Abstract
The imprinting control region (ICR) upstream of H19 is the key regulatory element conferring monoallelic expression on H19 and Igf2 (insulin-like growth factor 2). Epigenetic marks in the ICR regulate its interaction with the chromatin protein CCCTC-binding factor and with other control factors to coordinate gene silencing in the imprinting cluster. Here, we show that the H19 ICR is biallelically transcribed, producing both sense and antisense RNAs. We analyse the function of the non-coding transcripts in a Drosophila transgenic system in which the H19 upstream region silences the expression of a reporter gene. We show that knockdown of H19 ICR non-coding RNA (ncRNA) by RNA interference leads to the loss of reporter gene silencing. Our results are, to the best of our knowledge, the first to show that ncRNAs in the H19 ICR are functionally significant, and also indicate that they have a role in regulating gene expression and perhaps epigenetic marks at the H19/Igf2 locus.
Keywords: gene silencing, H19, imprinting control region, non-coding RNAs
Introduction
Genomic imprinting is an epigenetic phenomenon that results in the parent of origin-dependent monoallelic expression of a particular set of mammalian genes (Reik & Walter, 2001). The imprinted Igf2 (insulin-like growth factor 2) and H19 genes are expressed monoallelically from the paternal and maternal alleles, respectively. Two crucial regulatory elements control the monoallelic expression of H19 and Igf2 in endodermal tissues: a set of enhancers downstream of the H19 gene and the imprinting control region (ICR) upstream of H19 (Thorvaldsen et al, 1998). The H19 ICR is methylated on the paternal chromosome, whereas the insulator protein CCCTC-binding factor (CTCF) binds to the element on the maternal chromosome. CTCF establishes a chromatin boundary on the maternal allele, restricting the enhancer to act on the maternal H19 gene (Bell & Felsenfeld, 2000; Hark et al, 2000; Kaffer et al, 2000).
A growing body of evidence indicates an essential role for non-coding RNAs (ncRNAs) in the regulation of eukaryotic gene expression (Prasanth & Spector, 2007). The paternally expressed antisense Igf2r RNA (Air) and Kcnq1ot1 genes produce two ncRNAs with an essential role in genomic imprinting. Air represses the transcription of the paternal alleles of the imprinted genes Igf2r, Slc22a2 and Slc22a3 (Sleutels et al, 2002). Similarly, Kcnq1ot1 mediates transcriptional repression of multiple genes within the Kcnq1 imprinting locus (Mancini-DiNardo et al, 2006). Furthermore, in three other imprinting loci, deletion of ncRNA promoters leads to the loss of silencing of associated protein-coding genes, indicating that gene silencing mediated by ncRNAs might represent a widespread mechanism in imprinted clusters (Chamberlain & Brannan, 2001; Lin et al, 2003; Williamson et al, 2006). However, so far, the ICRs themselves have not been systematically analysed to see whether they are transcribed. This is of interest because several regulatory sequences in the genome, including polycomb response elements and locus control regions, are also transcribed and this is important for their function (Gribnau et al, 2000; Schmitt et al, 2005). Some preliminary observations indicate that there might be transcripts in the H19 ICR; however, they have not yet been characterized and nothing is known about their function (Drewell et al, 2002). Here, we show that in the mouse, the H19 ICR is biallelically transcribed, resulting in both sense and antisense transcripts. The non-coding H19 ICR transcripts are coexpressed with the H19 and Igf2 genes, and are retained in the nucleus. To address the potential function of the non-coding H19 ICR RNAs, we used a Drosophila transgenic H19 locus in which expression of the reporter gene mini-white is silenced by the H19 upstream region (Lyko et al, 1997). Remarkably, expression of an RNA interference (RNAi) hairpin construct directed against the transcribed H19 ICR relieved transcriptional repression of the reporter gene in the Drosophila transgene. Derepression of reporter gene transcription correlated with downregulation of the level of H19 ICR transcripts, indicating a role for ncRNAs in regulating gene silencing at the H19–Igf2 locus.
