Abstract
We present evidence that Escherichia coli RNA polymerase β subunit may be a transcriptional activator contact site. Stimulation of the activity of the pR promoter by DnaA protein is necessary for replication of plasmids derived from bacteriophage λ. We found that DnaA activates the pR promoter in vitro. Particular mutations in the rpoB gene were able to suppress negative effects that certain dnaA mutations had on the replication of λ plasmids; this suppression was allele-specific. When a potential DnaA-binding sequence located several base pairs downstream of the pR promoter was scrambled by in vitro mutagenesis, the pR promoter was no longer activated by DnaA both in vivo and in vitro. Therefore, we conclude that DnaA may contact the β subunit of RNA polymerase during activation of the pR promoter. A new classification of prokaryotic transcriptional activators is proposed.
Activation of transcription is a common way to regulate gene expression in both prokaryotes and eukaryotes (for reviews, see refs. 1 and 2). In bacterial cells, transcription activation at a given promoter is achieved usually by a direct contact between a transcriptional activator and RNA polymerase. Until now, there are four known activator contact sites on the Escherichia coli RNA polymerase holoenzyme: the C-terminal domain of the α subunit (for reviews, see refs. 3 and 4), the N-terminal domain of the α subunit (ref. 5; for a review, see ref. 6), the σ70 subunit (ref. 7; for a review, see ref. 2), and the β′ subunit (8). It was also reported that the σ54 subunit of RNA polymerase may be the contact site during transcription activation from distant sites known as prokaryotic enhancers (for review, see ref. 2).
The initiation of replication of plasmids derived from bacteriophage λ, known as λ plasmids (a map of the λ replication region present in standard λ plasmids is presented in Fig. 1), requires transcription at or near the origin of replication oriλ (for a review, see ref. 9). Transcription starting from the pR promoter and provides mRNA for production of the replication proteins O and P. Moreover, this transcription serves in the so called transcriptional activation of oriλ. It seems that the main regulatory role in the initiation of λ plasmid replication is played by the transcriptional activation of oriλ rather than by binding of the initiator protein O to oriλ (10–15). Therefore, regulation of the activity of the pR promoter is crucial for the control of λ plasmid DNA replication.
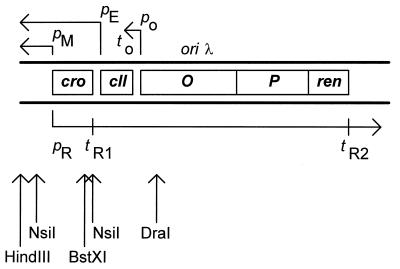
Figure 1.
Map of bacteriophage λ replication region present in standard λ plasmids (also those used in this work). Genes cro, cII, O, P, and ren are marked (these plasmids do not contain a gene coding for the CI repressor). All promoters (p) and terminators (t) are indicated. The pR promoter region contains the operator sites, and the pM promoter (whose activity could potentially play a role in transcription initiated at pR) is repressed by the cro gene product and cannot be activated in the absence of the CI protein. Transcripts are presented as arrows with arrowheads indicating directionality of transcription. Origin of λ DNA replication (oriλ) is located in the middle of the O gene. Restriction sites for HindIII, NsiI, BstXI, and DraI endonucleases (used for preparation of DNA fragments for templates in in vitro transcription reactions) are indicated.
It has been reported that E. coli DnaA protein is important for replication of bacteriophage λ (16) and λ plasmids (17, 18). Subsequent studies demonstrated that transcription starting at pR and proceeding through oriλ is depressed in certain dnaA mutants, which led to the conclusion that this promoter is positively regulated by DnaA (19). The above mentioned conclusion (18) were strengthened by observations that wild-type λ plasmids cannot replicate in certain temperature-sensitive dnaA mutants even at temperatures permissive for bacterial growth (30 or 37°C) and that this defect may be suppressed by a mutation of the π type in λ P gene. It was proposed that transcriptional activation of oriλ is coupled with the chaperone-mediated rearrangement of the λ preprimosomal complex (liberation of DnaB from λ P inhibition) and insertion of the preprimosome between transiently separated λ DNA strands (16, 18). Because the product of the P gene harboring a π mutation interacts significantly weaker with the host DnaB helicase than does the wild-type P protein (20), it was proposed that impaired transcriptional activation of oriλ observed in dnaA mutants is able to promote the coupled reaction only when λ plasmid harbors the π mutation (18).
