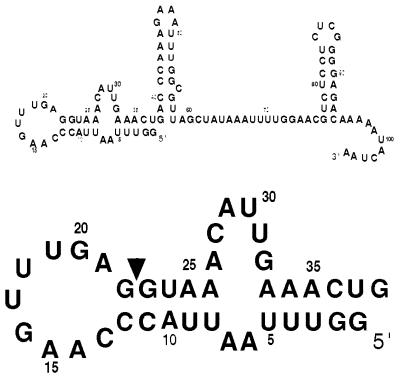Abstract
The role of spliced leader RNA (SL RNA) in trans-splicing in Caenorhabditis elegans has been studied through a combination of in vitro mutagenesis and in vivo complementation of rrs-1 mutant nematodes, which lack endogenous SL1 RNA. Three classes of mutant SL1 RNAs have been found—those that rescue the lethal phenotype at low concentration of transforming DNA, those that rescue at high but not low concentration, and those that do not rescue at all. These studies showed that some mutations in the otherwise highly conserved 22-nt spliced leader are tolerated for splicing and post-splicing events. A longer spliced leader also can be tolerated but only when present in high copy number. Changes in the first 16 nucleotides result in the appearance of no SL RNA, consistent with the in vitro studies by others showing that the SL1 RNA promoter partly resides within the spliced leader sequence.
Trypanosomes, trematodes, and nematodes are responsible for enormous health problems world-wide, particularly in tropical countries. All of these organisms process their mRNAs by trans-splicing. Trans-splicing is both essential and unique to these organisms. Therefore, it serves as a reasonable target for drug intervention. Trans-splicing is also of special interest for studying the mechanism and the evolution of pre-mRNA processing. Trans-splicing involves the transfer of the spliced leader (SL) from the donor molecule, the SL RNA, to the splice acceptor site upstream of the AUG initiation codon in pre-mRNAs. As a result, all trans-spliced mRNAs contain the SL on their 5′ ends. The SL RNA is a chimeric molecule containing a short exon, the spliced leader, covalently linked to an intron that resembles, both in base pairing and function, the U RNA molecules that assist in pre-mRNA splicing in higher organisms.
Trans-splicing originally was discovered in trypanosomes. In these protozoa, an ≈140-nt SL RNA donates a 39-nt SL to pre-mRNAs (see refs. 1 and 2 for reviews). Through trans-splicing, polycistronic pre-mRNAs in trypanosomes are processed to monocistronic messages containing uniform spliced leaders at their 5′ ends. The discovery of trans-splicing in Caenorhabditis elegans showed that the process exists in multicellular eukaryotes; however, there are differences between nematodes and trypanosomes (3). All mRNAs are trans-spliced in trypanosomes, but ≈30% of the messages in C. elegans are not trans-spliced (4). Whereas trypanosomes only carry out trans-splicing, nematodes carry out cis-splicing in addition to trans-splicing. In trypanosomes, all of the messages are synthesized as polycistronic transcripts, but in C. elegans, three-quarters of the messages are monocistronic and one-quarter are polycistronic (4). With the discovery of a second SL RNA (SL2 RNA) that donates its leader to a subset of messages, it became apparent that trans-splicing in C. elegans has an inherent specificity (5). SL2 RNA, which differs in sequence from SL1 RNA, is reserved for processing downstream mRNAs in polycistronic pre-mRNAs (6, 7). Several different SL RNA molecules now have been identified in nematodes, but SL1 RNA is by far the most abundantly used in trans-splicing (8). All nematodes show remarkable conservation in using the same 22-nt SL1 sequence for trans-splicing from SL1 RNA. Like U RNAs, SL RNAs contain consensus Sm binding sites and 2,2,7-trimethylguanosine caps (m3G) (9–12). As a result, SL-trans-spliced mRNAs acquire m3G caps at their 5′ ends.
The SL RNA secondary structure consists of three hairpin stem loops. The 5′ hairpin of the SL RNA contains the splice donor site in an A form helix (13). This base-paired structure resembles the pairing of the 5′ end of U1 RNA with the splice donor site in pre-mRNA, a structure that is essential for pre-mRNA cis-splicing (11). The relationship of the SL1 RNA sequence to its function has been studied in vitro by using Ascaris extracts and Ascaris SL1 RNA. Those experiments revealed that part of the promoter for the SL1 RNA gene is contained within the 5′ end of the SL RNA (14, 15). Altering these nucleotides severely reduced transcription. The in vitro trans-splicing studies also created a conundrum. Removing the 5′ 20 nucleotides of SL1 RNA that base pair with the trans-splice site did not eliminate trans-splicing despite its structural homology with the essential pairing of U1 with the pre-mRNA splice donor site (16). In contrast to the results of these Ascaris in vitro experiments, structure–function analyses performed in vivo with trypanosomes showed that the SL exon was needed for trans-splicing and that base pairing across the splice site was essential (17).