Results And Discussion
The H19 ICR produces sense and antisense transcripts
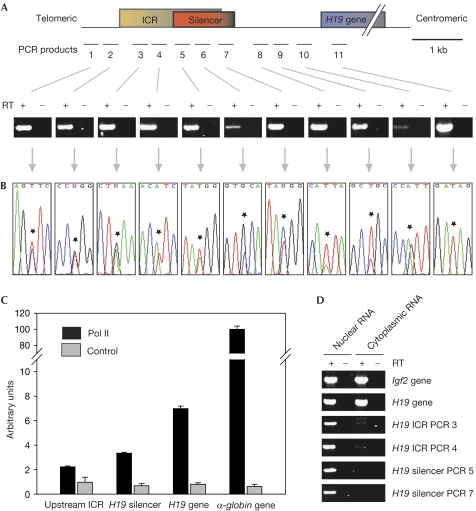
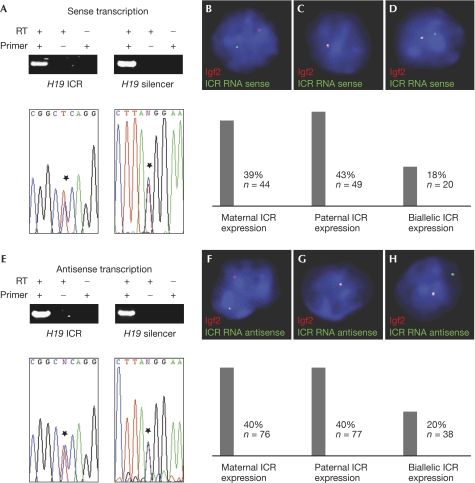
ICRs have been shown to harbour promoters for ncRNAs that control monoallelic gene expression (Smilinich et al, 1999; Sleutels et al, 2002). In addition, it has been estimated that more than 80% of all imprinted transcription units produce sense–antisense transcript pairs (Katayama et al, 2005). These findings prompted us to search for transcripts in the H19 ICR. Using reverse transcription reactions, we detected transcripts with all primer pairs that we had chosen to analyse the transcriptional status of the H19 upstream regulatory region, including those specific for the H19 ICR and the silencer region (Fig 1A). The RNA for the reverse transcription reactions was isolated from F1 generation fetal livers obtained from a cross between the C57Bl6 and SD7 strains, therefore single nucleotide polymorphisms between the strains allowed us to determine the parental origin of the transcripts. Sequencing of the complementary DNA PCR products showed that the H19 upstream region is biallelically transcribed (Fig 1B, PCRs 1–10), whereas a PCR product covering a part of the H19 transcribed region confirmed its monoallelic expression (Fig 1B, PCR 11). Next, we carried out quantitative chromatin immunoprecipitation (ChIP) experiments using an antibody raised against the transcribing form of RNA polymerase II (Pol II) to analyse Pol II occupancy within the H19 upstream region in mouse fetal liver cells. The Pol II antibody and an unspecific control antibody were used to pull down crosslinked fragments. The immunoprecipitates were then analysed using real-time PCR. We detected only a weak Pol II enrichment in the region upstream of the H19 ICR, whereas, surprisingly, the H19 silencer region was four- to fivefold enriched in the Pol II immunoprecipitates compared with the control immunoprecipitates (Fig 1C). Pol II binding to H19 silencer sequences was only twofold lower than to the H19 gene itself, and approximately 30-fold lower than to the highly transcribed α-globin gene. Next, we sought to determine the subcellular localization of the non-coding H19 ICR transcripts. We separated nuclear and cytoplasmic RNA fractions from mouse embryonic fibroblasts. Igf2 and H19 gene transcripts were found both in the nucleus and in the cytoplasm, whereas the ncICR transcripts were almost exclusively detected in the nuclear fraction (Fig 1D), which is consistent with a function in gene expression. We then addressed the question of the orientation of the transcripts by carrying out reverse transcription reactions with strand-specific primers in the cDNA synthesis. Sense and antisense transcripts were detected in the H19 ICR and the silencer region, originating from both parental chromosomes, as determined by sequencing of the cDNA PCR products (Fig 2A,E). To analyse whether the ncICR transcripts are expressed in the same cells as the genes encoding Igf2 and H19, we carried out double-labelled RNA fluorescence in situ hybridization (RNA-FISH) experiments on mouse fetal liver cells using strand-specific probes against the ncICR transcripts in combination with probes