In this work we investigated the mechanism of DnaA-mediated activation of the pR promoter. We found that DnaA directly activates the pR promoter in vitro. By using a simple assay that measures ability of λ plasmid DNA to replicate in E. coli, we found that the effect of certain dnaA mutations on λ plasmid replication can be suppressed by particular rpoB alleles. When a potential DnaA-binding sequence located several base pairs downstream of the pR promoter was scrambled by in vitro mutagenesis, the pR promoter was no longer activated by DnaA both in vivo and in vitro. Thus, on the basis of the allele-specific suppression and location of the DnaA-binding site important for transcription activation relative to the RNA polymerase binding site at pR, we conclude that the β subunit of RNA polymerase may be a transcriptional activator contact site. This and results reported by others (2–8) indicate that each subunit of RNA polymerase may interact with certain transcriptional activators during positive regulation of particular promoters.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Fusions.
A series of otherwise isogenic E. coli K-12 strains bearing either wild-type dnaA gene (CM732) or temperature-sensitive mutations in dnaA, dnaA46 (CM742), dnaA204 (CM746), and dnaA508 (CM2555), was described by Hansen and von Meyenburg (21) and Hansen et al. (22) (see also ref. 18). The dnaA46 allele specifies nucleotide changes C551 → T and C754 → T that result in Ala184Val and His252Tyr substitutions in the DnaA protein, respectively; dnaA204 allele specifies a nucleotide change T1166 → A that results in the Ile389Asn substitution in the DnaA protein; and dnaA508 allele specifies nucleotide changes C83 → T and C239 → T that result in Pro28Leu and Thr80Ile substitutions in the DnaA protein, respectively. Strain TC3478 bearing a dnaA-null allele (dnaA∷cm) was described by Ingmer and Atlung (23). The rpoB902 (TC3145), rpoB903 (TC1432), and rpoB904 (TC1404) mutant strains were described by Atlung (24). All of these rpoB mutations result in amino acid substitutions in the region of the β subunit of RNA polymerase responsible for rifampicin binding (F. Hansen, personal communication), and cause rifampicin-resistant phenotypes. For construction of strains harboring different combinations of dnaA and rpoB alleles, appropriate alleles were transferred by P1 transduction (25) into a CM732 genetic background. Plasmids pKB2 (17) and pCB104 (26) are wild-type λ plasmids harboring a kanamycin-resistance gene and a chloramphenicol-resistance gene, respectively. Plasmid pKB2π is a pKB2-derivative bearing the π422 mutation (17, 18). Plasmid pAW6 is a pCB104-derivative bearing the πA66 mutation (27). The previously described pR–lacZ fusion (13) and a newly constructed pR–lacZ fusion (pR–HG) in the pHG86 vector (28) were used. The pR–HG fusion was constructed by cloning the PCR fragment encompassing the bacteriophage λ pR promoter (amplified by using the external primers described below and cut with HindIII and BglI) into the HindIII–BamHI sites of pHG86. The DnaA box located immediately downstream of the pR promoter was scrambled in both fusions by PCR mutagenesis according to Langer et al. (29). Briefly, two PCRs were used for the separate amplification of two overlapping DNA fragments. External primers (i.e., primers flanking the whole amplified region, described below) and internal primers (described below) were used. Primers carrying the mutation (so called internal primers) were complementary to the same DNA fragment (each to a different DNA strand) at the region of desired mutation. Separately amplified DNA fragments were denatured, mixed, and cooled-down slowly to produce a double-stranded DNA fragment in the overlapping region. T4 DNA polymerase (1 unit) and all four dNTPs (each at 200 mM) were added to produce whole double-stranded DNA fragments. These fragments were then used as templates in PCRs with external primers. After final amplification, the DNA fragments were cut with HindIII and BglI and introduced into HindIII–BamHI sites of either pTL61T (30) or pHG86. All PCR amplifications were performed with following program: denaturation at 94°C for 2 min; 5 cycles of denaturation at 94°C for 15 sec, annealing at 58°C for 30 sec, and extension at 72°C for 45 sec; and then 35 cycles of denaturation at 94°C for 15 sec, annealing at 52°C for 25 sec, and extension at 72°C for 30 sec. The following primers were used: external primers, 5′-TTGAATTCATACGTTAAATCTATCACCGCA-3′ and 5′-GGCGACGTGCGTCCTCAAGCTGCTCTT-3′; internal primers, 5′-GTACTAACGAGTCTATAGCTAACAACGCATA-3′ and 5′-TATGCGTTGTTAGCTATAGACTCCTTAGTAC-3′.
Proteins.
DnaA protein was purified as described (31) (this purification was performed in Walter Messer’s laboratory at the Max-Planck Institute for Molecular Genetics, Berlin, Germany). RNA polymerase holoenzyme saturated with σ70 subunit was purchased from Epicentre Technologies (Madison, WI).
Transformation Efficiency.
Efficiency of transformation of E. coli strains with various λ plasmids was measured as described (18).
β-Galactosidase Activity.
Activity of β-galactosidase was measured as described by Miller (32).
In Vitro Transcription.