Recently, new opportunities emerged for genetically studying trans-splicing in C. elegans with the finding that a mutation, rrs-1 (e2482), deletes all 110 SL1 RNA genes (18). The rrs-1 mutant also is lacking 5S RNA because each unit of the SL1 RNA chromosomal tandem array carries a gene encoding 5S RNA. rrs-1 is an embryonic lethal mutation. Transforming rrs-1 mutants with 5S genes alone does not rescue the embryonic lethality, but transforming with 5S and SL1 RNA genes rescues the embryonic lethality, and fertile adult transformants are obtained. SL1 RNA alone rescues embryonic lethality, but the transformants die as larvae. We have used complementation of rrs-1 (-) lethality to perform a mutational analysis in vivo of the structure–function relationships in SL1 RNA.
MATERIALS AND METHODS
Genetic Rescue of rrs-1.
C. elegans cultures were maintained and genetic analyses were performed according to Brenner (19). Bristol strain N2 served as wild type. Transformation was done by microinjecting DNA into the germ line syncytium (20). The plasmid p31.21 was used in all of the transformation studies; it contains the 1-kb BamHI fragment of the repeat unit encoding both SL1 RNA and 5S rRNA (3). In vitro mutagenesis was done by using the QuickChange site-directed mutagenesis kit (Stratagene). A mutant plasmid (100 μg/ml or 10 μg/ml) was coinjected with 100 μg/ml of pRF4, a plasmid containing the cloned dominant rol-6 (su1006) gene serving as the transformation marker. The DNAs were injected into unc-76 (e911) rrs-1 (e2482)/unc-61(e228) dpy-21 (e428) hermaphrodites. Individual F1 rollers were picked and allowed to propagate. Among the F2, individual non-Unc-61, non-Unc-76 roller lines were established. Those producing dead embryos, presumably because of incomplete transmission of the extrachromosomal transforming DNA, were scored for rescue of rrs-1. Transgenic lines, heterozygous for rrs-1, were scored for their total number of progeny, number of arrested embryos, and number of Unc-76 animals. One-quarter of the progeny are expected to be unc-76 homozygotes, and all are expected to die if no rescue occurs. However, we expect 1.8% of the animals will be live Unc-76 non-Rrs-1 worms that arise because of recombination in the interval between unc-76 and rrs-1. Therefore, candidates for rescue of embryonic lethality were picked onto individual plates if >3% of the animals were Unc-76. (Because the roller phenotype is incompletely penetrant, individual Unc-76 worms from these rescue candidates were allowed to propagate to insure that Unc-76 roller offspring would be produced and to confirm that they were not recombinants.)
PCR, Reverse Transcriptase (RT) PCR, and 5′ Rapid Amplification of cDNA Ends.
One or two 35-mm Petri plates of transformed worms were grown until the bacterial lawn cleared. Worms were washed off the plates with M9 buffer and pelleted in a microcentrifuge for 30 s. The M9 buffer was removed and replaced with 1 ml of TRIzol reagent (GIBCO/BRL). The worms were homogenized with an Omni International (Waterbury, CT) 2000 homogenizer at maximum speed for 1 min. RNA and DNA then were prepared by using the TRIzol procedure (GIBCO/BRL), and the RNA was dissolved in 20 μl of water, and the DNA was dissolved in 50 μl of 8 mM NaOH. RNA was heated at 95° for 5 min and then put on ice for 15 min before the reverse transcription reaction. The RT-PCR and 5′ rapid amplification of cDNA ends reactions were carried out by using the GIBCO/BRL 5′ rapid amplification of cDNA ends kit, Version 2.0, with slight modifications. To obtain the RT-PCR product of SL1 RNA, 1 μ1 of the RNA was incubated with 1 μM SL RNA 3′ primer No. 1 (5′-TTT GTC TCC CCG AGA-3′), 1 mM dNTP, cDNA synthesis buffer (Boehringer Mannheim; 50 mM Tris⋅HCl/1 mM DTT, pH 8.5), 20 units of RNase inhibitor (Boehringer Mannheim), and 12.5 units of avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim) in a 20-μ1 reaction. The reaction mixture was incubated at room temperature for 10 min, 42° for 1 h, and 95° for 15 min. Excess primer was removed by diluting the reaction mixture with 2 ml of 0.1× (10 mM Tris·HCl/1 mM EDTA, pH 8.0) followed by centrifugation three times with a Centricon-30 (Amicon). Of the ≈50 μl of cDNA, 10 μ1 was tailed with poly dC, and the PCR was carried out by using the anchor primer (5′-GGC CAC GCG TCG ACT AGT ACG GGI IGG GII GGG IIG-3′) and the SL RNA 3′ primer No. 2 (5′-TTT ATA GCT AAC GCC AAA TTT CTT T-3′). The PCR product was ≈120 bp.