against the paternal Igf2 gene transcripts. Cells with monoallelic Igf2 expression and ICR transcripts were grouped into three categories: expression from the maternal chromosome (Fig 2B,F: one ICR signal non-overlapping with the Igf2 signal), expression from the paternal chromosome (Fig 2C,G: one ICR signal overlapping with the Igf2 signal) and biallelic expression (Fig 2D,H: two ICR signals, one of which overlaps with the Igf2 signal). Sense and antisense ICR transcripts were detected at comparable frequencies originating from both parental chromosomes. We confirmed these results by RNA-FISH experiments using probes against the ncICR transcripts in combination with a probe against the maternal H19 gene transcript (supplementary Fig 1 online). Together, our results indicate an unexpectedly high transcriptional activity within the H19 ICR.
Figure 1.
The mouse H19 upstream regulatory region is biallelically transcribed. (A) Schematic representation of the mouse H19 locus and the PCR amplicons used in reverse transcription reactions. The imprinting control region (ICR) extends from −4 to −2 kb in relation to the H19 gene transcription start site, overlapping with the silencer (−2.9 to −1.7 kb relative to the H19 transcription start site). PCR products shown below were amplified from complementary DNA obtained from an F1 generation fetal liver (embryonic day (E)15.5) of a C57Bl6/SD7 cross using random primers, with or without reverse transcriptase (RT). (B) Selected sequence traces of PCR products shown in (A). Asterisks indicate SNPs between the C57Bl6 and SD7 strains. (C) Pol II occupancy at the H19 region. Real-time PCR of ChIP material obtained with an antibody against Pol II or a control antibody. Sequences correspond to amplicons 2 (upstream ICR), 5 (H19 silencer) and 11 (H19 gene), and a control fragment from the α-globin gene. (D) Analysis of the subcellular localization of the ICR transcripts. RNA from mouse embryonic fibroblasts was fractionated and reverse transcription reactions were carried out on the nuclear and the cytoplasmic RNA pools, respectively. ChIP, chromatin immunoprecipitation; Pol II, RNA polymerase II; SNP, single-nucleotide polymorphism.
Figure 2.
The H19 imprinting control region produces sense and antisense transcripts. Reverse transcription reactions with RNA obtained from mouse embryonic day (E)15.5 fetal liver cells, using primers specific for the sense (A) or antisense (E) ICR transcripts. Complementary DNA synthesis reactions were carried out with or without reverse transcriptase (RT), and with or without strand-specific primers. PCR products correspond to amplicons 4 (H19 ICR) and 7 (H19 silencer) in Fig 1. Sequence traces of PCR products are shown below the gel pictures, with asterisks indicating SNPs. Detection of Igf2 in combination with ICR sense (B–D) and antisense (F–H) transcripts in mouse E14.5 fetal liver cells by double-labelled RNA-FISH. Green signals are ICR transcripts, red signals are Igf2 gene transcripts. DAPI staining is blue. Bars below the pictures show frequencies of the respective signals. DAPI, 4,6-diamidino-2-phenylindole; FISH, fluorescence in situ hybridization; ICR, imprinting control region; SNP, single-nucleotide polymorphism.