The in vitro transcription reaction was performed in total volume of 25 μl, in the buffer M [20 mM Hepes, pH 8.0/5 mM magnesium acetate/4 mM DTT/1 mM EDTA/1 mM ATP/BSA (5 mg/ml)/0.2% Triton X-100/5% glycerol]. Template DNA at 0.5 μg and DnaA protein at the amounts indicated were used. The binding reaction of DnaA to the template was carried out for 5 min at 37°C. Samples were then transferred into ice bath and 1 unit of RNA polymerase (Epicentre Technologies) was added. After addition of nucleotides (CTP and GTP to a final concentration of 150 μM, ATP to 1 mM, UTP to 15 μM, and [α-32P]UTP to 1 mCi/ml; 1 Ci = 37 GBq), the samples were incubated at 37°C for 12.5 min and the reaction was stopped by addition of equal volume of BSA (1.2 mg/ml)/0.1 mM EDTA, pH 8.0/5.1 M ammonium acetate and immediately transferred into ice bath. RNA was precipited with 2 vol of 96% ethanol in liquid nitrogen. The samples were centrifuged in the Microcentrifuge at maximal speed for 30 min, and the pellet was dried, resuspended in 20 μl of 98% formamide/0.25% bromophenol blue/0.25% xylene cyanol, and incubated at 65°C for 5 min. The samples were electrophoresed in a 6% polyacrylamide gel containing 46% urea in the TBE buffer (33) at 30 mA. The gel was dried, and RNA bands were visualized by autoradiography and quantified by densitometry.
RESULTS
DnaA-Mediated Activation of pR in Vitro.
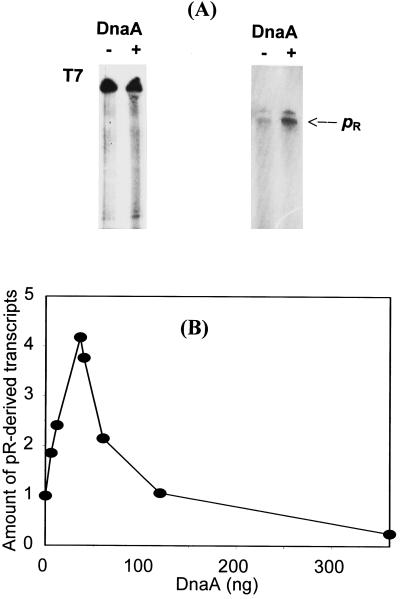
Activity of the bacteriophage λ pR promoter was found to be decreased in certain E. coli dnaA mutants (19). To determine whether this effect is from the direct activation of pR by DnaA protein or is the result of an indirect effect of dnaA mutations, we investigated in vitro transcription from this promoter in the presence and absence of DnaA. We found that pR activity increases in the presence of DnaA protein (Fig. 2). Addition of large amounts of DnaA resulted not only in lower activation of pR but also in repression of this promoter (this effect will be discussed below). Nevertheless, results presented in Fig. 2 indicate that DnaA directly activates the pR promoter.
Figure 2.
Activation of the pR promoter by DnaA protein in vitro. (A) Examples of autoradiograms after in vitro transcription experiments. (Left) In the control, 0.5 μg of bacteriophage T7 DNA was used as a template. (Right) In the experiment, 0.5 μg of a HindIII–DraI fragment of plasmid pKB2 was used as a template DNA. The reaction mixtures contained no DnaA (lanes −) or 35 ng (final concentration, 26.6 nM) of DnaA (lanes +). The position of pR-derived transcripts (812 nucleotides long) is indicated. (B) Average data from six experiments of in vitro transcription. Template DNA (0.5 μg) and DnaA protein (as indicated) were used, and appropriate bands on the autoradiograms were quantified by densitometry. Results very similar to those obtained with the HindIII–DraI fragment of pKB2 were observed when NsiI–NsiI fragment of pKB2 or whole pKB2 plasmid linearized with BstXI were used as DNA templates (the lengths of pR-derived transcripts were 288 and 269 nucleotides, respectively). The positions of appropriate restriction sites in λ plasmid DNA are depicted in Fig. 1.
Replication of λ Plasmids in E. coli dnaA and rpoB Mutants.
