Abstract
Bacterial Gre transcript cleavage factors stimulate the intrinsic endonucleolytic activity of RNA polymerase (RNAP) to rescue stalled transcription complexes. They bind to RNAP and extend their coiled-coil (CC) domains to the catalytic centre through the secondary channel. Three existing models for the Gre–RNAP complex postulate congruent mechanisms of Gre-assisted catalysis, while offering conflicting views of the Gre–RNAP interactions. Here, we report the GreB structure of Escherichia coli. The GreB monomers form a triangle with the tip of the amino-terminal CC of one molecule trapped within the hydrophobic cavity of the carboxy-terminal domain of a second molecule. This arrangement suggests an analogous model for recruitment to RNAP. Indeed, the β′-subunit CC located at the rim of the secondary channel has conserved hydrophobic residues at its tip. We show that substitutions of these residues and those in the GreB C-terminal domain cavity confer defects in GreB activity and binding to RNAP, and present a plausible model for the RNAP–GreB complex.
Keywords: transcription, Gre factors, RNA polymerase, crystal structure, binding site
Introduction
Transcript cleavage factors assist RNA polymerase (RNAP) in the hydrolytic removal of the 3′-proximal segment of nascent RNA (Orlova et al, 1995) in the backtracked transcription elongation complexes (TECs). This activity might contribute to productive initiation (Hsu et al, 1995), proofreading (Erie et al, 1993; Zenkin et al, 2006) and the rescue of arrested (Borukhov et al, 1993) or roadblocked (Kireeva et al, 2005) TECs. In bacteria, the transcript cleavage factor paralogues GreA and GreB are thought to have similar two-domain architectures (Stebbins et al, 1995; Koulich et al, 1997), but exhibit several differences. First, GreB binds to RNAP approximately 100-fold tighter than GreA (Koulich et al, 1997). Second, GreA-induced cleavage yields di- and trinucleotide products, whereas GreB stimulates the accumulation of excised products in a much wider size range (2–18 nucleotide long RNAs; Borukhov et al, 1993; Kulish et al, 2000). Third, unlike GreA, which is only able to prevent arrest, GreB can also rescue the already arrested TECs (Borukhov et al, 1993).
Substitution of the two crucial acidic residues at the tip of the extended amino-terminal coiled-coil (CC) domain of the Gre factor (Fig 1) resulted in a marked decrease in Gre-assisted cleavage (Laptenko et al, 2003; Opalka et al, 2003; Sosunova et al, 2003). This suggests a common mechanism in which the Gre-factor reaches into the secondary channel of the RNAP with its CC domain and donates the acidic side chains to the RNAP active site where, together with the RNAP catalytic residues, they coordinate the catalytic Mg2+ ions. The structural models of the RNAP–Gre complexes proposed by these three research groups agree in positioning the GreB CC tip near the RNAP catalytic centre, thereby providing one common point of reference for modelling. However, they disagree on the location of the globular carboxy-terminal domain of GreB, which is thought to constitute the main RNAP-binding site, and suggest three essentially distinct GreB orientations and subsequent GreB–RNAP-binding modes. In fact, in these models, the GreB C-domains are positioned approximately 20–40 Å apart from each other and interact with distinct, non-overlapping segments on the RNAP. The semi-empirical GreB–RNAP complex model, based on low resolution (∼15 Å) experimental electron density (Opalka et al, 2003), shows that although the GreB C-domain is located outside the secondary channel near the C-terminal β′-subunit CC (β′CC) it, in fact, ‘hangs in the air' and forms no direct interactions with the RNAP. In the other two theoretical models, the GreB C-domains are accommodated inside the secondary channel; however, they are attached to distinct structural elements in the RNAP located on the opposite sides of the secondary channel (Laptenko et al, 2003; Sosunova et al, 2003). So far, neither of these conflicting models has been supported by additional biochemical and/or structural data. Thus, the Gre-binding site on the RNAP—the precise localization of which is crucial for understanding the detailed mechanism of action of the Gre factors and also for understanding the general principles of transcription regulation through the secondary channel—remains obscure.
