Abstract
The arginine-rich motif provides a versatile framework for RNA recognition in which few amino acids other than arginine are needed to mediate specific binding. Using a mammalian screening system based on transcriptional activation by HIV Tat, we identified novel arginine-rich peptides from combinatorial libraries that bind tightly to the Rev response element of HIV. Remarkably, a single glutamine, but not asparagine, within a stretch of polyarginine can mediate high-affinity binding. These results, together with the structure of a Rev peptide-Rev response element complex, suggest that the carboxamide groups of glutamine or asparagine are well-suited to hydrogen bond to G-A base pairs and begin to establish an RNA recognition code for the arginine-rich motif. The screening approach may provide a relatively general method for screening expression libraries in mammalian cells.
Keywords: RNA-protein recognition, arginine-rich motif, HIV Rev-Rev response element, expression cloning, protoplast fusion
The problem of RNA recognition is fundamental to many biological systems and is still at a relatively early stage of study. One particularly well-studied class of RNA-binding proteins contains arginine-rich domains that can bind tightly to their cognate RNA sites as isolated peptides (1–5). Though classified together as a family, arginine-rich domains are in fact structurally diverse; for example, an HIV Rev peptide binds to the Rev response element (RRE) in an α-helical conformation whereas a bovine immunodeficiency virus (BIV) Tat peptide binds to the BIV transactivating response element (TAR) as a β-hairpin (6–9). In each case, few amino acids other than arginine provide specific contacts to the RNA, leading to the hypotheses that: (i) specific RNA-binding peptides could have evolved relatively easily beginning with polyarginine and (ii) rules for amino acid-base interactions might be found within the polyarginine context. Here we explore these hypotheses by using an HIV Tat-TAR system to screen RNA-binding libraries in mammalian cells.
Tat is a potent activator of viral gene transcription and is essential for viral replication. Tat activates transcription largely by enhancing the processivity of RNA polymerase II transcription complexes initiated at the HIV long terminal repeat (LTR) (10–14). To function, Tat must bind to TAR, an RNA hairpin located at the 5′ end of the nascent transcripts (15, 16). Tat contains a functionally defined activation domain (amino acids 1–48) and an arginine rich RNA-binding domain (amino acids 49–57) that also functions as a nuclear localization signal (17–19). The activation and RNA-binding domains are modular and separable. Tat can activate transcription when bound to the nascent transcript through heterologous RNA-protein interactions (20, 21) or even when bound to DNA (22). Thus, by replacing TAR with a “bait” RNA and fusing a library to Tat, it should be possible to screen for RNA binders by using an appropriate HIV LTR reporter. The method described here uses green fluorescent protein (GFP) as the reporter and fluorescence-activated cell sorting (FACS) to isolate positive members of a library.
Several systems recently have been described to screen libraries for RNA-binding molecules: phage display has been used to identify ribonucleoprotein (RNP) domain variants that bind to RNAs with altered specificities (23), bacterial assays have been devised in which RNA binding either promotes transcriptional antitermination or interferes with translation (24–27), and a yeast “three-hybrid” assay has been set up in which a bivalent RNA is used as the “bait” (28). The limitations of each system are not yet clear and no screen has been reported using mammalian cells where, for example, interactions may be studied in the context of the natural cellular environment or protein folding or posttranslational modification may be more efficient. Strategies for expression cloning in mammalian cells have improved substantially over the past decade. In particular, transient transfection methods have been developed in which simian virus 40 (SV40), polyoma, or Epstein–Barr virus origins are used to amplify transfected plasmids in replication-competent recipient cells or in which active plasmids are purified from mixed populations by subdividing active pools (29–32). Despite the advances, important limitations still remain, including restrictions on the types of cells that can be used (32). Here we describe an expression cloning strategy in which plasmids are amplified in bacteria and delivered into mammalian cells by protoplast fusion (29, 33–35), positive cells are isolated by using a GFP reporter and FACS, and plasmids are recovered without additional replication in the recipient cell, in principle allowing delivery to many different cell types. In part, the method is efficient because protoplast fusion is nearly clonal, resulting in delivery of thousands of copies of a single plasmid into a single mammalian cell.
MATERIALS AND METHODS
Plasmids and Cell Lines.