The presence of SL1 on a trans-spliced message was assayed by RT-PCR of the ges-1 mRNA by using GIBCO/BRL 5′ rapid amplification of cDNA ends kit, Version 2.0. To obtain the ges-1 RT-PCR product, reverse transcription used the ges-1 primer No. 2 (5′-ATT AAC TCC TTG TGA AGC ATA-3′). Tailed cDNA was amplified by using the anchor primer and the ges-1 primer No. 5 (5′-CGT AAC CAT GTT GAC TAG CC-3′), and the resultant product was reamplified by using AUAP primer (5′-GGC CAC GCG TCG ACT AGT AC-3′), which corresponds to the 5′ portion of the anchor primer, and ges-1 primer No. 4 (5′-CTC AAA TCA TCG ACT GG-3′). The resultant PCR product was ≈200 bp.
In the case of the mutant Add C23A24G25 transformant, the presence of the mutant SL1 on ges-1 mRNA was assayed by first reverse transcribing with ges-1 primer No. 2, followed by PCR with SL1 20 (5′-GGT TTA ATT ACC CAA GTT TG-3′) and ges-1 primer No. 5. The product was re-amplified with the same primers or with SL1 20 and primer No. 4. The resulting cDNA product was cloned into the Promega TGEM vector and sequenced with M13 forward and reverse primers.
Genomic and transforming DNA were amplified under the following conditions. The DNA (1 μ1) was incubated with 10 mM Tris⋅HCl (pH 8.3), 0.2 μM SL RNA 3′ primer No. 1 and SL primer up No. 2 (5′-GGT TGC CGA TCG TAG ACC TCG-3′), 0.2 mM dNTP, 3.5 mM MgCl2, 75 mM KCl, and 2.5 units of Taq DNA polymerase in a 100-μ1 reaction. Amplification was done by annealing at 52° for 30 s and extending at 72° for 40 s for 35 cycles, resulting in a 159-bp PCR product. All final PCR products were gel-purified by using QIAEX II Gel Extraction Kit (Qiagen, Chatsworth, CA) before using the single nucleotide primer extension assay (21).
All of the assessments of SL1 RNA levels were made relative to the amount of the mutant transforming DNA detected in the single nucleotide primer extension assays. In all of the lines of class III transformants, the same approximate amount of mutant transforming DNA was detected among the mixed progeny of the heterozygous parents, relative to wild-type genomic DNA, indicating that the number of transforming genes was approximately the same in the different lines.
RESULTS
Deletion of the rrs-1 gene cluster results in embryonic lethality in C. elegans (18). This lethality can be rescued by transforming the wild-type SL1 RNA and 5S rRNA genes into worms by microinjecting DNA at either high (100 μg/ml) or low (10 μg/ml) concentrations (Table 1). The process of SL1 trans-splicing itself is essential to C. elegans development because SL1 RNA molecules mutated at residues essential for trans-splicing, such as mutations in the trans-splice donor site (G22G23 to U22U23) (Table 1) and in the Sm binding site (18), fail to rescue the embryonic lethal phenotype. Therefore, the phenotypic rescue of rrs-1 can serve as a standard for measuring the sequence requirements for SL1 RNA function.
Table 1.
Rescue of embryonic lethality of unc-76 rrs-1/unc-61 dpy-21 by transformation with SL1 RNA genes
| 100 μg/ml | 10 μg/ml | SL RNA present | Transfer to ges-1 | ||
|---|---|---|---|---|---|
| Wild type | 3/3 (1210) | 5/5 (1194) | + | + | |
| G22G23 to U22U23 | 0/5 (1541) | ND | +† | −† | |
| Class I | Del A27C28A29 | 1/1 (852) | 2/3 (706) | +* | ND |
| G16 to C16 | ND | 3/3 (608) | +* | ND | |
| Class II | A21 to C21 | 4/4 (1384) | 0/1 (211) | +* | ND |
| A21 to U21 | 8/8 (2812) | 0/3 (1369) | +* | ND | |
| Add C23A24G25 | 6/6 (3051) | 0/7 (1874) | +* | + | |
| Class III | Del U3-A7 | 0/3 (847) | ND | −† | −† |
| Del A6A7 | 0/9 (2590) | 0/3 (848) | −† | −† | |
| Del U8-C12 | 0/6 (1377) | ND | −† | −† | |
| Del C13-U17 | 0/3 (953) | ND | −† | −† | |
| Del G16-G20 | 0/8 (2365) | ND | ±† | −† | |
| A7U8U9A10C11 to C7A8C9C10A11 | 0/6 (2282) | ND | −† | −† |
Transforming DNA (100 μg/ml or 10 μg/ml) was used. The number of rescued lines over the number of transformed lines analyzed is shown.
The presence of mutant SL RNA or mutant SL on ges-1 mRNA was assayed using clones of rescued animals.
The assays were performed on mixed populations (wild-type and rrs-1 heterozygotes) of transgenic worms.
Numbers in parentheses designate the total number of progeny scored. Del, deletion; ND, not determined; Add, addition of nucleotides. Classes are described in text.