Transcription at a Drosophila H19 ICR transgene
To characterize the molecular function of the ncRNAs in the H19 ICR, we used a transgenic fly line carrying an H19 ICR transgene (Lyko et al, 1997). In this fly strain, expression of the eye colour reporter gene mini-white is controlled by the 3.8 kb H19 upstream regulatory element, including the H19 ICR. Flies carrying the H19 ICR transgene show a yellow eye colour, which indicates a partial silencing of mini-white. Using Drosophila genetics, we have previously identified a silencer element within the H19 ICR (Lyko et al, 1997). The H19 silencer represses transcription of reporter genes in Drosophila (Lyko et al, 1997). Deletion of the silencer from the endogenous mouse locus results in loss of silencing of the paternal H19 allele (Drewell et al, 2000). This indicates that the Drosophila transgene can uncover evolutionarily conserved regulatory elements of the mice H19 ICR. Here, we first investigated whether the Drosophila transgene recapitulates the transcriptional activity of the ICR at the endogenous locus. Indeed, we found that the H19 ICR is bidirectionally transcribed in H19 ICR transgenic Drosophila (supplementary Fig 2 online), mirroring the transcriptional activity at the endogenous mouse locus. The finding that both the silencing activity and the transcriptional status of the H19 upstream region are conserved between mice and transgenic Drosophila strongly indicates a common underlying epigenetic mechanism and enables us to use the Drosophila ICR transgene as a model system to study the molecular function of the non-coding H19 ICR transcripts.
Silencing at the H19 ICR transgene is RNAi-independent
RNAi pathways are known to mediate transcriptional gene silencing in the nucleus (Matzke & Birchler, 2005). However, despite the presence of sense and antisense RNAs, we were unable to detect small interfering RNAs (siRNA) from the H19 ICR, nor did mutations in RNAi pathway genes relieve H19 ICR-conferred reporter gene repression. This argues against an RNAi mechanism mediating transcriptional repression at the transgenic Drosophila H19 ICR locus (supplementary Fig 3 online).
RNA-mediated silencing at the transgenic H19 ICR locus
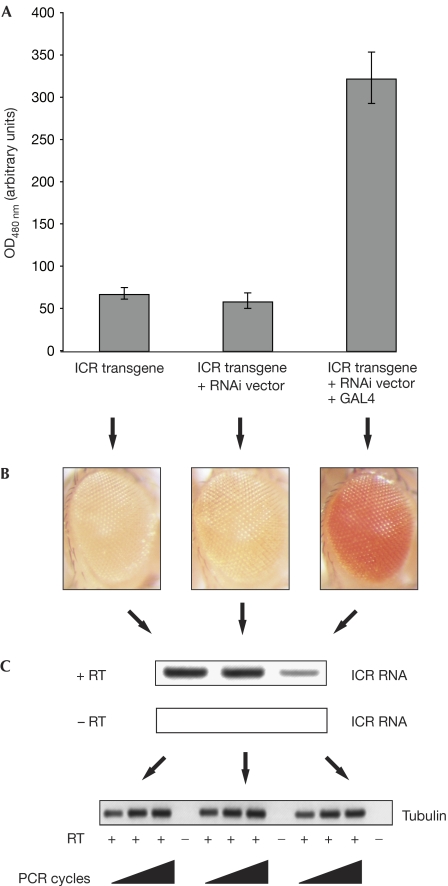
To eliminate the non-coding transcript in the ICR to potentially uncover its function, we decided to use the RNAi technique. We expressed a 430-bp fragment from the H19 ICR as an inverted repeat, which leads to the formation of a double-stranded RNA that is subsequently cleaved to siRNAs targeting the ncICR transcripts (supplementary Fig 3 online). We found that expression of the RNAi hairpin vector led to a substantially reduced level of H19 ICR transcripts (Fig 3C,D). Semiquantitative reverse transcription PCR (RT–PCR) showed that the level of H19 ICR RNA was reduced by more than 50% upon GAL4-induced expression of the RNAi vector, whereas it was not significantly reduced in H19 ICR transgenic flies in which expression from the RNAi vector was not induced (Fig 3C,D).
Figure 3.