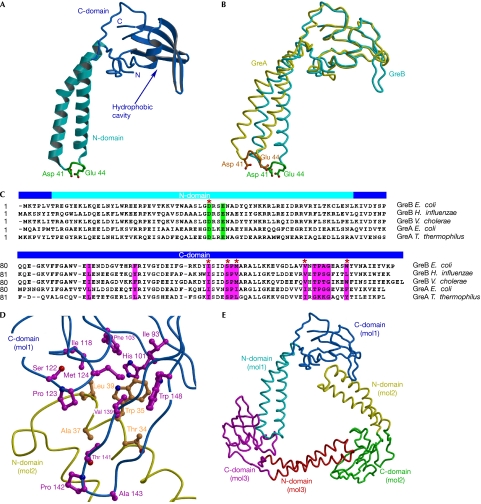
Figure 1.
Structure of the GreB protein. (A) Overall structure. The α-helical turn at the tip of the N-domain and the two principal acidic side chains are shown in green. (B) Structures of GreB and GreA superimposed by the C-domains. (C) Sequence alignment of the Gre factors. The principal acidic residues and the conserved hydrophobic residues lining the C-domain cavity are highlighted in green and magenta, respectively. We included serine and threonine, whose small polar side chains often appear in the hydrophobic cores of proteins, as residues that contribute to the hydrophobic intermolecular interface, as observed in the GreB trimers (D). GreB residues that were altered in the present study are marked by red asterisks. (D) Hydrophobic interface formed between two GreB molecules in the asymmetric unit. (E) Overall view of the trimer that the GreB monomers form in the crystal. C-domain, carboxy-terminal domain; N-domain, amino-terminal domain.
Here, we determined the crystal structure of Escherichia coli GreB at a resolution of 2.6 Å. The intermolecular interactions observed in the crystal suggested that GreB binds to RNAP through a hydrophobic cavity on its C-domain. Subsequent functional analysis supported this model and allowed us to identify the complementary binding site on RNAP. Together, these findings allowed us to build a plausible structural model of the RNAP–GreB complex.
Results
The GreB structure
The GreB molecule (Fig 1A) consists of two domains: the amino-terminal α-helical CC and the globular C-domain, which comprises a central four-stranded β-sheet flanked on one side by an α-helix and a hairpin loop. Although each of the GreB domains closely resembles the corresponding domain of GreA (Stebbins et al, 1995; r.m.s.d. values are 0.83 Å and 0.87 Å for the N- and C-domains, respectively), superimposition of the C-domains revealed a notable difference (∼14°) in the orientations of their CCs (Fig 1B). When superimposed by their C-domains, the six GreB molecules present in the asymmetric unit of the crystal also showed distinct orientations (∼4–9°) of the N-domains (supplementary Fig S1 online). This interdomain flexibility might serve to (i) fit surface-anchored Gre factors to the active site embedded deep in the secondary channel, (ii) adapt to conformational changes in TEC during catalysis, or (iii) switch the factor on and off, as was shown for the Gre-like regulator Gfh1 (Laptenko et al, 2006).
A notable structural feature of GreB—which is also present in GreA, but has not been commented on previously—is an open cavity in the C-domain lined in total by 11 hydrophobic residues (Fig 1C,D). This cavity seems to be more hydrophobic in GreB than in GreA (both from E. coli), as it contains more hydrophobic (Phe 103 versus Tyr 105, respectively) or bulky (Trp 148 versus Phe 150, and Met 124 versus Ile 126, respectively) exposed groups (Fig 1C).
The six GreB molecules in the asymmetric unit are arranged as a pair of identical trimers in which the individual protomers are related by a nearly perfect threefold rotational symmetry. In each trimer, the elongated GreB molecules form a triangle (Fig 1E) that is stabilized through intermolecular interfaces between several hydrophobic side chains from the tip of the N-domain CC of one molecule and the hydrophobic C-domain cavity of a second molecule (Fig 1D). We speculated that a similar type of interaction mediates Gre binding to RNAP. In support of this hypothesis, the β′ C-terminal CC (E. coli residues 645–703), which lies at the point of entry into the secondary channel and becomes strongly protected against hydroxyl radical cleavage by the bound GreB or GreA (Laptenko et al, 2003), has six conserved hydrophobic residues at its tip (Fig 2A). It is worth noting that this hypothesis did not emerge from the earlier structural analysis of GreA (Stebbins et al, 1995) probably because it forms monomers in the crystals and has a less pronounced hydrophobic cavity on its C-domain.