We were particularly interested in the structure-function relationship within the first stem loop of the SL1 RNA because it contains the splice donor site and the spliced leader itself (Fig. 1). We tested the significance of certain structural features of the first stem loop of SL1 RNA that were apparent from the NMR-derived model of the structure (13, 23). First, the 9-nt loop (C13-A21) appears to be stable because of two base pairs (A14U19 and C13G20) and base stacking on the 5′ side of the loop. To determine whether maintaining the loop structure is essential for SL1 RNA function, we expanded the loop by adding three residues at the end of the loop between G22 and G23 (Add C23A24G25). This insertion maintains the conserved AG dinucleotide sequence adjacent to the splice site. Second, in the stem loop of SL1 RNA, a pocket is formed near the splice site by a bulged adenosine (A21) 5′ to the splice site. This cavity with dimensions ≈ 5 Å ×6 Å appears as if it could be the docking site for another molecule that could assist in the trans-splicing reaction. An adenosine in this position is conserved among all SL RNA molecules in nematodes. To determine whether A21 is essential for SL1 RNA function, we changed A21 to C21 and U21. Third, a guanosine in the loop (G16) is found in the syn conformation instead of the usual anti-conformation typical of RNA. The chain reversal occurs 5′ to G16. We mutated G16 to C16 to see if altering it resulted in a change in SL1 RNA function. Fourth, we tested the importance of the internal loop (A27 through G32) downstream of the splice site in the first stem. It seemed that this internal loop might play a critical role in SL1 RNA splicing function because bulges and internal loops are frequently sites of protein–RNA interactions and because the secondary structure prediction of C. elegans SL2 RNA also contains a bulged C26 as well as a bulged A32 in this region (5). Our test construct produces SL1 RNA lacking half of the bulge (Del A27C28A29). Fifth, we made several five nucleotide block deletions along the spliced leader to determine which regions of the spliced leader were essential for pre-splicing events, e.g., transcription, or for trans-splicing, or for post-splicing functions.
Figure 1.
Sequence of C. elegans SL1 RNA. Arrowhead indicates the trans-splice site. Numbers are assigned relative to the 5′-terminal G in the SL1 RNA gene. The first stem loop of the SL1 RNA is enlarged in the lower part of the figure.
These mutant genes were transformed into unc-76 rrs-1/unc-61 dpy-21 worms and tested for their ability to rescue the embryonic lethality of the rrs-1 deletion mutant. The appearance of >3% hatched Unc-76 worms was taken as evidence for genetic rescue. Candidate Unc-76 rescued lines were then further tested by determining their molecular phenotype. The progeny from an Unc-76 roller were assayed by a single nucleotide primer extension assay for the absence of the wild-type SL1 RNA and the presence of the mutant SL1 RNA (21) (see Fig. 2; an example of the assay is shown in Fig. 3). Thus, both genetic and molecular phenotypes were required to deem that a line represented SL1 RNA-dependent rescue of the embryonic lethality and to confirm that the rescue was not a result of recombination in the heterozygous parent.
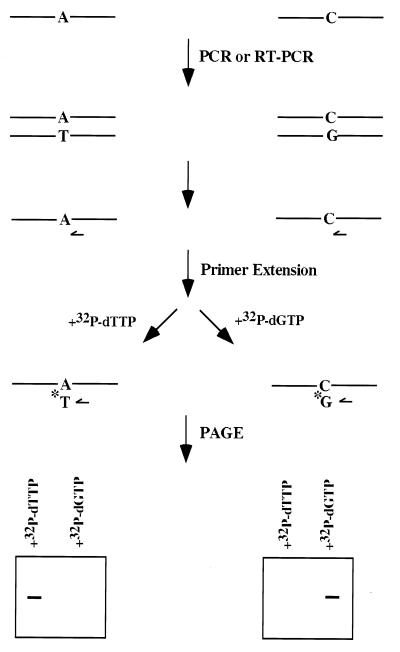
Figure 2.
Single nucleotide primer extension assay to detect the presence of wild-type and mutant SL1 RNA genes, SL1 RNAs, and ges-1 mRNA. DNA and total RNA were isolated from either Unc-76 or rrs-1 worms transformed with mutant SL1 RNA genes and amplified by PCR and RT-PCR as described in Materials and Methods. PCR products were gel-purified and subjected to single nucleotide primer extension reactions by incubating with a primer 5′ to the mutation, the 32P-labeled nucleoside triphosphate complementary to either the wild-type or the mutant base, and Taq DNA polymerase. The primer was allowed to anneal at 55° for 30 s, followed by extension at 72° for 1 min. Because the radiolabeled nucleotide is the only nucleotide in the reaction, it will be incorporated into the primer only when it is complementary to the base 3′ to the primer. The reaction mixture was then run on a denaturing polyacrylamide gel and analyzed by autoradiography or a Packard Instant Imager.