An HIV-1 LTR-GFP reporter plasmid (containing wild-type TAR) was constructed by inserting a gene encoding the Ala-65 GFP mutant (containing a GCC alanine codon in place of the TCT serine codon) between the HindIII and XhoI sites in pCDNA3 (Invitrogen), and by replacing the cytomegalovirus (CMV) promoter in pCDNA3 with the HIV-1 LTR from PHIV-CAT (15). BIV TAR and HIV-1 RRE IIB GFP reporters were constructed by replacing the HIV-1 LTR-TAR region with corresponding regions from BIV TAR and RRE IIB chloramphenicol acetyltransferase reporters (CAT) (3, 4). The U1 GFP reporter was constructed by replacing the upper part of the TAR hairpin (+20 to +40) with an oligonucleotide containing the 22-nucleotide U1 small nuclear RNA hairpin II (36). Tat-Rev14, Tat-Arg14, and selected Tat-peptide hybrids were constructed by cloning oligonucleotide cassettes encoding each peptide plus four alanines at the N terminus and four alanines and an arginine at the C terminus after amino acid 49 of Tat (using an EagI site at the end of the activation domain) in vectors derived from pSV2tat72 (37). The Tat-U1A fusion was constructed by fusing oligonucleotide cassettes encoding three glycines followed by residues 2–102 of U1A to Tat1–72. The CMV promoter was used to constitutively express GFP or human CD4 in pCDNA3 plasmids.
HeLa cell lines containing stably integrated HIV LTR-GFP reporters were selected by using neomycin (G418). Clones that showed low backgrounds in the absence of Tat and bright fluorescence when transfected with a corresponding Tat fusion, as judged by fluorescence microscopy, were chosen for expansion. Several GFP variants were examined by transient transfection and in stable cell lines, and Ala-65 displayed the best signal to noise.
Protoplast Fusion.
Plasmid-containing DH-5α cultures were grown at 37°C, plasmids were amplified with 250 μg/ml chloramphenicol for 16 hr, and protoplasts were prepared as described (34), monitoring conversion by phase-contrast microscopy (bacteria are rod-shaped whereas protoplasts are round). Protoplasts were diluted slowly with 20 ml of room temperature serum-free DMEM containing 10% sucrose and 10 mM MgCl2, and suspensions (≈1.5 × 109 protoplasts/ml) were kept at room temperature for 15 min. HeLa cells were grown to ≈70% confluence in six-well plates, medium was removed, and cells were washed with serum-free DMEM. Protoplast suspension (4 ml) was added (>1,000-fold excess over cells), plates were centrifuged at 1,650 × g for 10 min at 25°C, and supernatants were removed carefully by suction. Two milliliters of prewarmed 50% (vol/vol) PEG1000 or 50% (wt/vol) PEG1500 was added at room temperature, incubated for 2 min, and removed by suction. Cells were washed three times using 2 ml serum-free DMEM, and 4 ml DMEM containing 10% fetal bovine serum, penicillin, streptomycin, and kanamycin was added. Under these conditions, many protoplasts bind to each cell as judged by light microscopy. Medium was changed after 24 hr, and cells were grown for an additional 24 hr before examining fluorescence.
FACS.
Transfected or protoplast-fused cells were harvested by trypsinization after 48 hr and were resuspended at a concentration of 106 cells/ml in DMEM containing 10% cell dissociation buffer (GIBCO/BRL), 0.3% fetal bovine serum, and 1 μg/ml propidium iodide. Samples were analyzed or sorted by FACS by using an argon laser to excite cells at 488 nm and a 530 ± 15 nm band pass filter to detect GFP emission. For FACS scans, 10,000 cells were typically analyzed by using a FACScan flow cytometer (Becton Dickinson). FACS sorting (2,000–3,000 cells/sec) was performed by using the Howard Hughes Medical Institute (University of California, San Francisco) FACStarPlus cell sorter (Becton Dickinson). FACS data were analyzed using cellquest software.
Plasmid Recovery from Sorted Cells.
FACS-sorted cells were mixed with 20,000 HeLa cells, centrifuged, resuspended in 10 μl TE buffer (10 mM Tris, pH 8.0/1 mM EDTA) containing 0.2 mg/ml tRNA, and plasmids were prepared by alkaline lysis (38). Following phenol:chloroform (1:1) extraction, glycogen was added to 0.5 mg/ml and plasmid DNAs were ethanol precipitated. Electrocompetent cells were prepared as described (38) except that four water washes were performed and cells were resuspended at a high concentration (≈3 × 1011/ml) to obtain high efficiency competent cells (≈1–1.5 × 1011 colonies/μg were typically obtained when using 0.1–10 pg supercoiled pUC19 DNA).
Combinatorial Peptide Library Design and Screening.