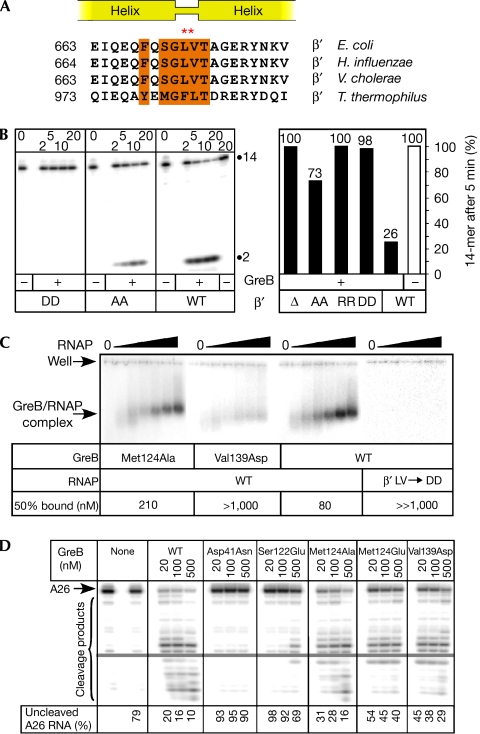
Figure 2.
Functional analysis of the putative β′-subunit coiled coil–GreB interface. (A) Sequence alignment of the tip of the β′CC. The conserved residues that might constitute the hydrophobic intermolecular interface with GreB are highlighted in orange. The double-substituted tip residues are indicated by red asterisks. (B) Effects of the β′CC substitutions on RNA cleavage in scaffold complexes (used because the deletion enzyme was incapable of initiation at the λPR promoter). Left panel: a 15% representative gel with the 3′-end-labelled 14-mer RNA and a 2-mer 3′-cleavage product indicated. TECs assembled with core enzymes were incubated at 37°C for the durations indicated above (in min) in the absence or presence of 100 nM of wild-type GreB. Right panel: quantification of the 14-mer RNA that remained uncleaved after 5 min incubation. The assay was repeated at least twice for each enzyme variant. (C) Gel shift assay with radiolabelled GreBs (at 8 nM) and increasing concentrations of core RNAP (0, 40, 80, 150, 300, 500 and 1,000 nM). Positions of the wells and the Gre–RNAP complexes are indicated. The core concentration at which approximately 50% of 32P-GreB was bound is shown; no binding was detected even at 1 μM of the DD core (as well as the RR and deletion variants; data not shown). (D) Effects of the selected GreB variants on RNA cleavage in halted radiolabelled A26 TECs. The fraction of A26 RNA remaining after 10 min incubation at 37°C is shown below each lane. The far left lane contains A26 RNA before 37°C incubation. AA, double alanine substitution; β′CC, β′-subunit coiled coil; DD, double aspartic acid substitution; RNAP, RNA polymerase; RR, double arginine substitution; TEC, transcription elongation complex; WT, wild-type.
β′CC is the main GreB-binding site on the RNAP
Having tentatively identified the β′CC and C-domain hydrophobic cavity as the respective RNAP- and GreB-derived parts of the TEC–GreB interface, we constructed double substitutions of the β′CC residues Leu 672 and Val 673 for alanine, aspartic acid or arginine, as well as the deletion of one helical turn on each side of the tip, which effectively shortens the β′CC. We then analysed the effects on endonucleolytic transcript cleavage in A26 TECs (Fig 2B) that are sensitive to GreB-assisted cleavage. Alanine substitutions (designated as an AA variant in Fig 2B) resulted in an expected reduction in GreB-dependent cleavage activity and affinity to GreB (Fig 2C). Substitutions with either aspartic acid or arginine at both positions (DD and RR variants, respectively) essentially prevented GreB-dependent enhancement of cleavage, as did the removal of the entire β′CC tip; the same was also true for GreA (Fig 2B; data not shown). The essential loss of activity (affinity to GreB) in the RNAP mutants with the charged substitutions at the β′CC tip and retention of the AA variant suggest that the RNAP–Gre binding relies largely on hydrophobic interactions. These results alone provide strong support for the structural prediction that the GreB C-domain hydrophobic cavity constitutes the docking site for RNAP as the GreB C-domain cavity is the only site on the GreB surface that complements the β′CC hydrophobic tip, both in its shape and chemical properties (consistent with the GreB crystal structure).
