as oncogenes in lung cancer (Allen et al, 2006).
Gro/TLE proteins contain five discernible domains: Q (oligomerization and protein interactions), GP (repression), CcN (nuclear localization), SP (repression) and WD (protein interactions, repression), of which the first and last are the most highly conserved (Stifani et al, 1992; Chen & Courey, 2000). The WD domain folds to form a β-propeller (Pickles et al, 2002), which mediates protein–protein interactions with several repressors, including Hairy (through a carboxy-terminal WRPW peptide motif) and Engrailed (through the ‘eh1' motif; Paroush et al, 1994; Fisher et al, 1996; Jimenez et al, 1997; Jennings et al, 2006).
Sequences within the glutamine-rich Q domain are predicted to form two amphipathic α-helical motifs—AH1 and AH2—that facilitate oligomerization of Gro/TLE molecules into tetramers in vitro (Pinto & Lobe, 1996; Chen et al, 1998; Song et al, 2004). Previous studies have indicated that oligomerization of the Q domain is required for Gro to function as a corepressor in cultured cells, and for ectopic Gro to cause developmental defects in the fly (Song et al, 2004). The Q domain also interacts with several repressors, including LEF1/TCF during Wnt signalling (Daniels & Weis, 2005).
Here, we describe the recovery and characterization of many new Drosophila gro alleles, including a null allele and two mutations that disrupt the Q domain. One of the latter mutations—groMB12—leads to an amino-terminal truncation that abolishes oligomerization. GroMB12 protein retains significant activity in vivo, indicating that homo-oligomerization of Gro is not essential for all its corepressor activities.
Results And Discussion
Isolation of new gro alleles
The Hairy basic helix–loop–helix (bHLH) protein is a transcriptional repressor that depends on maternally contributed Gro to establish spatial periodicity during Drosophila embryogenesis. Hairy is expressed in stripes in the unicellular blastoderm embryo in which it, in turn, establishes complementary stripes of one of its target genes, fushi tarazu (ftz; Howard & Ingham, 1986). Hairy also contributes to progressive retinal differentiation in the imaginal disc of the eye (Brown et al, 1991).
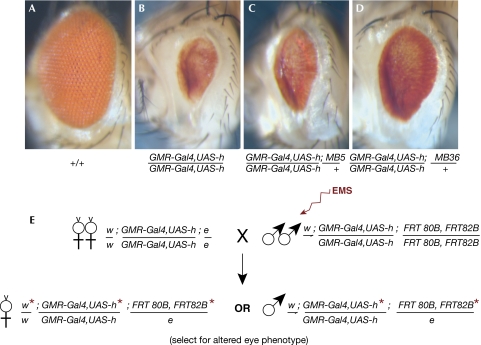
To identify new factors that interact with Hairy in vivo, we established a genetic screen for mutations that modulate the effects of ectopic Hairy expression in the developing eye (Fig 1). Driving Hairy expression in cells posterior to the morphogenetic furrow (GMR-GAL4;UAS-hairy; Hay et al, 1994) led to a marked reduction in the size of the eye and some loss of pigmentation (Fig 1B). A pilot screen for ethyl methane sulphonate-induced modifier mutations showed that a high proportion of the suppressor mutations on chromosome 3 were allelic to gro (supplementary information online).
Figure 1.
Genetic screen for modifiers of ectopic hairy expression in the eye of Drosophila. (A–D) Light photomicrographs showing dominant suppression of the eye phenotype by gro alleles; weak for MB5 and strong for the null allele MB36. Genotypes are as indicated below. (E) Schematic representation of the screening strategy. F1 progeny homozygous for GMR-Gal4,UAS-hairy and heterozygous for a mutagenized FRT 80B FRT 82B chromosome were scored for the suppression of the eye phenotype. e, ebony; EMS, ethyl methane sulphonate; Gro, Groucho; h, hairy; w, white.
From approximately 137,000 mutated chromosomes, we recovered 62 suppressors that mapped to chromosome 3, of which 50 were new gro alleles. Single-stranded conformation polymorphism analysis and DNA sequencing showed specific molecular lesions in 22 alleles (Table 1; supplementary information online). Most of the gro mutations represent nonsense or frameshift alleles encoding truncated Gro proteins.