A degenerate oligonucleotide (5′-ATCTCTTACGGCCGTGCCGCTGCAGCCXXYAGAXXYXXYAGGCGAXXYAGGAGACGGCGACGTCGCAGAGCTGCCGCCGCAAGATGACTCGAGACTAGTGGA-3′, where X is a A:G:C mixture at a 1:1:1 ratio and Y is a G:T mixture at a 1:1 ratio) was synthesized encoding the arginine-rich peptide library. A primer (5′-TCCACTAGTCTCGAG-3′) was annealed to the degenerate oligonucleotide, and double-stranded DNA was synthesized by using Sequenase 2.0 (United States Biochemical). The double-stranded product (≈0.1 μg) was digested with EagI and XhoI and ligated into 5 μg EagI-XhoI-digested pSV2tat72 to generate fusions to amino acid 49 of Tat. The encoded peptides contain four randomized positions within a stretch of 14 arginines, AAAAXRXXRRXRRRRRRRAAAAR, where X represents any of 12 amino acids in the boldface box in Fig. 4A. Protoplasts containing Tat-Rev14, Tat-Arg14, and library plasmids were fused to HeLa cells containing a stably integrated HIV LTR RRE IIB-GFP reporter. After 48 hr, 10,000 positive control cells (Tat-Rev14) and 10,000 negative control cells (Tat-Arg14) were analyzed by FACS to estimate fusion efficiency and to establish the sorting window. Library-fused cells (≈107) were sorted by FACS and positive cells were collected. Plasmids were recovered by alkaline-lysis phenol-extraction, and electroporated into DH-5α cells, and resulting colonies were used to prepare protoplasts for the next round of selection. The cycle was repeated for three rounds, until the fraction of GFP-positive cells was similar to that of the positive control.
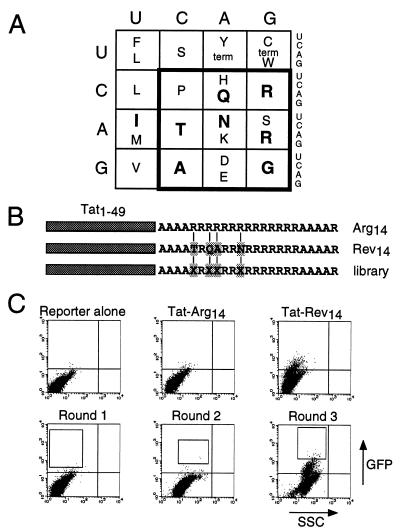
Figure 4.
Screening an arginine-rich combinatorial library for RRE binders. (A) The genetic code viewed from the perspective of arginine-rich peptides. Amino acids known to be important for specific RNA binding by HIV Rev, HIV Tat, BIV Tat, and λ N peptides are indicated in bold (see ref. 24). A restricted genetic code (boldface box) encodes all charged and hydrophilic residues, glycine, alanine, and proline, and contains all six arginine codons. Combinations of these amino acids in an arginine-rich context are expected to encode a variety of helical and nonhelical RNA-binding peptides. (B) Fourteen arginines (Arg14) or a 14-amino acid Rev peptide (Rev14 corresponds to residues 34–47 of Rev, with Trp-45→Arg and Glu-47→Arg substitutions, and specifically binds RRE IIB RNA; ref. 24) were fused to the HIV Tat activation domain (residues 1–49) in the context of surrounding alanines as shown. In the library, four residues corresponding to non-arginine positions in Rev (Xs) were randomized with the amino acids encoded by the boldface box in A. Arg14 served as a negative control and Rev14 as a positive control. (C) FACS analysis of the reporter alone, cells fused to negative and positive control protoplasts used to set sorting windows, and library-containing protoplasts carried through three rounds of sorting. Boxes show the windows used to collect cells in each round. Individual clones from rounds 2 and 3 were tested for activity and positive clones were sequenced.
CAT Assays, Peptides, and Gel Shift Assays.
Levels of activation by the Tat fusion proteins were assessed by cotransfecting 50 ng of an HIV LTR RRE IIB-CAT reporter plasmid (3) and 0.2–25 ng Tat expression plasmids into HeLa cells using lipofectin. Total plasmid DNA was adjusted to 1 μg with pUC19. CAT activities were assayed after 48 hr using an appropriate amount of cell extract (2), and activities were quantitated using a Molecular Dynamics PhosphorImager. Peptides were synthesized, purified, and quantitated, and gel shift assays were performed with radiolabeled RRE IIB and mutant RNAs as described (3, 5). All peptides contained four alanines at the N terminus and four alanines and one arginine at the C terminus to help stabilize α-helical conformations (3).
