Abstract
Telomeres play an important role in the immortalization of proliferating cells. The long tandem repeats of 5′-TTAGGG-3′ sequences in human telomeres are potential targets for the anticancer drug cisplatin, which forms mainly intrastrand d(GpG) and d(ApG) cross-links on DNA. The present study reveals that telomeres in cisplatin-treated HeLa cells are markedly shortened and degraded. A dose that killed 61% of the cells but allowed one round of cell division resulted in shortened telomeres before the induction of apoptosis. Higher doses of cisplatin halted cell cycle progression during the first S phase and triggered apoptosis followed by degradation of telomere repeats. A model in which both cell division with incomplete replication and induction of apoptosis by cisplatin could occur was devised to explain the drug-induced telomere loss.
Keywords: apoptosis, flow cytometry, human repetitive DNA
Cisplatin is one of the most effective and broadly used anticancer drugs. Apoptosis induced by cisplatin is generally considered to be mediated by the formation of covalent DNA adducts, which block replication and transcription (1). The detailed mechanism by which DNA damage triggers cell death remains unknown, however (2). The most common DNA adducts formed by cisplatin are 1,2-intrastrand d(GpG) cross-links (65%), 1,2-intrastrand d(ApG) cross-links (25%), and minor 1,3-intrastrand d(GpNpG) cross-links (3). These adducts are all removed by the nucleotide excision repair (NER) pathway (4), and cells from patients with xeroderma pigmentosum, a disease caused by a deficiency in NER, are extremely sensitive to cisplatin (5, 6). The transcribed strand is repaired more efficiently than the untranscribed strand because of transcription-coupled repair (7), and NER is influenced by the nucleosome structure and histones in vitro (8). It is not known, however, whether the formation of DNA adducts on transcribed genes is sufficient to induce cell death or whether a specific region of the genome is responsible for apoptosis. It is possible that apoptosis in repair-proficient cells is triggered by damage to DNA that is not transcribed, and poorly repaired, but important for survival.
Telomeres are short tandem repeats of DNA, comprising a G-rich strand and a complementary C-rich strand, which occur at the ends of the chromosome (9, 10). In some species the G-rich strand protrudes from the 3′ end of the duplex and forms an unusual structure (11, 12). The sequence of a repeating unit of human telomeres is 5′-TTAGGG-3′ (13, 14). Telomeres protect the chromosomes from DNA degradation, end-to-end fusions, rearrangements, and chromosome loss and maintain nuclear structure (15, 16). Human telomeres are tightly associated with the nuclear matrix and function as a nucleoprotein complex (17). In primary human fibroblasts, telomeric DNA is lost at a rate of 50–200 bp per doubling (18–20). When telomeres are critically shortened, these mortal cells stop dividing, become senescent, and finally die. One way that immortal cells and cancer cells compensate for telomere loss is with telomerase, a ribonucleoprotein that uses its RNA component as a template to synthesize the 5′-TTAGGG-3′ repeats at the ends of the chromosomes (21, 22). Telomerase activity can be detected in human cells (23, 24) and may also be required for the long-term growth of malignant cancer cells (25–27).
To maintain telomere length, cells have to replicate the repetitive sequence completely. The (TTAGGG)n telomeric sequence has the potential to be a good target for cisplatin, owing to its runs of purine nucleotides. It is possible that the platinated untranscribed telomeric repeats are not completely repaired during cell division and that replication blockage by unrepaired covalent cisplatin–DNA adducts results in substantial telomere shortening or loss. Recently it was reported that pyrimidine dimer sites in telomeres of UV-irradiated normal human fibroblasts were less repaired than transcriptionally active genes such as DHFR and in the overall genome (28). The precise correlation between telomere loss and apoptosis has not been defined. Here, we demonstrate rapid degradation of telomeres in cells exposed to cisplatin. This result and the dependence of cell proliferation on telomeric repeats suggest that this effect may contribute to cisplatin cytotoxicity.
MATERIALS AND METHODS
Cell Culture.
HeLa cells were maintained in monolayers on tissue culture dishes in DMEM (GIBCO/BRL), supplemented with 10% fetal bovine serum (GIBCO/BRL), 100 μg/ml of streptomycin, and 100 units/ml of penicillin.
Analysis of Terminal Restriction Fragment (TRF) Length.