Table 1.
groucho alleles for which molecular data were obtainable
| Type of lesion | Allele | Base change* | RNA/protein changes |
|---|---|---|---|
| Nonsense | |||
| MBB | G4624A | W550stop | |
| MB6 | C2315T | R190stop | |
| MB11 | C4769T | Q599stop | |
| MB25 | C4361T | Q463stop, as MB27 | |
| MB27 | C4361T | Q463stop, as MB25 | |
| MB34 | G4749A | W592stop | |
| MB42 | G4716A | W581stop | |
| Frameshift | |||
| MB13 | G2485A | Splice junction mutated, frameshift from residue 225 as intronic sequences transcribed, as MB37 | |
| MB15 | G138A | Ectopic splice acceptor site in intron, frameshift from residue 13, as MB36 | |
| MB18 | G266A | Splice junction mutated, frameshift from residue 45 as intronic sequences transcribed | |
| MB33 | Δ3782–3813 | 31 bp deletion; frameshift from residue 103 | |
| MB36 | G138A | as MB15 | |
| MB37 | G2485A | as MB13 | |
| Other | |||
| MB5 | T189C | P19S, Δ19–27 | |
| C190T | See Fig 2B for sequence | ||
| Δ191–214 | |||
| MB12 | G3A | Initiator ATG → ATA | |
| Missense† | |||
| MBD | A4613T | I547F | |
| MB19 | G4683A | C570Y | |
| MB21 | G4422A | R483H, as MB41 | |
| MB30 | A4977C | H646P | |
| MB31 | C5176T | L692F | |
| MB35 | G4326A | G451D | |
| MB41 | G4422A | R483H, as MB21 | |
| *Bases numbered from the start of translation, corresponding to 3R:21869066 Drosophila melanogaster genome Release 5.1. | |||
| †For characterization of missense alleles, refer to Jennings et al (2006). | |||
Seven of the gro mutations are missense mutations that largely abolish Gro activity and all lie within the WD domain (Jennings et al, 2006). The intensive screen led to the recovery of independent, identical mutations at four different sites (Table 1). Nevertheless, no missense mutations were recovered in the central GP, CcN or SP domains of Gro, suggesting that these domains are functionally dispensable in vivo or represent very small mutagenic targets.
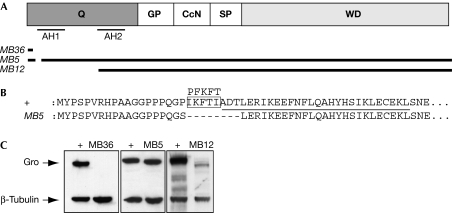
We also recovered a protein-null allele—groMB36—and two alleles containing deletions within the Q domain: groMB5 contains a small in-frame deletion, whereas groMB12 is a single-base-pair substitution in the initiator codon that encodes an N-terminal truncated protein. We describe these in more detail below.
groMB36 is a protein-null allele
MB36, and the independently recovered MB15 allele, represents a G–A mutation at position 101 in the first intron that generates an ectopic splice acceptor site. Sequencing MB36 complementary DNA shows that they include 31 bp of intronic sequences and thereby encode only the first 12 amino acids of Gro, with the following 104 amino acids being derived from intronic and frameshifted sequences (Table 1; Fig 2A). MB36 lacks all conserved Gro protein domains and should be inactive and probably unstable. No Gro protein was detected in MB36 embryos (embryos that were derived from homozygous MB36 germ cells thus contain only mutant protein; Fig 2C), although this might be because the truncated protein lacks an appropriate epitope. MB36 transcripts are indeed mis-spliced and underrepresented fivefold, presumably owing to nonsense-mediated messenger RNA decay (supplementary information online). We conclude that MB36 is a null allele of gro.
Figure 2.
Deletions recovered in gro. (A) Diagram of the domain structure of Drosophila Gro. The two predicted amphipathic α-helices in the Q domain (AH1 and AH2) are indicated. The coding regions of MB36, MB5 and MB12 transcripts are also shown. (B) Alignment of the amino-acid sequences of the mutated region in MB5 to the corresponding wild-type region (+). AH1 is underlined; amino-acid residues deleted in MB5 that lie outside AH1 are boxed with the corresponding sequence from hTLE1 shown above. (C) Western blots of extracts from gro mutant embryos (from gro mothers) run on SDS–polyacrylamide gel electrophoresis gels. MB36 is not detected and levels of MB12 are reduced. β-Tubulin is shown as a loading control. Gro, Groucho.