RESULTS
GFP Expression Occurs with Specific RNA-Protein Interactions.
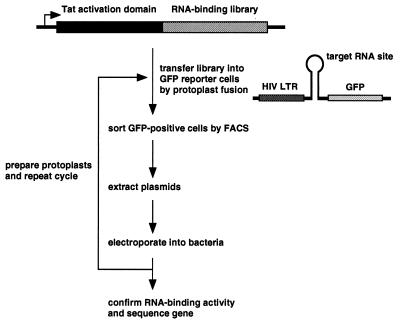
The basic protocol for screening RNA-binding libraries is outlined in Fig. 1. The Tat activation domain, or in some cases full-length Tat, is fused to a library and an HIV-LTR GFP reporter is constructed in which an RNA site of interest replaces the TAR site. The library is delivered into reporter-containing HeLa cells by protoplast fusion under conditions in which approximately one bacterium fuses to one cell. After two days, HeLa cells expressing high levels of GFP are sorted by FACS, and plasmids are extracted and electroporated into bacteria. The procedure is repeated using protoplasts from the enriched population until a high proportion of GFP-expressors is obtained and resulting plasmids are sequenced.
Figure 1.
Strategy for screening RNA-binding libraries. Libraries are fused to the activation domain of HIV Tat or to full-length Tat and are delivered into stable cells containing an appropriate GFP reporter by protoplast fusion. GFP-expressing cells are isolated by FACS, and plasmids are extracted by alkaline lysis and electroporated into bacteria. Protoplasts are made from the enriched population and the cycle is repeated until a large proportion of fused cells express GFP. Individual clones are tested for activity and positives are sequenced.
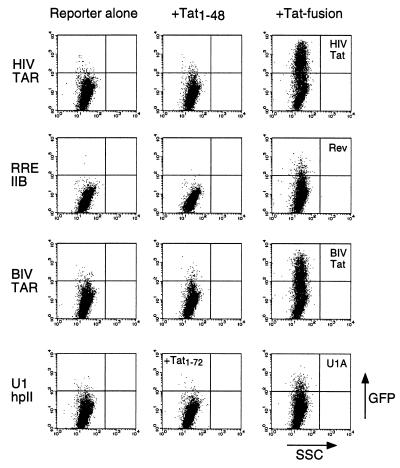
To test whether a Tat-GFP reporter system could be used to monitor specific RNA-protein interactions, we first constructed a set of reporters containing HIV TAR, RRE IIB (the high-affinity Rev binding site), BIV TAR, and U1 small nuclear RNA hairpin II (the U1A protein binding site), and a set of Tat fusions containing the RNA-binding domains from HIV Tat, Rev, BIV Tat, and the U1A protein (2–4, 24, 36). GFP reporters were transfected into HeLa cells either alone or with Tat fusions, and GFP expression was observed only with the cognate interactions. The activation domain of Tat alone (Tat1–48) did not activate through HIV TAR, RRE, or BIV TAR reporters, and full-length Tat (Tat1–72) did not activate through U1 hpII (Fig. 2). U1A was fused to full-length Tat to ensure nuclear localization (the arginine-rich domain of Tat functions as a nuclear localization signal) whereas the other RNA-binding domains were fused to just the activation domain because they provide their own nuclear localization signal. When reporters were cotransfected with the corresponding Tat fusion proteins, GFP expression increased ≈10–100-fold (Fig. 2). No activation was observed through noncognate RNAs (data not shown), indicating that the Tat-GFP reporter system accurately reflects specific RNA-protein interactions. As expected, the Tat-U1A fusion, which includes the RNA-binding domain of Tat, also functioned through HIV TAR (data not shown).
Figure 2.
Activation of GFP expression by Tat fusion proteins and corresponding RNA reporters. Cells were lipofected with HIV TAR, RRE IIB, BIV TAR, or U1 hairpin II GFP reporters (2 μg) alone (Left), along with Tat1–48 or Tat1–72 (Center), or along with full-length HIV Tat or Tat fusions to a Rev peptide, BIV Tat peptide, or U1A RNA-binding domain, respectively (4 μg) (Right). Rev and BIV Tat peptides were fused to Tat1–48 whereas the U1A domain was fused to Tat1–72 to ensure nuclear localization. Plots show relative GFP fluorescence on the y axis and relative side scatter (a measure of cell granularity) on the x axis for 10,000 cells.