Cells (about 2 × 106) in mid-log phase were seeded onto 100-mm dishes (Falcon) 20 h before an experiment and exposed continuously to various concentrations of cisplatin without a change in medium. For 10-day exposures, the cells were replated on the 5th and 9th day to remove dead cells and debris. At the times indicated, cells were trypsinized, collected into the cell culture media, and counted to monitor growth rate. Genomic DNA was isolated from the cell pellets and digested with HinfI and RsaI (GIBCO/BRL) to generate the TRF. A 2-μg portion of DNA was electrophoresed on a 0.36% agarose gel for 24 h at 2 V cm−1 and hybridized directly to a 5′-32P-labeled (CCCTAA)3 oligonucleotide probe (29).
Flow Cytometry.
About 2 × 105 HeLa cells were saved from the sample that was collected to prepare genomic DNA and then fixed in 70% ethanol. The cells were then incubated in 1 ml PBS containing 50 μg/ml propidium iodide (Sigma) and 250 μg/ml RNaseA (Boehringer Mannheim) at 37°C for 30 min to stain the DNA and eliminate RNA. The fluorescence intensity in the cell was measured by a FACScan (Becton Dickinson). For each sample, 20,000 cells were analyzed by using cell quest and modfit software.
Colony Counting.
The growth and survival of cisplatin-treated HeLa cells were determined by the colony-forming ability of single cells exposed to the drug. Cells seeded at a density of 100 per 60-mm dish (Falcon) 20 h before the experiment were exposed to various concentrations of cisplatin and cultured for 10 days without a change in medium. The cells were fixed and stained with 50% (vol/vol) methanol and 1% methylene blue. The number of colonies was counted and the average from triplicates was determined as a percentage of the number of colonies from untreated cells.
DAPI Staining.
Cells were seeded 20 h before cisplatin treatment onto a Lab-Tec chamber slide glass (Nunc), precoated with 1 mg/ml of poly-l-lysine (Sigma, Mr 3.7 × 104), at a concentration of 3 × 104 cells/cm2. The cells on the glass slide were rinsed in PBS and fixed with methanol. After another rinse, cells were stained with 1 μg/ml DAPI (4′,6-diamidino-2-phenylindole, Sigma) in PBS for 30 min. Then the cells were washed with PBS and mounted. The percentage of apoptotic cells was determined by evaluating nuclear morphology by fluorescence microscopy (Zeiss, Axioplan2).
In Vitro Platination of Telomeric Repeats.
Single-stranded 5′-(TTAGGG)3-3′ and double-stranded 5′-AAA(TTAGGG)5TTT-3′ were incubated at various platinum/strand ratios of cisplatin at 37°C for 24 h with or without 10 mM sodium phosphate (pH 6.8). All of the oligonucleotides were synthesized on a Cruachem PS250 DNA synthesizer and purified on polyacrylamide-urea denaturing gels. A double-stranded pentameric telomere repeat was constructed by PCR. The oligonucleotide used for the template was TG5, 5′-AATCCGTCGAGCAGAGTTTAAA(TTAGGG)5TTTAAAGTACTAGTCGACGCGTGGCC-3′. The primers were TS, 5′-AATCCGTCGAGCAGAGTT-3′ (24) and 3P, 5′-GGCCACGCGTCGACTAGTACT-3′ (GIBCO/BRL). The 78-nt PCR product was digested with DraI (GIBCO/BRL), and the middle 36-nt fragment, 5′-AAA(TTAGGG)5TTT-3′, was purified. Platinum concentrations were determined by atomic absorption spectroscopy, performed on a Varian AA-1475 spectrophotometer equipped with a GTA-95 graphite tube atomizer. Standard solutions of potassium hexachloroplatinate(IV) were used to calibrate the instrument, as described previously (30).
RESULTS
Telomere Length of Cisplatin-Treated HeLa Cells.