Nevertheless, we engineered a further eight amino-acid substitutions in GreB that disrupted the presumed interface with RNAP (Fig 2C; supplementary Fig S2 online). The effects of these variants fell in the range between the wild-type and the cleavage-deficient Asp41Asn variant (Laptenko et al, 2003), with Ser122Glu showing the strongest defect. Substitutions for alanine conferred minor defects, whereas substitutions of the same residues (for example, Met124 and Ile118) for glutamate produced much larger effects, which were particularly evident when the rate of cleavage was assayed (supplementary Figs S2, S3 online).
To ascertain whether the observed defects in transcript cleavage were due to defects in binding, we used the gel mobility shift (Loizos & Darst, 1999) and Fe-mediated cleavage (Laptenko et al, 2003) assays. We found that defective GreB variants (for example, Val139Asp in Fig 2C, and Ser122Glu in supplementary Fig S4 online) showed reduced affinity to RNAP, and the observed drops in affinity correlated with the concomitant defects in cleavage (Fig 2C; data not shown). However, we found that the ‘direct' binding assay was much less sensitive to small differences among the GreB variants when compared with the RNA cleavage assay.
Discussion
RNAP-binding determinants on GreB
Among the ten GreB residues that were subjected to alanine scanning (Loizos & Darst, 1999), substitutions of only two—Asp121Ala and Pro123Ala—resulted in a notable loss of affinity for RNAP. Although three of the ten residues—Pro 123, Val 139 and Pro 142—are located in the C-domain hydrophobic cavity, only the substitution of Pro 123 affected binding. Given the hydrophobic nature of the proposed interface, the modest effects of these variants might be due to the fact that alanine is also a hydrophobic residue. Conversely, the relatively small effect of the Asp121Ala substitution, which markedly changed both the chemical and the structural properties of the side chain, suggests that Asp 121 could contribute to GreB structural integrity, but does not have a direct role in binding to RNAP. In our studies, we combined alanine scanning with radical mutagenesis (Bryant et al, 1996); indeed, we found that only charged substitutions at positions 118, 122, 124 and 139 led to major functional defects (Fig 2B; supplementary Fig S2 online). Our analysis identifies these residues (and the adjacent Pro 123; Loizos & Darst, 1999) as a part of the GreB–RNAP interface and highlights the fact that alanine scanning often does not produce strong effects when applied to hydrophobic interfaces (as opposed to the hydrophobic interior of proteins), outside the so-called hot spots, the origins and rigorous thermodynamic interpretation of which remain elusive. This feature of alanine scanning could be an important impediment in the analysis of hydrophobic intermolecular interfaces.
A model for the GreB-bound TEC
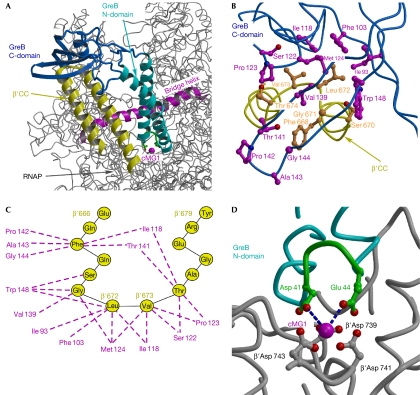
Identification of the β′CC tip as the crucial docking site for the Gre factors provides a second reference point—in addition to the RNAP active site—for building a high-confidence structural model of the RNAP–Gre complex (Fig 3A), in which the β′CC tip is inserted into the hydrophobic cavity of the GreB C-domain (Fig 3B,C), and the crucial GreB acidic residues are positioned close enough to the active site to coordinate the catalytic Mg2+ ions (Fig 3D). Our model involves no alterations to the RNAP and GreB structures, and produces no steric clashes within the complex. The network of interactions between the GreB and β′CC residues in the model seem to be entirely consistent with all the data obtained in our studies and those reported previously. Indeed, in the model, the β′CC Leu 672 and Val 673 are positioned at the centre of the C-domain cavity, making multiple van der Waals interactions with many surrounding GreB hydrophobic side chains. In excellent agreement with the model and proposed hydrophobic nature of the RNAP–GreB interface, substitutions of Ile 118, Ser 122, Met 124 and Val 139 to acidic residues (this work) and Thr 141 to bulky methionine (Laptenko et al, 2003) conferred the strongest GreB and GreA defects, respectively.