Introduction of Plasmids by Protoplast Fusion.
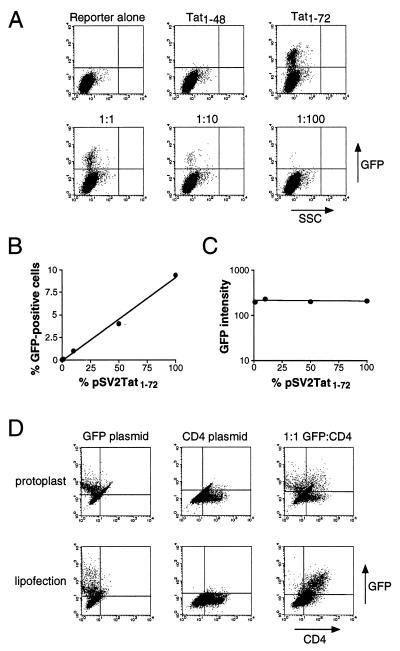
Bacterial protoplast fusion has been used in the expression cloning work of Seed and Aruffo (29) and is thought to deliver a large and relatively homogeneous population of plasmids into cells, in principle allowing detection of weak binders and reducing the background from cotransfected library members. To test whether protoplast fusion might be used in our system, we fused protoplasts containing amplified pSV2Tat1–72 or pSV2Tat1–48 plasmids into HeLa cells containing a stably integrated HIV LTR-GFP reporter. Approximately 10% of cells fused with pSV2Tat1–72 displayed high GFP fluorescence whereas fusion to pSV2Tat1–48 produced virtually no signal (Fig. 3A). Diluting pSV2Tat1–72 protoplasts with increasing amounts of inactive pSV2Tat1–48 protoplasts resulted in a proportional decrease in the number of GFP-expressing cells but not a proportional decrease in fluorescence intensity per positive cell (Fig. 3 A–C), suggesting that, statistically, few protoplasts delivered their contents into each HeLa cell. In contrast, the same plasmid ratios delivered by lipofection resulted in a proportional decrease in fluorescence intensity per positive cell and fluorescence was undetectable at 1% pSV2Tat1–72 (data not shown), as expected if cells were randomly sampling the distribution of plasmids in the transfection mixture. Two additional experiments suggest that protoplast fusion is near-clonal: First, an equal number of protoplasts containing either the HIV LTR-GFP reporter or pSV2Tat1–72 were fused to HeLa cells and few GFP-positives were detected by fluorescence microscopy (compared with >10% positives when pSV2Tat1–72 protoplasts were fused to stable GFP reporter cells), suggesting that the activator and reporter plasmids were rarely delivered into the same cell. Second, an equal number of protoplasts containing either a constitutively expressing GFP plasmid or CD4 plasmid were fused to HeLa cells, and the majority of positives expressed one or the other protein but not both (Fig. 3D); in contrast, introducing an equal mixture of the same plasmids by lipofection resulted in a high frequency of double-positive cells (Fig. 3D). Thus, one or very few protoplasts fuse per HeLa cell under these conditions.
Figure 3.
Plasmid delivery by protoplast fusion. (A) FACS analysis of cells containing a stably integrated HIV LTR-GFP reporter (reporter alone) or reporter cells fused with protoplasts containing pSV2Tat1–48 or pSV2Tat1–72 plasmids, or fused with protoplast mixtures containing pSV2Tat1–72 and pSV2Tat1–48 in 1:1, 1:10, or 1:100 ratios. (B) Plot of the percentage of GFP-expressing cells as a function of the proportion of pSV2Tat1–72 in the mixture, from A. Based on the percentage of positive cells obtained with pSV2Tat1–72 alone, we estimate that ≈10% of cells fused productively in this experiment. (C) Plot of GFP intensity as a function of the proportion of pSV2Tat1–72 in the mixture, from A. (D) Protoplasts were prepared containing CMV-GFP or CMV-CD4 expression plasmids and were fused to HeLa cells either separately or in a 1:1 mixture, and analyzed for protein expression by FACS (Upper). Plasmid DNAs also were introduced by lipofection and analyzed (Lower).