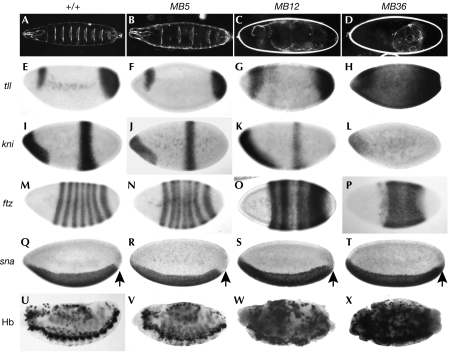
Maternal MB36 embryos show severe cuticle loss owing to ectopic neurogenesis at the expense of epidermal development (Fig 3D). This ‘neurogenic' phenotype is associated with the inability of the E(spl) bHLHs, targets of Notch signalling, to repress proneural transcription in the absence of Gro (Fig 3X; Paroush et al, 1994). Other patterning events are also affected in MB36 embryos such that their phenotype resembles that previously described for groE48 embryos (see below).
Figure 3.
Phenotypic analysis of gro alleles. (A–D) Cuticles of wild-type (+) and gro mutant embryos. MB5 embryos show a weaker phenotype (loss of two or three even-numbered denticle belts) than MB12 or MB36. MB12 embryos are neurogenic but retain more body mass than MB36. Expression of (E–H) tll, (I–L) kni and (M–P) ftz transcripts in blastoderm embryos, showing that MB5 and MB12 retain activity. tll expression is wild type in MB5, slightly expanded in MB12 and throughout the embryo in MB36, indicating residual activity in MB12. Expression of the central stripe of kni is reduced in most MB12 embryos, but not completely abolished as in MB36. Stripes of ftz expression are irregular in MB5, broadened in MB12 and completely fused in MB36. (Q–T) Expression of sna transcripts is expanded towards the posterior pole in MB12 or MB36 embryos (indicated by arrows) in a manner resembling the sna expression pattern in hkb mutants (Goldstein et al, 1999), indicating that repression mediated by Hkb is lost in MB12 embryos. (U–X) Staining of stage 13 wild type and gro for the neural marker Hb. MB12 and MB36 mutants show ectopic expression of Hb protein that is characteristic of excessive neural development. ftz, fushi tarazu; Gro, Groucho; Hb, Hunchback; Hkb, Huckebein; kni, knirps; sna, snail; tll, tailless.
Q domain mutations retain partial function
The two new gro alleles with lesions within the Q domain allowed us to study the roles of this domain in vivo. MB5 includes a complex deletion and inversion that leads to an 8-amino-acid deletion/1-amino-acid substitution lacking the first four amino acids of the AH1 helix (Table 1; Fig 2A,B). MB5 embryos show loss of alternate ventral denticle bands, resembling hairy mutant embryos (Fig 3B; Howard & Ingham, 1986). However, the segmentation phenotype is weak: usually only 2–3 even-numbered denticle belts are missing (0% wild type, 5% missing one belt, 27% missing two belts, 52% missing three belts, 16% missing more than three belts; n=110), and expression of ftz is only slightly perturbed (Fig 3N). MB5 embryos seem to be otherwise normal (Fig 3F,J,R,V; data not shown); however, the lesion must disrupt an essential Gro activity because homozygous and hemizygous adults were never recovered.
The second Q domain mutation, MB12, is a single-base-pair substitution in the initiator ATG codon. MB12 protein migrates slightly faster than Gro+ on denaturing gels (Fig 2C), indicating that it is an N-terminal truncated protein lacking the complete AH1 amphipathic α-helix, in which translation is initiated at the next in-frame start codon, 58 codons downstream (Fig 2A; Chen et al, 1998; Song et al, 2004).
N-terminal truncation clearly disrupts Gro activity. MB12 embryos have a moderate to strong hypomorphic neurogenic phenotype, as judged by the lack of embryonic cuticle and by ectopic expression of the Hunchback (Hb) neural marker in stage 12 embryos (Fig 3C,W). The mutation abolishes terminal repression of snail (sna) transcription by Huckebein (Hkb), so that sna is expressed ectopically at the termini of the embryo, as in MB36 and E48 embryos (Fig 3S,T; Goldstein et al, 1999).