Because DnaA directly activates the pR promoter, we wanted mutants that could suppress the effects of dnaA mutations. It had been demonstrated that wild-type λ plasmids cannot replicate in dnaA46, dnaA204, and dnaA508 mutants even at 30 and 37°C (temperatures permissive for their growth) due to impaired pR-initiated transcriptional activation of oriλ (18). We found that replication of these plasmids is possible in dnaA mutants when they contain an additional mutation in the rpoB gene, coding for the β subunit of RNA polymerase (Table 1). In fact, these rpoB mutants were isolated long ago as suppressors of certain dnaA alleles that caused bacterial growth to be thermosensitive (24). The suppression of λ plasmid replication was allele-specific; i.e., λ plasmids could be introduced only to dnaA46 rpoB902, dnaA204 rpoB903, and dnaA508 rpoB904 strains; other combinations of dnaA and rpoB mutant alleles were not permissive for λ plasmid replication (Table 1 and data not shown). It is interesting that with the dnaA+ rpoB902 host, we observed a significant decrease in the efficiency of transformation by λ plasmids compared to the dnaA+ rpoB+ strain. In control experiments we observed transformation of all tested strains by λ plasmids bearing π422 or πA66 mutation (data not shown). These results may suggest a direct interaction between DnaA and RNA polymerase β subunit during activation of the pR promoter.
Table 1.
Efficiency of transformation of various dnaA and rpoB mutants with wild-type λ plasmids (pKB2 and pCB104) at 30°C
| Host | Efficiency of transformation
|
|
|---|---|---|
| pKB2 | pCB104 | |
| dnaA+rpoB+ | 1 × 105 | 7 × 104 |
| dnaA46 rpoB+ | <1 × 101 | <1 × 101 |
| dnaA+ rpoB902 | 4 × 102 | 7 × 102 |
| dnaA46 rpoB902 | 6 × 104 | 5 × 104 |
| dnaA204 rpoB+ | <1 × 101 | <1 × 101 |
| dnaA204 rpoB902 | <1 × 101 | <1 × 101 |
| dnaA204 rpoB903 | 1 × 105 | 1 × 105 |
| dnaA508 rpoB+ | <1 × 101 | NT |
| dnaA508 rpoB902 | <1 × 101 | NT |
| dnaA508 rpoB904 | 6 × 104 | NT |
Efficiency of transformation is presented as number of transformants per 1 μg of plasmid DNA. NT denotes “not tested” due to the fact that strain CM2555 (dnaA508) and its derivatives are sensitive to chloramphenicol even when containing a plasmid bearing the cat gene (18), like pCB104.
Identification of the DnaA Box Responsible for DnaA-Mediated Activation of pR.
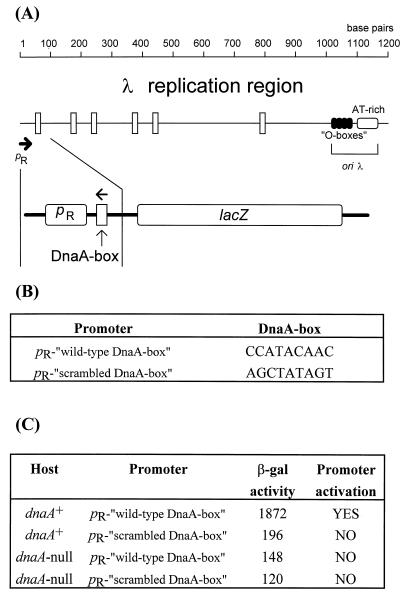
DnaA protein binds to 9-nucleotide-long DNA sequences called DnaA boxes. There are more or less stringent DnaA-box consensus sequences proposed on the basis of results obtained with different experimental systems (for a review, see ref. 34). When we subjected the fragment of λ DNA present in λ plasmids used in this work to a computer-mediated search, we found several potential DnaA-box sequences of the relaxed consensus 5′-YYHTMCRHM (where Y is T or C; H is A, T, or C; M is A or C; and R is A or G), defined by Schaefer and Messer (35) between the pR promoter and the oriλ region (Fig. 3). To identify the DnaA box important for DnaA-mediated activation of pR, we used in vitro mutagenesis to replace the wild-type DnaA-binding region with a scrambled DnaA box (Fig. 3), containing a sequence that was completely unable to bind DnaA protein (29). From the locations of all potential DnaA-binding sequences in the λ replication region, we presumed that if pR were activated by DnaA from a nearby site, the only DnaA box that could ensure direct contact of DnaA protein with RNA polymerase was that placed immediately downstream of the promoter (the center of this DnaA box is at position +18 relative to the pR transcription start site). We constructed pR–lacZ fusions containing either the wild-type sequence or the scrambled DnaA box. We found that the wild-type fusion was active in the dnaA+ host, but its activity was significantly decreased in the dnaA-null strain (Fig. 3). We conclude that because of the lack of DnaA-mediated activation of pR in the dnaA-null host, the latter result reflects the residual activity of the pR promoter. When the fusion containing the scrambled DnaA box located just downstream of the pR promoter was used, we observed a low activity of the fusion in both dnaA+ and dnaA-null hosts (Fig. 3). The residual activity of the pR promoter was only slightly decreased with this fusion relative to the fusion containing the wild-type sequence. These results indicate that the DnaA-box sequence whose center is at nucleotide position +18 relative to the pR transcription start site is crucial during activation of the pR promoter by DnaA protein.