RNA interference-mediated downregulation of H19 imprinting control region non-coding RNA leads to loss of reporter gene silencing. (A) Quantification of reporter gene expression by photometric measurement of eye colour pigmentation in Drosophila lines carrying the transgene combinations indicated below the respective bars. (B) Shown here are the eyes of the Drosophila lines carrying the H19 ICR transgene (left panel), the H19 ICR transgene combined with the RNAi vector (middle panel), and the H19 ICR transgene combined with the RNAi vector and GAL4 (right panel). (C) Reverse transcription (RT) reaction with primers in the H19 ICR with (+RT, upper panel) or without (−RT, lower panel) reverse transcriptase. RNA samples are obtained from heads of the respective Drosophila lines shown in (B). (D) Reverse transcription reaction with primers against β-tubulin. RNA samples used for the reverse transcription reaction are the same as in (C). ICR, imprinting control region; OD, optical density; RNAi, RNA interference.
Remarkably, expression of the RNAi hairpin construct directed against the transcribed H19 ICR in Drosophila relieves transcriptional repression of the reporter gene mini-white, which is controlled by the H19 ICR (Fig 3A,B). Flies carrying the H19 ICR transgene have a yellow eye colour, which indicates silencing of mini-white (Fig 3B, left). Combining the H19 ICR transgene with the RNAi vector transgene did not alter the expression of mini-white, as the RNAi vector transgene fails to be expressed in the absence of the transactivator GAL4 (Fig 3B, middle). However, on GAL4-induced expression of the RNAi transgene in flies carrying the H19 ICR transgene, mini-white silencing is lost resulting in orange- to red-eyed flies (Fig 3B, right). Quantification of mini-white expression by photometric pigment measurements indicated that expression of the RNAi vector leads to a more than fivefold increase of mini-white gene function. We showed that this effect is specific as it depends on the presence of the H19 ICR sequence targeted by the RNAi vector (supplementary Fig 4 online).
In conclusion, the transgenic expression of an RNAi vector directed against the transcribed region of the H19 ICR leads to the loss of silencing of an H19 ICR-controlled reporter gene in Drosophila. Furthermore, the loss of transcriptional repression correlates with a marked reduction in the level of RNA from the H19 ICR. These findings indicate an active role for non-coding transcripts from the H19 ICR in the transcriptional repression of a reporter gene controlled by the H19 upstream regulatory region in transgenic Drosophila.
The expression status of the transgenic locus in Drosophila is most comparable with that of the paternal allele in the mouse. Here, the H19 gene is repressed, which requires DNA methylation of the ICR. Importantly, even in the presence of methylation, deletion of the region that corresponds to the Drosophila silencer element (and is transcribed in mice as shown here) relieves repression of H19 (Drewell et al, 2000). Therefore, the ncRNAs might be required for heterochromatic silencing in conjunction with DNA methylation, involving potentially H3K9 methylation. Other ncRNAs that direct gene silencing to imprinted regions have recently been shown to attract repressive histone modifications to adjacent genes (Lewis et al, 2004; Umlauf et al, 2004). We have previously found that mutations in some genes encoding heterochromatin components, such as the histone methyltransferase Su(var)3–9, partly relieve ICR-mediated silencing of mini-white in ICR transgenic flies (Schoenfelder & Paro, 2004), indicating that the H19 ICR confers silencing to reporter genes through the formation of a heterochromatin-like structure that could be targeted by the ncRNAs (supplementary Fig 5 online). The recent observation of strikingly similar patterns of repressive histone marks at the endogenous paternal H19 ICR and at pericentric heterochromatin has highlighted the molecular parallels between both chromosomal regions (Delaval et al, 2007). Remarkably, an RNA component has been shown to contribute to the maintenance of the higher-order structure at pericentric heterochromatin (Maison et al, 2002). A related mechanism controls gene expression in a ribosomal gene cluster in which ncRNAs are required to maintain a silent, heterochromatin-like state (Mayer et al, 2006). Experimental downregulation of the repressive ncRNAs leads to increased expression of the neighbouring ribosomal genes in the locus (Mayer et al, 2006), which is strikingly similar to the effect on reporter gene transcription that we describe here on knockdown of the ncRNAs in the H19 transgene system.