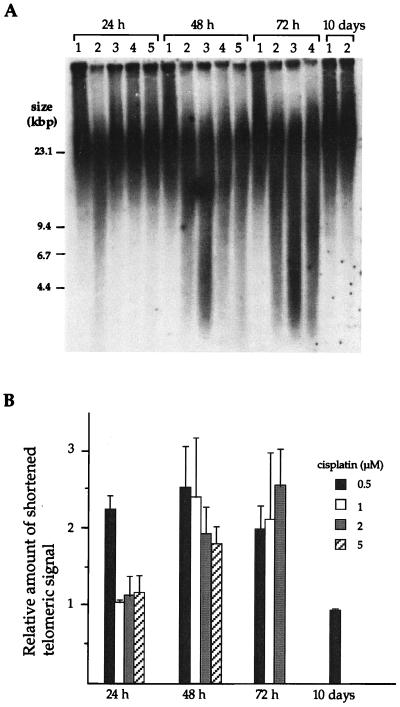
To investigate the effect of cisplatin on telomere length, the size of the TRF from HeLa cells treated with the drug was examined. The HeLa cells used in this study have very long telomeres, greater than 20 kbp (17). Fig. 1A displays the telomeric signals of HeLa cells treated with cisplatin. The distribution of shortened telomere fragments in the 4.4–14.6 kbp range was measured, and the relative amount of signal in each sample is plotted as a ratio of the signal in untreated cells (Fig. 1B). For cells exposed to 0.5 μM (and 0.7 μM, data not shown) cisplatin, the amount of shortened telomeric fragments reached a level of 2.25 after 24 h (Fig. 1A, lane 2), which was retained after 48 h and 72 h of exposure. Telomeric repeats in cells surviving 10 days of treatment returned to the untreated level of 1.0. In contrast, cells exposed to 1, 2, and 5 μM cisplatin exhibited no increase in the fraction of shortened telomeres after 24 h. After 48 h, however, the signals corresponding to shortened telomeres increased. Only at the lowest dose of cisplatin was degradation of telomere repeats observed after 24 h. Because the material analyzed was a mass of asynchronously growing cells and the cells were treated continuously with cisplatin, data were taken only at 24-h intervals.
Figure 1.
Degradation of telomeric DNA in cisplatin-treated HeLa cells. (A) Southern assay of TRF in HeLa cells cultured with or without cisplatin. Lanes: 1, untreated; 2, 0.5 μM; 3, 1 μM; 4, 2 μM; 5, 5 μM. (B) The hybridization signal from 4.4 to 14.6 kbp was quantitated in each fraction. The relative amount of shortened telomeric DNA in each fraction is shown normalized to the signal from untreated cells. Each signal was quantitated by PhosphorImager analysis. The average of the three independent experiments is shown. Error bars represent one standard deviation.
Cell Cycle Progression of Cisplatin-Treated HeLa Cells.
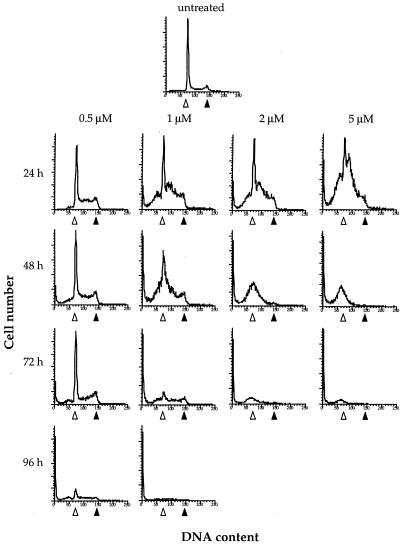
The cytotoxicity and cell cycle progression during cisplatin treatment were monitored to correlate these effects with telomere loss. Data for the relative growth and survival rates are reported in Table 1, and progression through the cell cycle was analyzed by flow cytometry, as plotted in Fig. 2. At 0.5 μM cisplatin, most of the cells completed one round of cell division after 24 h, but only 39% survived to form colonies (Table 1). As indicated in Fig. 2, there was some accumulation of cells in S phase, but no G2 arrest nor sub-G1 fraction appeared after 24 h. After 96 h, most of the cells had accumulated in the sub-G1 population, with only a small proportion progressing through the cell cycle.
Table 1.
Survival and growth rate of HeLa cells treated with cisplatin
| Cisplatin, μM | 10-day survival, % | Growth rate
|
|
|---|---|---|---|
| 24 h | 48 h | ||
| Untreated | 100 | 2.06 ± 0.02 | 3.95 ± 0.17 |
| 0.5 | 39 | 1.74 ± 0.03 | 1.41 ± 0.05 |
| 1 | 4 | 1.14 ± 0.19 | 0.85 ± 0.08 |
| 2 | 0 | 0.96 ± 0.04 | 0.43 ± 0.09 |
| 5 | 0 | 0.83 ± 0.06 | 0.30 ± 0.07 |
The 10-day survival data come from colony counting. The growth rate is presented as a ratio of the number of cells at a given time point to that at the start of cisplatin treatment. The numbers reported are mean values ±1 SD from three independent experiments.