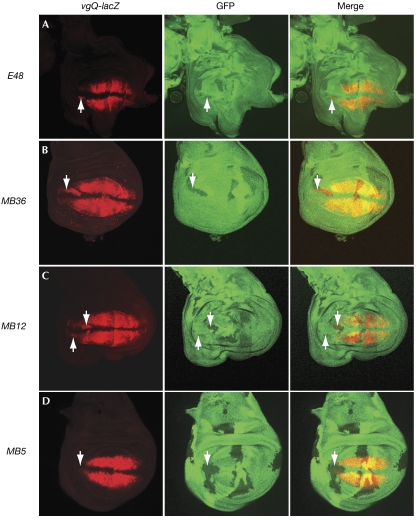
Gro oligomerization has been implicated in the repression of the vgQ-lacZ reporter by Brinker (Brk; Hasson et al, 2001); thus, vgQ-lacZ is inhibited by overexpression of Gro+, but not by the oligomerization-defective mutation GroL38D,L87D (Song et al, 2004). We found that vgQ-lacZ transcription was upregulated in null (MB36) clones at the anterior and posterior edges of the wing pouch, as previously reported for the E48 allele (Fig 4A,B; Hasson et al, 2001), showing that vg is indeed a target of Gro-mediated repression. vgQ-lacZ expression was also upregulated in MB12 clones, but not in MB5 clones (Fig 4C,D), indicating that MB12 is unable to support Gro-mediated repression by Brk in the wing imaginal disk.
Figure 4.
Expression of vgQ-lacZ in gro mutant clones. Third instar imaginal wing disks (anterior to the left and dorsal to the top) stained for GFP and β-galactosidase. Loss of GFP (green) marks clones of homozygous gro mutant cells. Ectopic vgQ-lacZ expression is detected in clones of (A) E48, (B) MB36 and (C) MB12 at the anterior and posterior edges of the wing pouch but not in clones of (D) MB5. Arrows indicate positions of gro clones at the edge of the wing pouch; GFP, green fluorescent protein; Gro, Groucho.
Nevertheless, MB12 maintains repressive activity in some contexts. Mutant embryos retain more body mass than null MB36 embryos (Fig 3C,D), and the broadened ftz stripes retained distinct periodicity, unlike gro null mutant embryos in which all ftz stripes are completely merged (Fig 3O,P; Paroush et al, 1994). More strikingly, Gro-mediated terminal repression is almost normal in MB12 embryos. tailless (tll) expression is restricted to the ends of wild-type embryos by the Gro-dependent repressor Capicua (Cic), thereby allowing expression of knirps (kni) and Krüppel (Kr) in a central domain (Paroush et al, 1997; Jimenez et al, 2000). A total of 93% of the MB12 embryos showed wild-type or only slightly expanded tll expression (n=22; Fig 3G), and most mutant embryos still expressed the gap genes kni (96%; n=45) and Kr centrally, although at reduced levels (Fig 3K; data not shown). By contrast, Cic activity is completely lost in MB36, leading to expanded tll expression and loss of expression of the central domains of kni and Kr (Fig 3H,L; data not shown).
Levels of Gro protein in MB12 embryos are greatly reduced (<5% of wild type; Fig 2C; supplementary information online). MB12 transcripts are translated as efficiently as wild type in vitro (Fig 5A,E,F), indicating that the low in vivo levels of MB12 probably resulted from protein instability. Reduced protein levels might also contribute to the MB12 phenotype, reinforcing the view that the N-terminal truncated protein retains significant WRPW-mediated repressive activity.
Figure 5.