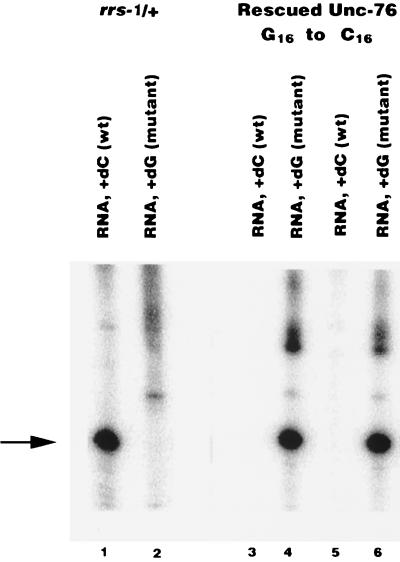
Figure 3.
Confirmation of genetic rescue by C16 mutant with the single nucleotide primer extension assay. Individual Unc-76 animals were propagated on separate plates. Total DNA and RNA were extracted. The presence of wild-type and mutant SL1 RNA was assayed by the single nucleotide primer extension assay by using 5′-GTT TCA ATG TTT ACC TCA AA-3′ as the primer. 32P-labeled dGTP or dCTP is incorporated into the primer in the presence of mutant or wild-type RNA, respectively. The arrow indicates the primer extended products. The parental strain contains only wild-type SL1 RNA (lane 1) and no mutant SL1 RNA (lane 2), whereas two lines of rescued Unc-76 worms contain only mutant SL1 RNA (lanes 3 and 4 and lanes 5 and 6).
We found three classes of mutant SL1 RNAs according to their ability to rescue (Table 1): those able to rescue when transformed at low DNA concentration (class I), those able to rescue at high but not at low concentration (class II), and those unable to rescue at high concentration (class III).
Class I mutant SL1 RNAs rescue as well as wild-type SL1 RNA. Therefore, these changes do not interfere with SL1 RNA function within the limits of our detection. G16 to C16 and Del A27C28A29 belong to this class of mutations. Class II mutants rescue only when transformed at high DNA concentration; apparently, these mutations render SL1 RNAs or the spliced leaders partially defective. For example, these mutations may cause the SL1 RNA to have a reduced affinity for splicing components, which can be overcome by increasing the mutant SL1 RNA concentration. Or, the mutations may interfere with transcription or stability of the SL RNA, which can be overcome by high gene dosage. A21 to C21, A21 to U21, and the expanded loop (Add C23A24G25) mutants belong to this class. The 22-nt spliced leader length is conserved across species, so we particularly were interested in knowing whether this longer mutant spliced leader was transferred intact to mRNAs. We chose to assay the ges-1 mRNA for these studies because it is well characterized, SL1 trans-spliced, and expressed at virtually all stages of worm development (22). Therefore, cDNA was made from ges-1 mRNA from each of the three rescued lines and sequenced. Each line contained ges-1 with the 25-nt long mutant SL (data not shown).
Class III mutations fail to rescue the embryonic lethality even at a high concentration of transforming DNA. These mutations, therefore, must alter essential sites in SL1 RNAs. All deletions in the spliced leader (Del U3U4U5A6A7, Del U8U9A10C11C12, Del C13A14A15G16U17, Del G16U17U18U19G20, and Del A6A7) or mutations altering the 5′ end of the spliced leader (A7U8U9A10C11 to C7A8G9C10A11) result in loss of function of the SL1 RNA. We reasoned that the in vivo functional defects in class III mutants could be at three different steps: (i) SL1 RNA transcription and/or stability, (ii) trans-splicing, or (iii) the post-splicing function(s) of the SL1 spliced leader. To distinguish between these three possibilities, we used the single nucleotide primer extension assays (Fig. 2; ref. 21) to detect the presence of a mutant SL1 RNA gene, a mutant SL1 RNA, and a mutant SL1 spliced leader on a message (ges-1) in transformed strains (22). These assays were performed on mixed populations of worms consisting of transformed heterozygous and wild-type worms because no rescued lines could be obtained. As a control for defective splicing, we chose to assay the splice site mutation G22G23 to U22U23. As anticipated, the mutant spliced leader was not transferred to the ges-1 message, but we were able to detect the mutant SL1 RNA gene and the mutant SL1 RNA (Fig. 4).
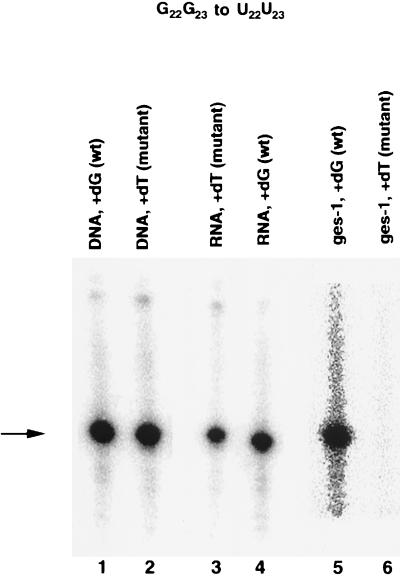
Figure 4.