Figure 3.
(A Upper) Map of the replication region of bacteriophage λ (also present in λ plasmids) from the pR promoter to oriλ. The scale is given in base pairs, the pR promoter is marked by the thick arrow, putative DnaA boxes are represented by vertical rectangles, and sequences recognized by the O replication initiator (O boxes) and the A+T-rich region are also indicated. (A Lower) The fragment of λ DNA present in the lacZ fusions is indicated, and the orientation of the DnaA-box is marked by arrow (this part of the figure is not drawn to scale). (B) Sequences of the wild-type and scrambled DnaA boxes present in appropriate fusions. (C) The activity of β-galactosidase per single copy of the fusion is presented (in a table form) for both fusions in dnaA+ and dnaA-null hosts. It seems that pR is activated only in the wild-type host harboring the “wild-type” fusion (YES in the table) and that the lack of either functional DnaA protein or DnaA-binding site immediately downstream of pR results in no activation of the promoter (NO in the table), and the obtained values reflect residual pR activity.
Allele-Specific Suppression of dnaA Mutations by rpoB Mutations During Activation of the pR Promoter in Vivo.
We measured the activation of the pR promoter in different dnaA and rpoB mutants by using the pR–lacZ fusions described above. The activity of the wild-type fusion in dnaA+ rpoB+ strain was considered as 100% activation, and the activity of the same fusion but bearing the scrambled DnaA box (whose activity should reflect residual activity of the pR promoter) was considered as 0% activation. With this system we found that the activation of the pR promoter is impaired in certain dnaA mutants, but this defect can be suppressed in the allele-specific manner by particular rpoB mutations (Table 2). The allele specificity of this suppression was exactly the same as that observed for λ plasmid replication (Table 1); i.e., dnaA46 was suppressed only by rpoB902, dnaA204 was suppressed only by rpoB903, and dnaA508 was suppressed only by rpoB904 (Table 2 and data not shown). These data support the proposal that DnaA and the RNA polymerase β subunit may interact during pR activation.
Table 2.
Activation of the pR promoter in various dnaA and rpoB mutants
| Host | Activation of the pR promoter |
|---|---|
| dnaA+ rpoB+ | +++ |
| dnaA46 rpoB+ | − |
| dnaA+ rpoB902 | + |
| dnaA46 rpoB902 | ++ |
| dnaA204 rpoB+ | − |
| dnaA204 rpoB902 | − |
| dnaA204 rpoB903 | ++ |
| dnaA508 rpoB+ | − |
| dnaA508 rpoB902 | − |
| dnaA508 rpoB904 | +++ |
Activation of the pR promoter was estimated on the basis of β-galactosidase activities measured in strains harboring fusions that consist of the lacZ gene under control of either pR–“wild-type DnaA box” or pR–“scrambled DnaA box” (see Fig. 2 for details). The value obtained with the wild-type fusion in the dnaA+ rpoB+ host (see Fig. 2) was considered as maximal (100%) activation, the value obtained with the fusion bearing the scrambled DnaA box was considered as no (0%) activation, and the values depicted in the table reflect these values (for each strain, activity of both fusions was measured and symbols presented in the table represent the activation of the wild-type fusion relative to the maximal activation). +++, 75–100% of maximal activation; ++, 50–75% of maximal activation; +, 25–50% of maximal activation; −, 0–25% of maximal activation. The experiments were performed at 30°C and 37°C, and very similar results were obtained at both temperatures.
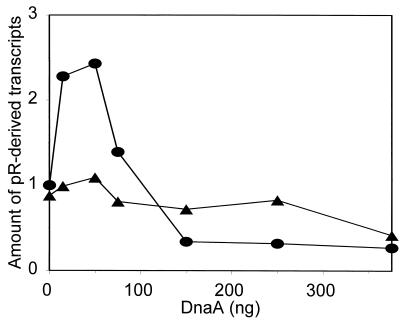
pR Does Not Respond to DnaA in Vitro in the Absence of the DnaA Box Downstream of the Promoter.
We investigated a role for the DnaA box located immediately downstream of pR in DnaA-mediated activation of the promoter in vitro. As templates we used DNAs derived from plasmids harboring pR and either the wild-type or scrambled DnaA-box sequences. We found that removal of the DnaA-binding sequence prevents DnaA-mediated activation of the pR promoter (Fig. 4). These results once again support the conclusions presented above, based on in vivo experiments.
Figure 4.