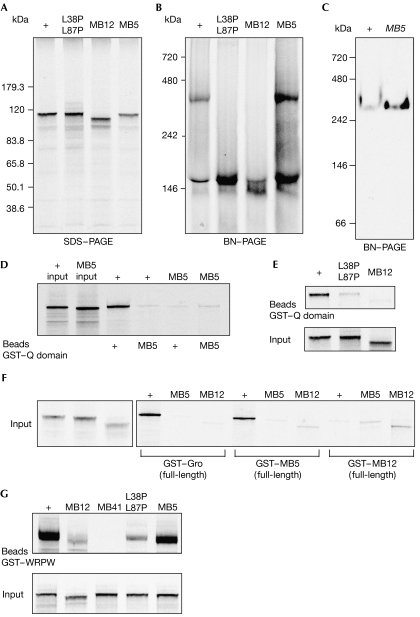
Q-domain-mediated oligomerization of Gro variants. (A) Autoradiograph of 35S-Met-labelled Gro mutant proteins run on SDS–polyacrylamide gel electrophoresis. MB12 migrates slightly faster than wild type (+), which is consistent with an amino-terminal truncation. MB5 and L36P,L87P variants run similar to wild type. (B) Autoradiograph of the translation reactions shown in (A) run on BN-PAGE. MB12 and L36P,L87P do not form a tetrameric complex. (C) Western blot of wild type and MB5 extracts run on BN-PAGE. MB5 forms a high-molecular-weight, tetrameric band. (D,E) Pull-down analysis of 35S-Met-labelled full-length, translated Gro+ and (D) MB5 or (E) L36P,L87P or MB12 protein on GST–wild-type and GST–MB5 Q domains. Neither MB5 nor MB12 binds to an isolated Q domain in this assay. (F) Pull-down analysis of translated Gro+, MB5 and MB12 proteins on GST fusions of full-length Gro+, MB5 and MB12 proteins, showing that immobilized, full-length MB5 retains the ability to bind to translated Gro+. (G) Disruption of oligomerization in two different Q domain mutants, MB12 and L38P,L87P, reduces binding to the WRPW motif. MB41 is a missense mutation in the WD domain that abolishes binding to WRPW (Jennings et al, 2006). Control input tracks represent 10% of labelled Gro protein. BN-PAGE, blue native-polyacrylamide gel electrophoresis; Gro, Groucho; GST, glutathione-S-transferase.
MB12 abolishes Q-domain-mediated oligomerization
The residual activity of MB5 and MB12 suggests that Gro oligomerization is not absolutely required for Gro corepressor activity. We tested whether these mutations are still able to oligomerize by measuring binding of [35S]Gro to glutathione-S-transferase (GST) fusions of full-length Gro or to the Q domain alone (GST–Q). We also used blue-native polyacrylamide gel electrophoresis (BN-PAGE) to visualize oligomerization of translated and embryonic full-length Gro (supplementary information online). This latter assay shows two main bands of in vitro-translated Gro+: a fast-migrating, monomeric band and a slow-migrating band that represents a Gro tetramer as indicated by its molecular weight and its absence from a non-oligomerizing Gro mutant, GroL38P,L87P (Fig 5B; Chen et al, 1998).
These assays show that the short deletion in MB5 reduces but does not abolish Q domain dimerization. GST–QMB5 binds to neither translated Gro+ nor MB5 protein (Fig 5D), and GST–Gro (full-length) is unable to bind to [35S]MB5 (Fig 5F). However, GST–GroMB5 (full-length) still binds to [35S]Gro+ (Fig 5F), showing that the binding activity is not completely lost. Indeed, native gel assays show a band of tetrameric MB5 both after in vitro translation (Fig 5B) and also in vivo, as detected by western blot analysis of extracts from maternal MB5 embryos (Fig 5C).
These results are consistent with previous analysis of Gro oligomerization in vitro, in which mutating an individual amino acid mediating contact between the two AH domains is insufficient to abolish oligomerization (Pinto & Lobe, 1996; Chen et al, 1998; Song et al, 2004). Our failure to find point mutations within the Q domain similarly points to a robustness of the Gro oligomerization interface. The deletion in MB5 removes four evolutionarily conserved amino acids that lie outside helix AH1 (Fig 2B) that might act as an interface for recognition by other proteins. Nevertheless, it remains possible that MB5 lethality is a consequence of reduced Gro oligomerization.
By contrast, removal of AH1 in MB12 completely prevents oligomerization in vitro. Translated MB12 lacks the slow oligomeric band on BN-PAGE (Fig 5B) and does not interact with the wild-type Q domain in a GST pull-down assay (Fig 5E). Oligomerization of MB12 is probably also abolished in vivo, although we could not test this directly because of the greatly reduced levels of MB12 protein in the mutant embryos. An inability to homo-oligomerize probably explains the strong phenotype of MB12, but its residual activities in certain Gro-dependent processes indicate that Gro also has oligomerization-independent activities.