The ncRNAs are also expressed from the maternal allele. On the maternal allele, the ICR is unmethylated, binds to CTCF and H19 is transcribed. Notably, deletion of the CTCF-binding sites or point mutations within the H19 ICR that abolish CTCF binding lead to DNA methylation of the maternal ICR and result in a marked reduction of H19 expression (Pant et al, 2003; Schoenherr et al, 2003; Fedoriw et al, 2004; Engel et al, 2006). One scenario we could predict is that CTCF acts to neutralize the action of the ncRNAs in attracting silencing. In addition, we cannot exclude a functional role of the maternal transcripts, for example in the establishment of an open chromatin conformation at the locus or in loop formation between the ICR and DMR1 in Igf2 (Murrell et al, 2004; Kurukuti et al, 2006).
Our results are the first, to our knowledge, that describe nuclear non-coding transcripts in the H19 ICR, and indeed in any paternally methylated ICR, and provide evidence that such transcripts have a role in gene silencing. Further insight into their function in the mouse requires detailed developmental studies and genetic manipulation.
Methods
RNA isolation, reverse transcription and PCR. Livers were collected from C57BL6/SD7 F1 progeny at an age of embryonic day (E)15.5. RNA was obtained by double purification with Qiagen (Crawley, UK) columns and dual DNase I treatment. cDNA was synthesized with Superscript III (Invitrogen, Paisley, UK). Nuclear and cytoplasmic RNA fractions were obtained from E14.5 mouse embryonic fibroblasts using Paris columns (Ambion, Ambion/Applied Biosystems, Warrington, UK). PCRs were carried out with Taq DNA polymerase and PCR optimizer buffers (Invitrogen).
RNA-FISH. RNA-FISH was carried out as described previously (Osborne et al, 2004). ICR transcripts were visualized with digoxigenin-labelled single-stranded DNA probes and fluorescein isothiocyanate detection. Dinitrophenol-labelled single-stranded DNA probes combined with Texas Red detection were used to visualize H19 and Igf2 transcripts.
Chromatin immunoprecipitation. ChIP was carried out as described previously (Umlauf et al, 2004). Antibodies used were Abcam (Cambridge, UK) Ab5131 (against RNA Pol II) and Rabbit anti-Goat IgG (G4018; Sigma, Gillingham, UK) as control.
Drosophila stocks and genetic crosses. The H19 ICR transgenic and the H19 ICR transgene Δ silencer line were originally described as P(hzh) and P(hzh)Δ4, respectively (Lyko et al, 1997). The H19 ICR RNAi transgene plasmid was obtained by cloning a 430-bp fragment from the H19 ICR (−2915 to −2486 relative to the H19 transcriptional start site) in inverted orientation into P(UASTyellow). The H19 ICR RNAi transgenic line was generated using standard techniques. To express the ICR RNAi transgene, transgenic flies were crossed to flies carrying the P(GMR-GAL4w-) transgene, which drives strong GAL4 expression in the developing and adult Drosophila eye.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank J. Birchler, R. Carthew, S. Henikoff, C. Osborne, L. Perrin and D. Smith for fly strains and plasmids; L. Chakalova and J. Mitchell for expert help with real-time PCR and ChIP; K. Arney for help during the initial stages of the project; W. Dean for help and advice with mouse experimental work; and P. Chandra for critical review of the manuscript. G.S. was supported by the International Human Frontier Science Program Organization. W.R. acknowledges grant funding from the Biotechnology and Biological Sciences Research Council, the Medical Research Council and the European Union NoE The Epigenome. P.F. is a Senior Fellow of the Medical Research Council, UK. This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB-Transregio 5) and the Fonds der Chemischen Industrie (FCI) to R.P.