Figure 2.
Cell cycle progression of HeLa cells treated with cisplatin. Histograms of DNA content per cell number of HeLa cells untreated and treated with 0.5, 1, 2, or 5 μM are shown. Cells were maintained in the presence of cisplatin for 24, 48, 72, or 96 h without a change in medium and collected at the times indicated. The DNA content at G1 (▵) and G2 (▴) in untreated cells is indicated.
At higher doses of cisplatin, 1, 2, and 5 μM, most of the cells failed to divide by 24 h. No increase in cell number occurred even after 48 h, and few or no colonies were formed (Table 1). At a 1-μM dose, which yielded 4% survival, there was much more cell cycle arrest than at 0.5 μM (Fig. 2). At 24 and 48 h, most of the cells were in G1 or early S phase with sub-G1 accumulation. After 48 h, the sharp peak of cells arresting at G1 disappeared, and the slope of the plot extended into the sub-G1 range, indicating fragmentation of the nucleus. After 96 h, there were few cells progressing through the cell cycle. When cells were grown in the presence of 2 μM or 5 μM cisplatin, most remained in G1 or early S phase. After 48 h, few (2 μM) or no (5 μM) cells entered the G2 phase, and a large fraction of sub-G1 material appeared. Cells containing a normal range of DNA content decreased after 48 h.
Nuclear Condensation in Cisplatin-Treated Cells.
To obtain morphological evidence for apoptosis, cells treated with cisplatin were stained with DAPI (Fig. 3). As indicated in Fig. 3A, at 24 h very few of the untreated cells and of the cells treated with a low dose (0.5 μM) of cisplatin revealed chromosome condensation. In contrast, 2.5, 13.9, and 36.0% of cells treated with 1, 2, and 5 μM cisplatin, respectively, revealed chromosome condensation indicative of apoptosis. After 48 h, nuclear condensation was observed even in cells treated with 0.5 μM cisplatin. Representative morphological changes of cells 24 and 48 h after treatment with cisplatin are depicted in Fig. 3B. These results are consistent with the FACScan analysis, which demonstrated nuclear fragmentation of cells treated with cisplatin (Fig. 2). At 24 h after treatment, only cells that failed to divide because of exposure to the higher doses, 1, 2, and 5 μM, of cisplatin underwent nuclear fragmentation and condensation, indicating apoptosis. There was no evidence of apoptosis in cells that completed one round of DNA replication and cell division at 24 h after treatment with 0.5 μM cisplatin.
Figure 3.
DAPI staining of HeLa cells cultured with or without cisplatin. DAPI-stained cells were viewed by fluorescence microscopy using iplab spectrum p software. (A) Percentage of cells having a condensed nucleus. A total of 200 cells was scored in each view, and three views were obtained for each sample. The average from three independent experiments is shown. Error bars represent one standard deviation. (B) Representative cells 24 h (Upper) and 48 h (Lower) after treatment. (Left) Untreated; (Center) 0.5 μM cisplatin; (Right) 5 μM cisplatin.
In Vitro Platination of a Telomeric DNA Sequence.
The human telomeric duplex, (TTAGGG)n, has a 3′ overhang of 130–210 bases (12). To investigate whether or not cisplatin can bind to telomere repeats, the compound was incubated with either double-stranded 5′-AAA(TTAGGG)5TTT-3′ or single-stranded 5′-(TTAGGG)3-3′ oligonucleotides, and the platinum bound per DNA strand was measured by atomic absorption spectroscopy (Table 2). Duplex DNA containing five telomeric repeats treated with cisplatin at formal platinum/strand ratios of 5 or 10 in water was platinated with efficiencies of 91.0% and 76.4%, respectively. Sodium phosphate buffer had no effect on the platinum levels. The single-stranded (TTAGGG)3 oligonucleotide was difficult to platinate in buffer, but much higher platinum levels were achieved in pure water (Table 2). The 3′ overhang of telomeric repeats forms a quadruplex structure in vitro that is stabilized by monovalent cations such as Na+ or K+ (10, 31). The reaction of cisplatin with the single-stranded telomeric sequence in sodium phosphate may be inhibited by the formation of such a structure, which would block access to the guanine N7 atoms, the major target of platinum binding on DNA.
Table 2.