Although it is clear that the WRPW and eh1 repressor peptides bind directly to the WD domain in vivo and in vitro (Paroush et al, 1994; Fisher et al, 1996; Jimenez et al, 1997; Jennings et al, 2006), such binding is also affected by mutations that impair Gro oligomerization (Fig 5G). The MB12 and L38P,L87P variants do not oligomerize (Fig 5B,E), and show greatly reduced binding to the WRPW and eh1 peptides in the GST pull-down assay (Fig 5G; data not shown). Similarly, Song et al (2004) reported reduced binding of GST–Brk, GST–Hkb and GST–Hairy to L38P,L87P. Most probably, these results reflect stronger binding of dimeric GST fusions to oligomerized corepressors, although we cannot exclude the possibility that the Q domain helps to structure the WD domain in solution for optimal peptide binding. Nevertheless, the in vivo relevance of dimeric peptide–Gro interactions remains to be determined. bHLH proteins act as dimers, but there is no current evidence that presentation of two WRPW or eh1 peptide repressor motifs is required for Gro-mediated repression.
Gro represses transcription through multiple mechanisms
In vivo, Gro can mediate ‘dominant' repression, causing the silencing of all linked enhancers to a gene (Barolo & Levine, 1997). Gro has also been described as a ‘long-range' repressor that can inhibit transcriptional initiation while bound to a distant (>1 kb away) enhancer element (Barolo & Levine, 1997). These observations, together with that of Gro oligomerization through the Q domain, have fuelled the predominant ‘spreading' model for Gro function, in which Gro oligomerizes along the DNA through the Q domain and thereby directs heterochromatic silencing and epigenetic changes in chromatin structure (Chen et al, 1998; Chen & Courey, 2000; Song et al, 2004).
However, Gro-dependent repression does not always cause the dominant silencing of linked enhancers within a complex cis-regulatory region (Nibu et al, 2001). Moreover, Gro-mediated repression during animal development is frequently dynamic and rapidly reversible. Striped expression of Drosophila segmentation genes such as hairy and ftz evolves and decays within a period of approximately 30 min (Edgar et al, 1986), serial production of Drosophila embryonic neuroblasts relies on five short pulses of E(spl)-mediated repression that occur within 4 h (Doe, 1992; Jennings et al, 1994; Paroush et al, 1994), and cyclic repression by Hes proteins during zebrafish somitogenesis has a periodicity of 20–30 min (Oates & Ho, 2002).
Although our results indicate that simple Q-domain-directed oligomerization of Gro is not obligatory for its activity in vivo, Song et al (2004) found that the effects of overexpressing Gro in wing imaginal disks depend on oligomerization. Our results can be reconciled if, as seems possible, Gro mediates repression through more than one distinct molecular mechanism, with varying requirements for an intact Q domain according to the different transcription factor complexes assembled at different promoters. Local repression might predominate in dynamic developmental contexts that make use of rapidly reversible transcriptional inhibition.
Methods
Drosophila culture and mutagenesis used standard conditions and protocols. Germ-line clone analysis, in situ hybridization, immunohistochemistry and GST pull-downs were carried out as described previously (Jennings et al, 2006). In vitro protein analysis was carried out using the XCell SureLock gel system (Invitrogen; www.invitrogen.com) with appropriate pre-cast gels and buffers. Blue-native gels were run with 0.1% Coomassie Blue-G250 in the cathode buffer (Schagger & von Jagow, 1991).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank M. Bienz, C. Dillon, C. Hill, C. Rallis and N. Tapon for comments on the manuscript. This work has been supported by Cancer Research UK.