Figure 3.
Structural model of the RNAP–GreB complex. (A) Overall view of the complex. The structure of the Thermus thermophilus RNAP (Vassylyev et al, 2002), in which the β′CC tip residues were substituted for the Escherichia coli counterparts, was used for the modelling. (B) Three-dimensional view of the hypothetical hydrophobic interface and (C) a schematic drawing of possible van der Waals contacts (dashed lines) between the GreB (magenta) and β′CC (yellow) residues. (D) The principal acidic residues (green) at the GreB N-domain tip are positioned at the interacting distance with the active site. The blue dashed lines indicate the shortest distances between the GreB Asp 41 (3.7 Å) and Glu 44 (3.4 Å) and the major (high affinity) catalytic Mg2+ ion (cMG1), suggesting that these side chains would be able to coordinate the second catalytic Mg2+ ion, the precise position of which in the active site of RNAP has not yet been determined. β′CC, β′-subunit coiled coil; C-domain, carboxy-terminal domain; cMG1, a high-affinity catalytic Mg2+ ion in the RNAP active site; N-domain, amino-terminal domain; RNAP, RNA polymerase.
Implications for transcriptional regulation
Altogether, our experimental data and modelling produces several important implications. First, the overall similarity between the GreA (Stebbins et al, 1995) and GreB (Fig 1B) structures, and, in particular, the nearly identical configuration of their N-domain tips that contain the two principal acidic side chains, suggest the same mode of recruitment to RNAP and similar structural mechanisms of stimulating the RNA cleavage activity. Second, the β′CC constitutes the major and, most likely, the only Gre-binding site on RNAP. Third, binding of the Gre factors to RNAP is probably dominated by hydrophobic interactions. The configuration of the C-domain cavity (which is apparently more hydrophobic in GreB; Fig 1C and see above) might account, in part, for the higher affinity of GreB to RNAP (Koulich et al, 1997). Fourth, in our model, the side chains that form the ‘basic patch' (Kulish et al, 2000), a prominent feature located on one side of the N-domain in GreB (but largely missing in GreA), are all exposed towards the open space in the secondary channel, which might become occupied by nascent RNA on RNAP backtracking, thereby supporting the idea that its large basic patch might allow GreB (in contrast to GreA) to recognize and mediate excision of long extruded RNAs (Kulish et al, 2000). Finally, the secondary channel becomes substantially occluded by bound GreB, suggesting that Gre factors might prevent transcriptional arrest by blocking RNA extrusion through the channel; this steric occlusion might be sufficient for the observed anti-arrest activity of GreA (Borukhov et al, 1993).
Methods
The crystals of E. coli GreB were obtained by a sitting-drop vapour diffusion technique followed by macro-seeding (Perederina et al, 2006). The crystals belong to the P43 space group, with unit cell dimensions of a=b=148.3, c=116.8 Å, and have perfect merohedral twinning with the twinning operator {h,−k,−l}. The details of the structure determination are provided in the supplementary Table S1 online.
Substitutions in the E. coli GreB and the β′-subunit (supplementary Table S2 online) were constructed by site-directed mutagenesis; proteins were purified as described previously (Perederina et al, 2006; http://www.osumicrobiology.org/faculty/documents/ExpressionvectorforE.coliRNApolymerase.htm). Halted A26 TECs were created with linear DNA template (80 nM), E. coli holo RNAP (100 nM), ApU (100 μM), 2.5 μM ATP, GTP and UTP, and 300 nM [α32P]GTP in 20 mM Tris-acetate, 20 mM Na-acetate, 2 mM Mg-acetate, 5% glycerol, 0.1 mM EDTA and 14 mM 2-mercaptoethanol (pH 8.0), for 15 min at 37°C, and purified through G50 columns. Scaffold EC13 complexes were assembled as described by Temiakov et al (2005), and RNA was extended with 150 nM [α32P]GTP. On the addition of GreB, the reactions were incubated at 37°C as indicated in Fig 2, stopped by the addition of urea (to 5 M) and EDTA (to 25 mM), and analysed on 15% denaturing gels (19:1) as described previously (Artsimovitch et al, 2003). GreB-RNAP gel shift assays were carried out with radiolabelled GreB as described by Loizos & Darst (1999), except that the samples were analysed on 2% NuSieve agarose gels in 1 × TBE at 5 V/cm.