Analysis of the SL1 RNA gene, SL1 RNA, and ges-1 mRNA of the splice site mutant, G22G23 to U22U23. In worms transformed with the G22G23 to U22U23 mutant SL1 RNA gene, the mutant SL1 RNA gene (lane 2) and SL1 RNA (lane 4) are detected. Only wild-type ges-1 (lane 5) is detected in these worms. The absence of mutant ges-1 (lane 6) indicates that the mutant SL1 RNA is defective in trans-splicing. The arrow indicates the primer extended products.
We were able to detect the presence of the mutant SL1 RNA genes in all class III transformed worms by using the single nucleotide primer extension assay. In contrast to the control strain in which the G22G23 was mutated, we failed to detect mutant SL1 RNA in the class III transformants except in the case of Del G16-G20 mutant lines. (Table 1; an example is shown in Fig. 5). Failure to detect SL1 RNA could result from defects in transcription or RNA stability. Variable levels of mutant SL1 RNA were found in the Del G16-G20 transformants. Two transformed lines of Del G16-G20 contained very low levels of mutant SL1 RNA whereas two others contained significant levels of the RNA. Despite the presence of the RNA, neither transformed line was viable as an rrs-1 homozygote. These variable levels of SL1 RNA indicate that this deletion may cause some promoter interference or perhaps increased SL1 RNA degradation. No mutant SL1 trans-spliced ges-1 mRNA was detected in this Del G16-G20 mutant nor in any of the other class III transformants.
Figure 5.
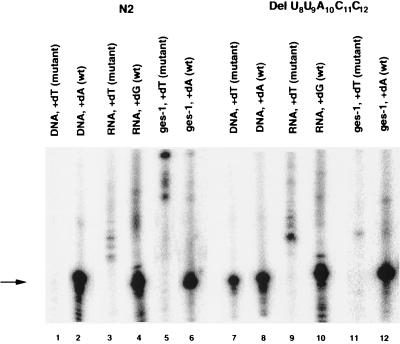
Analysis of SL1 DNA, SL1 RNA, and ges-1 mRNA of Del U8-C12. The single nucleotide primer extension assay was performed by using 5′-GTT TCA ATG TTT ACC TCA TT-3′ for the analysis of the SL RNA gene and SL RNA and 5′-CAG AGC CAA GGA ATA TCC GCA TC-3′ for the analysis of the ges-1 message. Wild-type N2 worms contain only the wild-type SL1 RNA gene (lane 2), SL1 RNA (lane 4), and ges-1 message (lane 6). In rrs-1/+ worms transformed with the mutant gene, both wild-type and mutant SL1 RNA genes (lane 7 and 8) are detected. However, only wild-type SL1 RNA (lane 10) and ges-1 (lane 12) are detected in these worms. The absence of mutant SL1 RNA (lane 9) and mutant ges-1 (lane 11) indicates that the mutant SL1 RNA transcription or stability is eliminated. The arrow indicates the primer extended products.
DISCUSSION
These studies have shown that, although the SL1 spliced leader is conserved evolutionarily among all nematodes, certain changes, including base substitutions or elongation of the spliced leader, can be tolerated for in vivo trans-splicing and the functioning of SL1. All nematodes that have been studied have the same 22-nt SL1 sequence. Such a high degree of conservation may reflect an essential function for the leader sequence or perhaps that the leader serves several different functions involving interactions with other components so that multiple second site mutations, each affecting a separate component, would be necessary to revert a single mutation in the leader. Earlier studies on trans-splicing in vitro found that nearly all 22 nucleotides of the SL1 could be deleted from the SL1 RNA without eliminating trans-splicing (23). These in vitro studies could not address whether such severe truncations affected the subsequent post-splicing functions of SL1. Our in vivo results, albeit with far subtler changes to the SL1, indicate that some altered SL1s can still function as spliced leaders.
The change from G16 to C16 in the leader appears benign because rescue was achieved at a low concentration of transforming DNA. The structural studies on the first stem loop of the SL1 RNA showed that G16 has two noteworthy features. It is located at the apex of the loop where the backbone changes direction, and the base, which is not stacked over neighboring bases, is in the syn instead of the usual anti-conformation (23). Attempts to model a rotation of the G16 guanosine resulted in a collision with the neighboring uridine (13). It is clear that, despite these structural features, substituting C16 for G16 allows both trans-splicing and SL1 post-splicing functions to occur efficiently enough for viability.
Changing A21 to U21 or C21 also was tolerated as rescue of embryonic lethality occurred and viable offspring were obtained. However, the mutations apparently render the SL1 RNA molecule partially defective because rescue was obtained only at the tenfold higher concentration of transforming DNA. In the wild-type RNA, the NMR structure shows an amino proton of A21 hydrogen bonded to the 2′ hydroxyl of the ribose of A14. A similar H-bond would be highly unlikely to form in the case of U21. Although the H-bond may assist in stabilizing the loop structure, it appears unnecessary and the higher concentration may provide enough SL RNA of the correct conformation. In any case, the SL1 post-splicing functions remain intact when U21 or C21 replaces A21, indicating that A21 is not critical for the subsequent post-splicing functions.