References
- Bell AC, Felsenfeld G (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485 [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Brannan CI (2001) The Prader–Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics 73: 316–322 [DOI] [PubMed] [Google Scholar]
- Delaval K, Govin J, Cerqueira F, Rousseaux S, Khochbin S, Feil R (2007) Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J 26: 720–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewell RA, Brenton JD, Ainscough JF, Barton SC, Hilton KJ, Arney KL, Dandolo L, Surani MA (2000) Deletion of a silencer element disrupts H19 imprinting independently of a DNA methylation epigenetic switch. Development 127: 3419–3428 [DOI] [PubMed] [Google Scholar]
- Drewell RA, Arney KL, Arima T, Barton SC, Brenton JD, Surani MA (2002) Novel conserved elements upstream of the H19 gene are transcribed and act as mesodermal enhancers. Development 129: 1205–1213 [DOI] [PubMed] [Google Scholar]
- Engel N, Thorvaldsen JL, Bartolomei MS (2006) CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Hum Mol Genet 15: 2945–2954 [DOI] [PubMed] [Google Scholar]
- Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS (2004) Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science 303: 238–240 [DOI] [PubMed] [Google Scholar]
- Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P (2000) Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol Cell 5: 377–386 [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489 [DOI] [PubMed] [Google Scholar]
- Kaffer CR, Srivastava M, Park KY, Ives E, Hsieh S, Batlle J, Grinberg A, Huang SP, Pfeifer K (2000) A transcriptional insulator at the imprinted H19/Igf2 locus. Genes Dev 14: 1908–1919 [PMC free article] [PubMed] [Google Scholar]
- Katayama S et al. (2005) Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566 [DOI] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R (2006) CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA 103: 10684–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W (2004) Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet 36: 1291–1295 [DOI] [PubMed] [Google Scholar]
- Lin SP, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC (2003) Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1–Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet 35: 97–102 [DOI] [PubMed] [Google Scholar]
- Lyko F, Brenton JD, Surani MA, Paro R (1997) An imprinting element from the mouse H19 locus functions as a silencer in Drosophila. Nat Genet 16: 171–173 [DOI] [PubMed] [Google Scholar]
- Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G (2002) Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet 30: 329–334 [DOI] [PubMed] [Google Scholar]
- Mancini-DiNardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM (2006) Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev 20: 1268–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA (2005) RNAi-mediated pathways in the nucleus. Nat Rev Genet 6: 24–35 [DOI] [PubMed] [Google Scholar]
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R (2006) Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 22: 351–361 [DOI] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W (2004) Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet 36: 889–893 [DOI] [PubMed] [Google Scholar]
- Osborne CS et al. (2004) Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36: 1065–1071 [DOI] [PubMed] [Google Scholar]
- Pant V, Mariano P, Kanduri C, Mattsson A, Lobanenkov V, Heuchel R, Ohlsson R (2003) The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev 17: 586–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL (2007) Eukaryotic regulatory RNAs: an answer to the ‘genome complexity' conundrum. Genes Dev 21: 11–42 [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J (2001) Genomic imprinting: parental influence on the genome. Nat Rev Genet 2: 21–32 [DOI] [PubMed] [Google Scholar]
- Schmitt S, Prestel M, Paro R (2005) Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev 19: 697–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S, Paro R (2004) Drosophila Su(Hw) regulates an evolutionarily conserved silencer from the mouse H19 imprinting control region. Cold Spring Harb Symp Quant Biol 69: 47–54 [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Levorse JM, Tilghman SM (2003) CTCF maintains differential methylation at the Igf2/H19 locus. Nat Genet 33: 66–69 [DOI] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP (2002) The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415: 810–813 [DOI] [PubMed] [Google Scholar]
- Smilinich NJ et al. (1999) A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith–Wiedemann syndrome. Proc Natl Acad Sci USA 96: 8064–8069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsen JL, Duran KL, Bartolomei MS (1998) Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev 12: 3693–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R (2004) Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet 36: 1296–1300 [DOI] [PubMed] [Google Scholar]
- Williamson CM et al. (2006) Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat Genet 38: 350–355 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information