References
- Allen T, van Tuyl M, Iyengar P, Jothy S, Post M, Tsao MS, Lobe CG (2006) Grg1 acts as a lung-specific oncogene in a transgenic mouse model. Cancer Res 66: 1294–1301 [DOI] [PubMed] [Google Scholar]
- Barolo S, Levine M (1997) Hairy mediates dominant repression in the Drosophila embryo. EMBO J 16: 2883–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Sattler CA, Markey DR, Carroll SB (1991) hairy gene function in the Drosophila eye: normal expression is dispensable but ectopic expression alters cell fates. Development 113: 1245–1256 [DOI] [PubMed] [Google Scholar]
- Chen G, Courey AJ (2000) Groucho/TLE family proteins and transcriptional repression. Gene 249: 1–16 [DOI] [PubMed] [Google Scholar]
- Chen G, Nguyen PH, Courey AJ (1998) A role for Groucho tetramerization in transcriptional repression. Mol Cell Biol 18: 7259–7268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, Weis WI (2005) β-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol 12: 364–371 [DOI] [PubMed] [Google Scholar]
- Doe CQ (1992) Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development 116: 855–863 [DOI] [PubMed] [Google Scholar]
- Edgar BA, Weir MP, Schubiger G, Kornberg T (1986) Repression and turnover pattern fushi tarazu RNA in the early Drosophila embryo. Cell 47: 747–754 [DOI] [PubMed] [Google Scholar]
- Fisher AL, Ohsako S, Caudy M (1996) The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein–protein interaction domain. Mol Cell Biol 16: 2670–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperowicz M, Otto F (2005) Mammalian Groucho homologs: redundancy or specificity? J Cell Biochem 95: 670–687 [DOI] [PubMed] [Google Scholar]
- Goldstein RE, Jimenez G, Cook O, Gur D, Paroush Z (1999) Huckebein repressor activity in Drosophila terminal patterning is mediated by Groucho. Development 126: 3747–3755 [DOI] [PubMed] [Google Scholar]
- Hasson P, Muller B, Basler K, Paroush Z (2001) Brinker requires two corepressors for maximal and versatile repression in Dpp signalling. EMBO J 20: 5725–5736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM (1994) Expression of baculovirus P35 prevents cell death in Drosophila. Development 120: 2121–2129 [DOI] [PubMed] [Google Scholar]
- Howard K, Ingham P (1986) Regulatory interactions between the segmentation genes fushi tarazu, hairy, and engrailed in the Drosophila blastoderm. Cell 44: 949–957 [DOI] [PubMed] [Google Scholar]
- Jennings B, Preiss A, Delidakis C, Bray S (1994) The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development 120: 3537–3548 [DOI] [PubMed] [Google Scholar]
- Jennings BH, Pickles LM, Wainwright SM, Roe SM, Pearl LH, Ish-Horowicz D (2006) Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol Cell 22: 645–655 [DOI] [PubMed] [Google Scholar]
- Jimenez G, Paroush Z, Ish-Horowicz D (1997) Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev 11: 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez G, Guichet A, Ephrussi A, Casanova J (2000) Relief of gene repression by Torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev 14: 224–231 [PMC free article] [PubMed] [Google Scholar]
- Nibu Y, Zhang H, Levine M (2001) Local action of long-range repressors in the Drosophila embryo. EMBO J 20: 2246–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates AC, Ho RK (2002) Hairy/E(spl)-related (Her) genes are central components of the segmentation oscillator and display redundancy with the Delta/Notch signaling pathway in the formation of anterior segmental boundaries in the zebrafish. Development 129: 2929–2946 [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley RL Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D (1994) Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79: 805–815 [DOI] [PubMed] [Google Scholar]
- Paroush Z, Wainwright SM, Ish-Horowicz D (1997) Torso signalling regulates terminal patterning in Drosophila by antagonising Groucho-mediated repression. Development 124: 3827–3834 [DOI] [PubMed] [Google Scholar]
- Pickles LM, Roe SM, Hemingway EJ, Stifani S, Pearl LH (2002) Crystal structure of the C-terminal WD40 repeat domain of the human Groucho/TLE1 transcriptional corepressor. Structure 10: 751–761 [DOI] [PubMed] [Google Scholar]
- Pinto M, Lobe CG (1996) Products of the grg (Groucho-related gene) family can dimerize through the amino-terminal Q domain. J Biol Chem 271: 33026–33031 [DOI] [PubMed] [Google Scholar]
- Schagger H, von Jagow G (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199: 223–231 [DOI] [PubMed] [Google Scholar]
- Song H, Hasson P, Paroush Z, Courey AJ (2004) Groucho oligomerization is required for repression in vivo. Mol Cell Biol 24: 4341–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stifani S, Blaumueller CM, Redhead NJ, Hill RE, Artavanis-Tsakonas S (1992) Human homologs of a Drosophila Enhancer of split gene product define a novel family of nuclear proteins. Nat Genet 2: 119–127 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information