In vitro platination of telomeric repeats
| Sequence | rf* | rb†
|
|
|---|---|---|---|
| Sodium phosphate | H2O | ||
| Single strand, | 3 | 0.91 | 2.48 |
| (TTAGGG)3 | 6 | 1.38 | 2.91 |
| Double strand, | 5 | 4.70 | 4.55 |
| AAA(TTAGGG)5TTT | 10 | 7.62 | 7.64 |
Formal platinum-to-strand ratio.
Bound platinum-to-strand ratio.
DISCUSSION
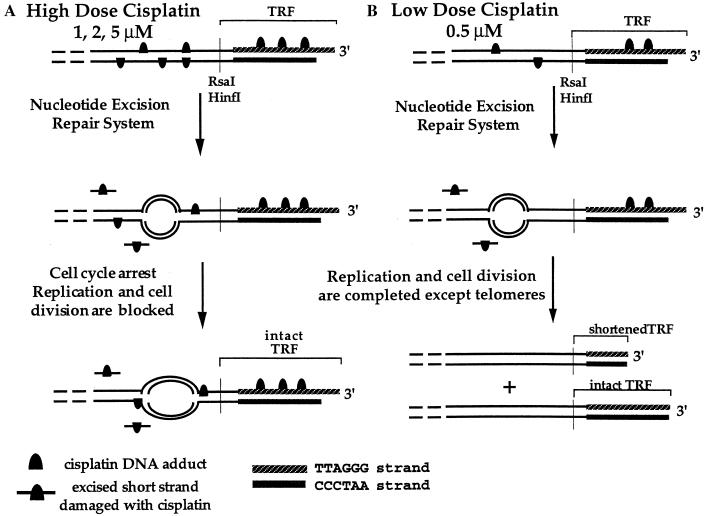
The results presented in this report indicate that long repeats of double-stranded telomeric sequence, which include triplets of guanosine residues, are good targets for cisplatin. Continuous exposure of cells to high doses of the drug (1, 2, or 5 μM) halts DNA replication during early S phase and kills the cells before division. In eukaryotes, transcriptionally active genes are replicated during early S phase (32), whereas telomeres are replicated in late S phase, with the replication origin located near telomeres being activated in late S phase (32, 33). DNA replicons therefore are likely to be blocked by cisplatin–DNA adducts before they reach the telomeres, as schematically represented in Fig. 4A. This model accounts for the lack of shortened telomeres after 24 h in cells treated with cisplatin at these doses. The increase of shortened telomeric signals after 48 h correlates with apoptosis as revealed by the greater sub-G1 fraction relative to the normal range of DNA content (Fig. 2), an increase of condensed nuclei (Fig. 3), and the diminution in viable cells (Table 1).
Figure 4.
Schematic representation of the TRF loss in the first 24 h, caused by high doses (1, 2, and 5 μM, A) and a low dose (0.5 μM, B) of cisplatin. Linear DNA double strands are shown as lines, and the bubbles indicate replication folks.
Although HeLa cells cultured with 0.5 μM cisplatin afforded no apoptotic sub-G1 fraction at 24 h, as judged by flow cytometry (Fig. 2) and DAPI staining (Fig. 3), shortened telomeric signals were observed in the Southern assay (Fig. 1). Most of these cells completed cell division at this low dose (Table 1). These results indicate that the telomere repeats were lost after completion of the first round of DNA replication and persisted after cell division. Because apoptosis was observed at 48 h by flow cytometry (Fig. 2) and DAPI staining (Fig. 3) after this low dose, the shortened telomeres present at this point may be signals for programmed cell death. Telomere shortening at 24 h in cells exposed to 0.5 μM cisplatin cannot be a consequence of nonspecific, damage-induced DNA degradation because shortened telomere fragments were not observed at 24 h in dying cells treated with higher doses of the drug.
Because no transcription products from telomeres have been reported, it is unlikely that damage to telomeric DNA will be repaired by the transcription-coupled NER system. Unrepaired cisplatin adducts on the telomere repeats will block DNA replication (34, 35). At a 0.5-μM dose of cisplatin, where survival levels are low after cell division, the significant shortening of the TRFs indicates incomplete replication with a concomitant loss of telomere repeats (Fig. 4B). Some new telomeric repeats might be synthesized by using the CCCTAA strand as template, affording intact TRFs, because only the TTAGGG strand is a strong target for cisplatin. In agreement with this model is the finding that UV-damaged telomeric DNA is less well repaired than a transcriptionally active gene (28). This same study found telomere damage to be repaired more efficiently than that of an inactive, X chromosome-associated region of the genome. These observations were made for pyrimidine dimer sites on the CCCTAA strands of telomeres, however, and it is unclear whether cisplatin adducts on TTAGGG strand would be processed similarly.