Activation of the pR promoter by DnaA protein in vitro in the presence (•) and absence (▴) of the DnaA-binding sequence downstream of the promoter. Template DNA (0.5 μg) and indicated amounts of DnaA protein were used. Similar results were obtained when different templates, all derived from pR–HG and its analogue bearing the scrambled DnaA box, were used: EcoRI–Bsu36I fragments, the plasmids linearized with Bsu36I, and the plasmids linearized with ClaI (the lengths of pR-derived transcripts were 512, 512 and 1112 nucleotides, respectively). EcoRI site is located upstream of pR and Bsu36I and ClaI sites are located in the lacZ gene of plasmid pR–HG and its analogue. Average data from seven experiments are presented; appropriate bands on the autoradiograms were quantified by densitometry.
DISCUSSION
We have demonstrated that E. coli DnaA protein directly activates the bacteriophage λ pR promoter, because it stimulates transcription from this promoter in vitro and the activity of a pR–lacZ fusion was decreased in the dnaA mutants. One might assume that DnaA could not only activate the initiation of transcription at pR but also affect transcriptional termination or antitermination. In fact, it was reported that DnaA can significantly influence transcription termination in vivo (35). However, a hypothesis that DnaA may stimulate pR-initiated transcription by inhibition of termination or enhancement of antitermination at the tR1 terminator is unlikely to be true at least in our experiments with the pR–lacZ fusions and in vitro transcription because the fusions did not contain the tR1 terminator (compare Figs. 1 and 3) and the N antitermination protein (absolutely required for antitermination at tR1) was absent in our in vitro reactions. Although it is theoretically possible that DnaA could act as an antitermination factor in manner similar to that of the phage λ Q protein at the pR′ promoter (the binding site for Q is just downstream of the pR′ promoter), such a type of regulation of the pR activity has not been observed to date.
The activation of pR, which stimulates the process of transcriptional activation of oriλ controlling frequency of λ plasmid replication initiation, is impaired in certain dnaA mutants but the effect of these mutations can be suppressed in an allele-specific manner by particular alleles of the rpoB gene, coding for the β subunit of RNA polymerase. We identified the DnaA-box sequence that is necessary for activation of pR by DnaA both in vivo and in vitro. Nevertheless, we cannot exclude a role for other DnaA-box sequences present in the λ replication region (see Fig. 3) in the regulation of the pR promoter activity and in the regulation of λ plasmid replication. Both allele-specific suppression of dnaA mutations by rpoB mutations and location of the DnaA-box sequence required for the activation close to and downstream of the promoter led us to the conclusion that RNA polymerase β subunit is a contact site for DnaA to act as a transcriptional activator at pR (Fig. 5). In fact, a direct contact between DnaA and RNA polymerase was hypothesized some time ago when the allele-specific suppression of temperature-sensitive growth in certain dnaA mutants by rpoB mutations was discovered (24); however, the β subunit was not considered as a transcriptional activator contact site. Recent reports (as summarized in ref. 36) indicate that DnaA, besides its crucial role in the initiation of E. coli chromosome replication, acts as a transcriptional activator not only at pR but also at some other promoters. However, putative DnaA-binding sequences were found upstream of these promoters rather than downstream (36). Nevertheless, it is tempting to speculate that impaired DnaA-mediated stimulation of transcription at certain promoters may be at least a part of the mechanism of temperature sensitivity in dnaA mutants, a phenotype that can be suppressed by particular rpoB mutations. On the other hand, the same pattern of rpoB-mediated suppression of dnaA mutations revealed by the bacteriophage λ pR promoter activation leading to transcriptional activation of oriλ (this work) and by the growth of E. coli dnaA mutants at the otherwise restrictive temperature (24) is striking. It suggests that transcriptional activation of oriC may occur through a molecular mechanism very similar to that found in λ plasmids.
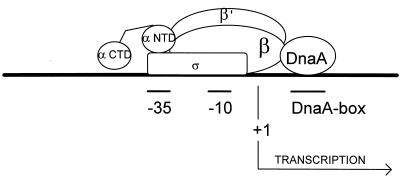
Figure 5.
Model for activation of the pR promoter by DnaA protein that assume a direct interaction between DnaA and RNA polymerase β subunit. The regions −35 and −10 of the promoter and location of the DnaA box important for the activation are marked. The scheme is not drawn to scale.