Most intriguing was the rescue provided by the mutant SL1 RNA containing an insertion of three extra nucleotides in the spliced leader sequence. The Add C23A24G25 insertion is 5′ to the splice donor site, but it maintains the partially conserved AG dinucleotide sequence preceding the splice site, as well as the absolutely conserved GU past the splice site. The Add C23A24G25 mutant molecule was capable of rescue only at the high concentration of transforming DNA, indicating that the molecule is more subject to nucleases or that it does not splice as well as wild-type. That trans-splicing occurs indicates that the exact structure of the stem loop need not be maintained. It further indicates that the distance between the loop and the splice site need not be maintained. The insertion occurs on the 3′ side of the stem loop where the structural studies noted little base stacking in wild-type SL1 RNA. As was suggested earlier, perhaps this side of the loop flexibly can accommodate changes in structure; therefore, the loop may function simply to hold together the bases at the end of the A-helix stem, which contains the splice site (13). The mutant spliced leader resulting from the insertion is 25-nt long, as opposed to the wild-type 22 nucleotides, because the insertion is 5′ to the splice donor site.
C. elegans SL1 and SL2 are each 22 nucleotides long; other C. elegans SLs are 21, 22, or 23 nucleotides long (8). The intervening sequences between pre-mRNA trans-splice acceptor sites and initiation codons vary in length in different mRNAs from 0 to ≈50 nucleotides. The insertion adds three extra nucleotides to this 5′ untranslated interval, and their presence does not generate a new AUG within the spliced leader. Thus, it is reasonable to expect that the effect of the three nucleotide addition would be relatively benign on the trans-spliced mRNA.
Our in vivo studies on the class III mutants corroborate the in vitro findings that part of the promoter for SL1 RNA resides within the SL1 sequence itself. C. elegans and Ascaris contain the same SL1 22-nt sequence. Ascaris uses RNA polymerase II for SL RNA transcription, and C. elegans most likely does also (14, 24). Class III mutant SL1 RNAs failed to rescue even at the high concentration of transforming DNA. In six different cases, no mutant SL1 RNA was present, probably because of a disruption of the promoter by the mutations. Five of the six cases represent deletions within the first 15 nucleotides of the SL1 RNA. In one deletion, Del G16-G20, the levels of the mutant SL1 RNA were found to be variably lower in two lines but near normal in two other lines. Perhaps this partial activity reflects the position of the deletion at the distal border of the promoter. Similar results were found in in vitro transcription studies of SL RNA genes in Ascaris extracts (14). In the Ascaris studies, mutating nucleotides 2–11 as a block, as well as changing nucleotides 12–22, severely reduced transcription of the SL1 RNA gene by RNA polymerase II. The trypanosome SL RNA gene also appears to have part of its promoter embedded in the corresponding 5′ region of the SL RNA coding sequence. Inserting six nucleotides near the 5′ end of a trypanosome SL RNA transgene diminished its level of in vivo expression, and base substitutions within the first 18 nucleotides similarly depressed expression (17).
Deleting half of the 5′ side of the internal loop in the first stem of the C. elegans SL1 RNA did not alter its ability to rescue the rrs-1 mutant worms. Because the deletion occurs within the intron beyond the SL1 exon, the change will affect only trans-splicing and not subsequent SL1 functions. A corresponding internal loop is also present in the predicted secondary structure of SL2 RNA from C. elegans but is missing in the SL1 RNA from Ascaris. Internal loops and bulges are favored sites of protein–RNA interactions, such as TFIIIA binding to loop E of 5S RNA, R17 coat protein to R17 RNA, or Tat to TAR in HIV (25–27). It is tempting to speculate that the internal loop in the first stem of the SL1 RNA of C. elegans serves as a similar site for protein binding. However, our in vivo rescue data would indicate that its function is dispensable or redundant.
Our results indicate that some changes in the first stem loop of the SL1 RNA can be tolerated in vivo for trans-splicing but that the 5′ region of the SL1 RNA comprises part of the promoter; both of these features had been observed earlier in in vitro studies. In addition, our in vivo studies have shown that changes also are tolerated by the post-splicing functions of the spliced leader, although the exact role of the downstream functions remains unresolved.
Acknowledgments
This research was supported by Public Health Service Grant R01 GM37823. We are especially grateful to Barth Grant for his generous help in analyzing specific ges-1 messages. We thank Sarah Paul and Laura Pedraza for excellent technical assistance. We also thank Nancy Greenbaum, Iva Greenwald, Vera Irikura, Anna Pyle, and Yinhua Zhang for reading the manuscript and Joel Rothman for providing the rrs-1 (e2482). Several C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources.