Because cell division occurred at a low dose (0.5 μM) of cisplatin, it would appear that no abortive lesions were present during replication that were able to halt the cell cycle and block cell division. Eventually, however, 61% of the cells exposed even to this low dose of cisplatin died, despite a successful first round of division. We therefore conclude that these cells had been lethally damaged. A candidate for the cause of cell death in this population of the cells is their substantial telomere loss (>6 kbp), which occurred in the absence of apoptosis after 24 h of low-dose drug treatment. It is known that cells can divide even when their telomeres are critically shortened. Substantial telomere loss will destroy the nuclear structure, and, eventually, the cells enter a crisis stage and die (15, 36). Cells treated with an antisense oligonucleotide against telomerase RNA eventually died in such a manner (22). Although the possibility remains that some internal, unrepaired cisplatin lesions are responsible for the delayed apoptosis observed in the present experiments, telomere shortening alone after low-dose treatment with the drug may be sufficient for cell death.
Selective telomere damage by cisplatin at the lowest dose could be a result of more efficient platination as well as heterogeneity of nucleotide excision repair. The human telomere repeat sequence is 5′-TTAGGG-3′, an especially good target for cisplatin. The structure or location of telomeres in the nucleus may also influence the efficiency of platination compared with other internal chromosome targets. In addition, p53 may interrupt the cell cycle in response to DNA platination (37–39) and induce apoptosis (40). These p53-dependent processes, which might not be activated in the first cycle owing to the low level of DNA damage, may recognize the abortive population of telomeres in the daughter cells, triggering apoptosis after the second replication cycle. The p53 gene is wild type in HeLa cells, but owing to the presence of the human papillomavirus E6 protein (41) may be inactive, unless stablized by cisplatin, as occurs elsewhere (42, 43).
After being cultured for 10 days in the presence of 0.5 μM cisplatin, the surviving cells did not have shortened telomeric signals (Fig. 1). This fact indicates that only cells having intact or repaired telomeres were capable of long-term survival. These data strongly suggest that substantial loss of telomeric repeats during cell division has lethal consequences for the cells if not repaired. Also, in other HeLa cell lines with shorter telomere repeats (≈4 kbp) and in HT1080, a fibrosarcoma cell line, we observed telomere loss resulting from treatment with a dose of cisplatin that was lethal but allowed a single round of cell division to occur before apoptosis (data not shown).
The correlation between telomere length and the sensitivity of cells to cisplatin generally is not known. Our results indicate that cells sensitive to low doses of the drug may die as a consequence of telomere loss, a phenomenon that could contribute to the success of cisplatin as an anticancer agent. Telomeric repeat sequences should be good candidates as targets for designing new anticancer drugs.
Acknowledgments
We thank Dr. C. M. Counter for technical advice and valuable discussions. The HeLa cell line was a gift from T. de Lange. This work was supported by Grant CA34992 from the National Cancer Institute.