The location of the transcriptional activator binding site downstream of the promoter, as for DnaA and pR, is rather unusual in prokaryotes. This allows for direct interaction between DnaA and RNA polymerase β subunit (Fig. 5), but on the other hand, it might result in creating a potential problem during initiation of transcription because a protein bound downstream of the promoter could be a barrier for RNA polymerase movement. Therefore, one may predict that binding of the activator downstream of the promoter should be relatively weak, so that removal of the activator could be possible after transcription initiation. In fact, DnaA binds weakly to the DnaA-box sequence located downstream of pR because we did not observe any significant DnaA-mediated protection of this sequence during a DNase I footprinting analysis and only a weak retardation of the DNA fragment containing pR and the downstream DnaA box was found during gel electrophoresis in the presence of DnaA protein (A.S.-P., C. Weigel, G.W., C. Speck, J. Marszałek, W. Messer, and K.T., unpublished results). According to the above explanation, we found that addition of excess of DnaA to the in vitro transcription reaction resulted in repression of pR rather than activation (Figs. 2 and 4), probably due to halting of RNA polymerase movement. In the light of this proposal, it is worth noticing that although DnaA-mediated activation of the pR promoter devoid of the downstream DnaA-binding site is abolished, this construct is significantly less sensitive to repression of transcription by excess DnaA protein in vitro than is the wild-type pR promoter (Fig. 4).
Until now, the following four activation targets had been identified on the E. coli RNA polymerase holoenzyme: the C-terminal domain of the α subunit (for a review, see ref. 4), the N-terminal domain of the α subunit (for a review, see ref. 6), the σ70 subunit (for a review, see ref. 2), and the β′ subunit (8). Herein we demonstrate that the β subunit may also be a transcriptional activator contact site, indicating that each RNA polymerase subunit may be an activation target. This is in accordance with recent prediction of Niu et al. (5) that nearly any exposed surface of the transcription machinery can serve as a target for an activator protein.
The classification of prokaryotic transcriptional activators that is currently used is based mainly on the localization of the activator binding site on DNA relative to the transcription start site (ref. 37, but see also ref. 6). Generally, class I activators bind to DNA upstream of the −35 region of the target promoter, and class II activators bind at the core promoter (the binding site overlaps the −35 sequence). However, in the light of additional data, this classification is not broad enough to group all known transcriptional activators. For example, DnaA seems to bind downstream of the target promoter pR (this work), and bacteriophage N4 SSB protein can activate transcription at the late phage promoters (through interaction with RNA polymerase β′ subunit) even without DNA binding (8). Therefore, we propose a new simple classification of prokaryotic transcriptional activators based on the RNA polymerase subunit with which the activator interacts. We propose to group activators interacting with the α subunit (the rpoA gene product) into class A, activators interacting with the β subunit (the rpoB gene product) into class B, activators interacting with the β′ subunit (the rpoC gene product) into class C, and activators interacting with the σ70 subunit (the rpoD gene product) into class D. This classification is open, and new classes of activators can be introduced; for example, if an activator interacting with the σ32 subunit (the rpoH gene product) were discovered, it should belong to class H. Moreover, if an activator interacting with two RNA polymerase subunits, for example α and σ70, were considered, it would be called class AD activator. The new classification should not replace the old one, especially, because promoters positively regulated by CAP–cAMP acting as class I or class II activator are called class I or class II promoters, respectively, and recent reports indicate that the old classification may be based not only on the position of the DNA site for an activator but also on the corresponding mechanisms for transcription activation (6). Thus, both classifications can be used independently, especially because the names of the classes in these two systems are different.
Acknowledgments
This work was supported by the Polish State Committee for Scientific Research (Projects 6 P04A 051 08, 6 P04A 059 09, and 6 P04A 065 10).