Data deposition. The coordinates and structure factors for the crystal structure of the GreB protein have been deposited in the Protein Data Bank under ID code 2P4V.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary data
Acknowledgments
We thank S. Wakatsuki, N. Igarashi, N. Matsugaki and M. Suzuki for their assistance in data collection at the NW12 Synchrotron beam line (Photon Factory, Japan). This work was supported by the National Institutes of Health grants GM74252 and GM74840 (to D.G.V.), and GM67153 (to I.A.). Use of the Advanced Photon Source was supported by the Department of Energy, Office of Energy Research, under contract number W-31-109-Eng-38.
References
- Artsimovitch I, Chu C, Lynch AS, Landick R (2003) A new class of bacterial RNA polymerase inhibitor affects nucleotide addition. Science 302: 650–654 [DOI] [PubMed] [Google Scholar]
- Borukhov S, Sagitov V, Goldfarb A (1993) Transcript cleavage factors from E. coli. Cell 72: 459–466 [DOI] [PubMed] [Google Scholar]
- Bryant GO, Martel LS, Burley SK, Berk AJ (1996) Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes Dev 10: 2491–2504 [DOI] [PubMed] [Google Scholar]
- Erie DA, Hajiseyedjavadi O, Young MC, von Hippel PH (1993) Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science 262: 867–873 [DOI] [PubMed] [Google Scholar]
- Hsu LM, Vo NV, Chamberlin MJ (1995) Escherichia coli transcript cleavage factors GreA and GreB stimulate promoter escape and gene expression in vivo and in vitro. Proc Natl Acad Sci USA 92: 11588–11592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M (2005) Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell 18: 97–108 [DOI] [PubMed] [Google Scholar]
- Koulich D, Orlova M, Malhotra A, Sali A, Darst SA, Borukhov S (1997) Domain organization of Escherichia coli transcript cleavage factors GreA and GreB. J Biol Chem 272: 7201–7210 [DOI] [PubMed] [Google Scholar]
- Kulish D, Lee J, Lomakin I, Nowicka B, Das A, Darst S, Normet K, Borukhov S (2000) The functional role of basic patch, a structural element of Escherichia coli transcript cleavage factors GreA and GreB. J Biol Chem 275: 12789–12798 [DOI] [PubMed] [Google Scholar]
- Laptenko O, Lee J, Lomakin I, Borukhov S (2003) Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J 22: 6322–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laptenko O, Kim SS, Lee J, Starodubtseva M, Cava F, Berenguer J, Kong XP, Borukhov S (2006) pH-dependent conformational switch activates the inhibitor of transcription elongation. EMBO J 25: 2131–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loizos N, Darst SA (1999) Mapping interactions of Escherichia coli GreB with RNA polymerase and ternary elongation complexes. J Biol Chem 274: 23378–23386 [DOI] [PubMed] [Google Scholar]
- Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA (2003) Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell 114: 335–345 [DOI] [PubMed] [Google Scholar]
- Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S (1995) Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci USA 92: 4596–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina AA, Vassylyeva MN, Berezin IA, Svetlov V, Artsimovitch I, Vassylyev DG (2006) Cloning, expression, purification, crystallization and initial crystallographic analysis of transcription elongation factors GreB from Escherichia coli and Gfh1 from Thermus thermophilus. Acta Crystallogr F 62: 44–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosunova E, Sosunov V, Kozlov M, Nikiforov V, Goldfarb A, Mustaev A (2003) Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc Natl Acad Sci USA 100: 15469–15474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Borukhov S, Orlova M, Polyakov A, Goldfarb A, Darst SA (1995) Crystal structure of the GreA transcript cleavage factor from Escherichia coli. Nature 373: 636–640 [DOI] [PubMed] [Google Scholar]
- Temiakov D et al. (2005) Structural basis of transcription inhibition by antibiotic streptolydigin. Mol Cell 19: 655–666 [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S (2002) Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417: 712–719 [DOI] [PubMed] [Google Scholar]
- Zenkin N, Yuzenkova Y, Severinov K (2006) Transcript-assisted transcriptional proofreading. Science 313: 518–520 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data