ABBREVIATIONS
- NER
nucleotide excision repair
- TRF
terminal restriction fragment
- DAPI
4′,6-diamidino-2-phenylindole
References
- 1.Pil P, Lippard S J. Cisplatin and Related Drugs. San Diego: Academic; 1997. pp. 392–410. [Google Scholar]
- 2.Chu G. J Biol Chem. 1994;269:787–790. [PubMed] [Google Scholar]
- 3.Lepre C A, Lippard S J. Nucleic Acids Mol Biol. 1990;4:9–38. [Google Scholar]
- 4.Zamble D B, Mu D, Reardon J T, Sancar A, Lippard S J. Biochemistry. 1996;35:10004–10013. doi: 10.1021/bi960453+. [DOI] [PubMed] [Google Scholar]
- 5.Fraval H N A, Rawlings C J, Roberts J J. Mutat Res. 1978;51:121–132. doi: 10.1016/0027-5107(78)90014-3. [DOI] [PubMed] [Google Scholar]
- 6.Dijt F J, Fichtinger-Schepman A M J, Berends F, Reedijk J. Cancer Res. 1988;48:6058–6062. [PubMed] [Google Scholar]
- 7.Mellon I, Hanawalt P C. Nature (London) 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 8.Sugasawa K, Masutani C, Hanaoka F. J Biol Chem. 1993;268:9098–9104. [PubMed] [Google Scholar]
- 9.Zakian V A. Annu Rev Genet. 1989;23:579–604. doi: 10.1146/annurev.ge.23.120189.003051. [DOI] [PubMed] [Google Scholar]
- 10.Blackburn E H. Nature (London) 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 11.Henderson E R, Blackburn E H. Mol Cell Biol. 1989;9:345–348. doi: 10.1128/mcb.9.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makarov V L, Hirose Y, Langmore J P. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 13.Moyzis R K, Buckingham J M, Cram L S, Dani M, Deaven L L, Jones M D, Meyne J, Ratliff R L, Wu J-R. Proc Natl Acad Sci USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyne J, Ratliff R L, Moyzis R K. Proc Natl Acad Sci USA. 1989;86:7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Counter C M. Mutat Res. 1996;366:45–63. doi: 10.1016/s0165-1110(96)90006-8. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa F. Biochem Biophys Res Commun. 1997;230:1–6. doi: 10.1006/bbrc.1996.5928. [DOI] [PubMed] [Google Scholar]
- 17.de Lange T. EMBO J. 1992;11:717–724. doi: 10.1002/j.1460-2075.1992.tb05104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allsopp R C, Vaziri H, Patterson C, Goldstein S, Younglai E V, Futcher A B, Greider C W, Harley C B. Proc Natl Acad Sci USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harley C B, Futcher A B, Greider C W. Nature (London) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 20.Levy M Z, Allsopp R C, Futcher A B, Greider C W, Harley C B. J Mol Biol. 1992;225:951–960. doi: 10.1016/0022-2836(92)90096-3. [DOI] [PubMed] [Google Scholar]
- 21.Greider C W, Blackburn E H. Nature (London) 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 22.Feng J, Funk W D, Wang S-S, Weinrich S L, Avilion A A, Chiu C-P, Adams R R, Chang E, Allsopp R C, Yu J, et al. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 23.Morin G B. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 24.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L C, Coviello G M, Wright W E, Weinrich S L, Shay J W. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 25.Counter C M, Hirte H W, Bacchetti S, Harley C B. Proc Natl Acad Sci USA. 1994;91:2900–2904. doi: 10.1073/pnas.91.8.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Lange T. Proc Natl Acad Sci USA. 1994;91:2882–2885. doi: 10.1073/pnas.91.8.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shay J W, Bacchetti S. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 28.Kruk P A, Rampino N J, Bohr V A. Proc Natl Acad Sci USA. 1995;92:258–262. doi: 10.1073/pnas.92.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Counter C M, Avilion A A, LeFeuvre C E, Stewart N G, Greider C W, Harley C B, Bacchetti S. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandman K E, Fuhrmann P, Lippard S J. J Biol Inorg Chem. 1998;3:74–80. [Google Scholar]
- 31.Williamson J R, Raghuraman M K, Cech T R. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 32.Fangman W L, Brewer B J. Cell. 1992;71:363–366. doi: 10.1016/0092-8674(92)90505-7. [DOI] [PubMed] [Google Scholar]
- 33.McCarroll R M, Fangman W L. Cell. 1988;54:505–513. doi: 10.1016/0092-8674(88)90072-4. [DOI] [PubMed] [Google Scholar]
- 34.Pinto A L, Lippard S J. Proc Natl Acad Sci, USA. 1985;82:4616–4619. doi: 10.1073/pnas.82.14.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Comess K M, Burstyn J N, Essigmann J M, Lippard S J. Biochemistry. 1992;31:3975–3990. doi: 10.1021/bi00131a013. [DOI] [PubMed] [Google Scholar]
- 36.Harley C B. Mutat Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 37.Lane D P. Nature (London) 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 38.Jacks T, Weinberg R A. Nature (London) 1996;381:643–644. doi: 10.1038/381643a0. [DOI] [PubMed] [Google Scholar]
- 39.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 40.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. Nature (London) 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 41.Scheffner M, Münger K, Byrne J C, Howley P M. Proc Natl Acad Sci USA. 1991;88:5523–5527. doi: 10.1073/pnas.88.13.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang N, Song Q, Lu H, Lavin M F. Oncogene. 1996;13:655–659. [PubMed] [Google Scholar]
- 43.Fritsche M, Haessler C, Brandner G. Oncogene. 1993;8:307–318. [PubMed] [Google Scholar]