References
- 1.Tjian R, Maniatis T. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 2.Gralla J D. Curr Opin Genet Dev. 1996;6:526–530. doi: 10.1016/s0959-437x(96)80079-7. [DOI] [PubMed] [Google Scholar]
- 3.Busby S, Ebright R H. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 4.Ebright R H, Busby S. Curr Opin Genet Dev. 1995;5:197–203. doi: 10.1016/0959-437x(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 5.Niu W, Kim Y, Tau G, Heyduk T, Ebright R H. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busby S, Ebright R H. Mol Microbiol. 1997;23:853–859. doi: 10.1046/j.1365-2958.1997.2771641.x. [DOI] [PubMed] [Google Scholar]
- 7.Li M, Moyle H, Susskind M M. Science. 1994;263:75–77. doi: 10.1126/science.8272867. [DOI] [PubMed] [Google Scholar]
- 8.Miller A, Wood D, Ebright R H, Rothman-Denes L B. Science. 1997;275:1655–1657. doi: 10.1126/science.275.5306.1655. [DOI] [PubMed] [Google Scholar]
- 9.Taylor K, Węgrzyn G. FEMS Microbiol Rev. 1995;17:109–119. doi: 10.1111/j.1574-6976.1995.tb00192.x. [DOI] [PubMed] [Google Scholar]
- 10.Szalewska A, Węgrzyn G, Taylor K. Mol Microbiol. 1994;13:469–474. doi: 10.1111/j.1365-2958.1994.tb00441.x. [DOI] [PubMed] [Google Scholar]
- 11.Szalewska-Pałasz A, Węgrzyn A, Herman A, Węgrzyn G. EMBO J. 1994;13:5779–5785. doi: 10.1002/j.1460-2075.1994.tb06916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szalewska-Pałasz A, Węgrzyn G. Biochem Biophys Res Commun. 1994;205:802–806. doi: 10.1006/bbrc.1994.2736. [DOI] [PubMed] [Google Scholar]
- 13.Szalewska-Pałasz A, Węgrzyn G. Acta Biochim Pol. 1995;42:233–240. [PubMed] [Google Scholar]
- 14.Węgrzyn A, Węgrzyn G. Biochem Biophys Res Commun. 1995;214:978–984. doi: 10.1006/bbrc.1995.2382. [DOI] [PubMed] [Google Scholar]
- 15.Węgrzyn A, Węgrzyn G, Herman A, Taylor K. Genes Cells. 1996;1:953–963. doi: 10.1046/j.1365-2443.1996.830283.x. [DOI] [PubMed] [Google Scholar]
- 16.Węgrzyn G, Węgrzyn A, Konieczny I, Bielawski K, Konopa G, Obuchowski M, Helinski D R, Taylor K. Genetics. 1995;139:1469–1481. doi: 10.1093/genetics/139.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kur J, Górska I, Taylor K. J Mol Biol. 1987;198:203–210. doi: 10.1016/0022-2836(87)90306-8. [DOI] [PubMed] [Google Scholar]
- 18.Węgrzyn G, Węgrzyn A, Pankiewicz A, Taylor K. Mol Gen Genet. 1996;252:580–586. doi: 10.1007/BF02172404. [DOI] [PubMed] [Google Scholar]
- 19.Węgrzyn G, Szalewska-Pałasz A, Węgrzyn A, Obuchowski M, Taylor K. Gene. 1995;154:47–50. doi: 10.1016/0378-1119(94)00849-n. [DOI] [PubMed] [Google Scholar]
- 20.Konieczny I, Marszałek J. J Biol Chem. 1995;270:9792–9799. doi: 10.1074/jbc.270.17.9792. [DOI] [PubMed] [Google Scholar]
- 21.Hansen F G, von Meyenburg K. Mol Gen Genet. 1979;175:135–144. doi: 10.1007/BF00425529. [DOI] [PubMed] [Google Scholar]
- 22.Hansen E B, Atlung T, Hansen F G, Skovgaard O, von Meyenburg K. Mol Gen Genet. 1984;196:387–396. doi: 10.1007/BF00436184. [DOI] [PubMed] [Google Scholar]
- 23.Ingmer H, Atlung T. Mol Gen Genet. 1992;232:431–439. doi: 10.1007/BF00266248. [DOI] [PubMed] [Google Scholar]
- 24.Atlung T. Mol Gen Genet. 1984;197:125–128. doi: 10.1007/BF00327932. [DOI] [PubMed] [Google Scholar]
- 25.Silhavy T J, Berman M L, Enquist L W. Experiments with Gene Fusions. Plainview, NY: Cold Spring Harbor Lab. Press; 1984. [Google Scholar]
- 26.Boyd A C, Sherratt D J. Gene. 1995;154:57–62. doi: 10.1016/0378-1119(94)00788-t. [DOI] [PubMed] [Google Scholar]
- 27.Węgrzyn A, Węgrzyn G, Taylor K. Mol Gen Genet. 1995;247:501–508. doi: 10.1007/BF00293153. [DOI] [PubMed] [Google Scholar]
- 28.Giladi H, Koby S, Gottesman M E, Oppenheim A B. J Mol Biol. 1992;224:937–948. doi: 10.1016/0022-2836(92)90461-r. [DOI] [PubMed] [Google Scholar]
- 29.Langer U, Richter S, Roth A, Weigel C, Messer W. Mol Microbiol. 1996;21:301–311. doi: 10.1046/j.1365-2958.1996.6481362.x. [DOI] [PubMed] [Google Scholar]
- 30.Linn T, Pierre R S. J Bacteriol. 1990;172:1077–1084. doi: 10.1128/jb.172.2.1077-1084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaper S, Messer W. J Biol Chem. 1995;270:17622–17626. doi: 10.1074/jbc.270.29.17622. [DOI] [PubMed] [Google Scholar]
- 32.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 34.Messer W, Weigel C. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R, Ingraham J III, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1579–1601. [Google Scholar]
- 35.Schaefer C, Messer W. Mol Gen Genet. 1991;226:34–40. doi: 10.1007/BF00273584. [DOI] [PubMed] [Google Scholar]
- 36.Messer W, Weigel C. Mol Microbiol. 1997;24:1–6. doi: 10.1046/j.1365-2958.1997.3171678.x. [DOI] [PubMed] [Google Scholar]
- 37.Ishihama A. J Bacteriol. 1993;175:2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]