ABBREVIATIONS
- SL
spliced leader
- RT
reverse transcriptase
References
- 1.Borst P. Annu Rev Biochem. 1986;55:701–732. doi: 10.1146/annurev.bi.55.070186.003413. [DOI] [PubMed] [Google Scholar]
- 2.Agabian N. Cell. 1990;61:1157–1160. doi: 10.1016/0092-8674(90)90674-4. [DOI] [PubMed] [Google Scholar]
- 3.Krause M, Hirsh D. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zorio D A R, Cheng N N, Blumenthal T, Spieth J. Nature (London) 1994;372:270–272. doi: 10.1038/372270a0. [DOI] [PubMed] [Google Scholar]
- 5.Huang X-Y, Hirsh D. Proc Natl Acad Sci USA. 1989;86:8640–8644. doi: 10.1073/pnas.86.22.8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spieth J, Brooke G, Kuerston S, Lea K, Blumenthal T. Cell. 1993;73:521–532. doi: 10.1016/0092-8674(93)90139-h. [DOI] [PubMed] [Google Scholar]
- 7.Blumenthal T. Trends Genet. 1995;11:132–136. doi: 10.1016/s0168-9525(00)89026-5. [DOI] [PubMed] [Google Scholar]
- 8.Ross L H, Freedman J H, Rubin C S. J Biol Chem. 1995;270:22066–22075. doi: 10.1074/jbc.270.37.22066. [DOI] [PubMed] [Google Scholar]
- 9.Thomas J D, Conrad R C, Blumenthal T. Cell. 1988;54:533–539. doi: 10.1016/0092-8674(88)90075-x. [DOI] [PubMed] [Google Scholar]
- 10.Van Doren K, Hirsh D. Nature (London) 1988;335:556–559. doi: 10.1038/335556a0. [DOI] [PubMed] [Google Scholar]
- 11.Bruzik J, Van Doren K, Hirsh D, Steitz J. Nature (London) 1988;335:559–562. doi: 10.1038/335559a0. [DOI] [PubMed] [Google Scholar]
- 12.Van Doren K, Hirsh D. Mol Cell Biol. 1990;10:1769–1772. doi: 10.1128/mcb.10.4.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenbaum N L, Radhakrishnan I, Patel D J, Hirsh D. Structure. 1996;4:725–733. doi: 10.1016/s0969-2126(96)00078-0. [DOI] [PubMed] [Google Scholar]
- 14.Hannon G J, Maroney P A, Ayers D G, Shambaugh J D, Nilsen T W. EMBO J. 1990a;9:1915–1921. doi: 10.1002/j.1460-2075.1990.tb08318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hannon G J, Maroney P A, Denker J A, Nilsen T W. Cell. 1990b;61:1247–1255. doi: 10.1016/0092-8674(90)90689-c. [DOI] [PubMed] [Google Scholar]
- 16.Maroney P A, Hannon G J, Shambaugh J D, Nilsen T W. EMBO J. 1991;10:3869–3875. doi: 10.1002/j.1460-2075.1991.tb04956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lücke S, Xu G-L, Paifi Z, Cross M, Bellofatto V, Bindereif A. EMBO J. 1996;15:4380–4391. [PMC free article] [PubMed] [Google Scholar]
- 18.Ferguson K C, Heid P J, Rothman J H. Genes Dev. 1996;10:1543–1556. doi: 10.1101/gad.10.12.1543. [DOI] [PubMed] [Google Scholar]
- 19.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mello C, Fire A. In: Caenorhabditis elegans: Modern biological analysis of an organism. Epstein H F, Shakes D C, editors. San Diego: Academic; 1995. pp. 451–482. [Google Scholar]
- 21.Singer-Sam J, LeBon J M, Dai A, Riggs A D. PCR Methods Appl. 1992;1:160–163. doi: 10.1101/gr.1.3.160. [DOI] [PubMed] [Google Scholar]
- 22.Kennedy B P, Aamodt E J, Allen F L, Chung M A, Heschl M F P, McGhee J D. J Mol Biol. 1993;229:890–908. doi: 10.1006/jmbi.1993.1094. [DOI] [PubMed] [Google Scholar]
- 23.Greenbaum N L, Radhakrishnan I, Hirsh D, Patel D J. J Mol Biol. 1995;252:314–327. doi: 10.1006/jmbi.1995.0499. [DOI] [PubMed] [Google Scholar]
- 24.Maroney P A, Hannon G J, Nilsen T W. Proc Natl Acad Sci USA. 1990;87:709–713. doi: 10.1073/pnas.87.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romaniuk P J, de Stevenson I L, Wong H-H A. Nucleic Acids Res. 1987;15:2737–2755. doi: 10.1093/nar/15.6.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu H-N, Uhlenbeck O C. Biochemistry. 1987;26:8221–8227. doi: 10.1021/bi00399a030. [DOI] [PubMed] [Google Scholar]
- 27.Summer-Smith M, Roy S, Barnett R, Reid L S, Kuperman R, Pelling U, Sonenberg N. J Virol. 1991;65:5196–5202. doi: 10.1128/jvi.65.10.5196-5202.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]