Previous expression cloning strategies have generally relied on plasmid replication in recipient cells to recover transfected plasmids, most often using an SV40 origin and T-antigen-expressing cells (such as COS) to amplify plasmids episomally, thus restricting cloning to relatively few cell types (32). We tested whether plasmids could be recovered directly from protoplast-fused cells by sorting GFP-positive cells, isolating plasmids by alkaline lysis, and transforming into highly electrocompetent bacteria. We obtained approximately one colony per sorted cell, sufficient for library screening, whereas plasmids isolated from Hirt supernatants (39) yielded only about one-tenth the number of colonies. To mimic the situation encountered in a library screen, we tested whether a small number of active plasmids could be recovered from a large pool of inactive plasmids. Protoplasts containing pSV2Tat1–72 and pSV2Tat1–48 were mixed in a 1:105 ratio and fused to HeLa GFP reporter cells. GFP-positive cells were sorted from ≈107 cells, plasmids were isolated, retransformed into bacteria, and protoplasts were prepared for subsequent rounds. PCR analyses indicated that 10% of the clones contained pSV2Tat1–72 after just two rounds of sorting and 60% contained pSV2Tat1–72 after a third round.
Identification of Tight RRE-Binding Peptides from a Combinatorial Library.
Given the structural diversity of the arginine-rich motif and the observation that few amino acids other than arginine appear to provide specific contacts to the RNA (see Fig. 4A), we wished to examine the hypothesis that specific RNA-binding peptides could be relatively easily evolved in a polyarginine framework. We designed a combinatorial library in which four residues within a stretch of 14 arginines were randomized using 12 hydrophilic or charged amino acids (Fig. 4 A, B). The randomized positions correspond to non-arginine residues in Rev (Fig. 4B), and the library contains 184 (≈1 × 105) codon sequences encoding 124 (≈2.1 × 104) peptides. The library was fused to the Tat activation domain in the context of flanking alanines to help stabilize α-helical conformations (3, 40) and was screened for tight RRE binders using a HeLa cell line containing a stably integrated HIV LTR-RRE IIB-GFP reporter.
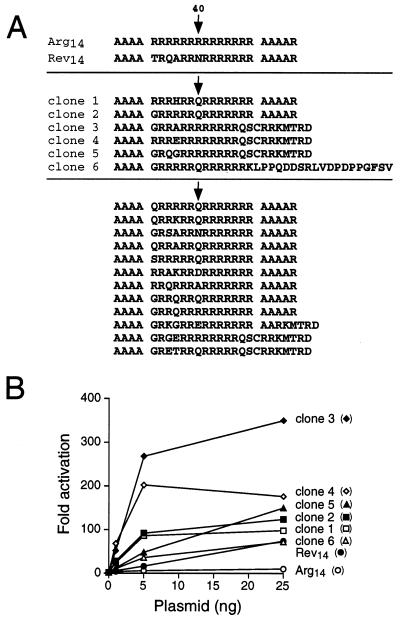
Protoplasts containing Tat-Rev14 and Tat-Arg14 were used as positive and negative controls (Fig. 4C), and the sorting window was set to identify fusion proteins with higher activities than Tat-Rev14, presumably reflecting tighter binding to the RRE IIB site. Three rounds of screening were performed; 7 × 106 cells were sorted in the first round and 800 positive cells were collected, and increasing numbers of strong GFP expressors were observed in the two subsequent rounds (Fig. 4C). Plasmids from 56 individual clones (20 from the second round and 36 from the third round) were tested for activation of the RRE IIB-GFP reporter and 35 showed high level GFP expression by fluorescence microscopy. Six different sequences were found (Fig. 5A), three containing glutamine at position 7 (clones 1, 2, and 6) and three containing frameshifts near the C terminus of the peptide that introduced a glutamine followed by additional arginine residues (clones 3–5). An additional screen was performed in which the sorting window was set slightly lower and 12 new RRE binders were identified, nine containing at least one glutamine, predominantly at position 7 (Fig. 5A).
Figure 5.
(A) Sequences of selected RRE binders. In one screen, a total of 35 GFP-positive clones were identified and six different sequences were found (clones 1–6). In a second screen, 12 new sequences were found after four rounds of sorting using a slightly lower window. Many additional clones were obtained from the second screen but were not sequenced. The position corresponding to Asn-40 of Rev is indicated by arrows. (B) Activities of Tat fusion proteins on an HIV LTR RRE IIB-CAT reporter. The fusion plasmids shown (1, 5, and 25 ng) were cotransfected with the reporter (50 ng) and CAT activities were measured after 48 hr. Fold activation was calculated as the ratio of activities with and without the Tat-expression plasmids.
RNA-binding activities of the six selected Tat fusions were measured using an HIV LTR RRE IIB-CAT reporter, which shows a tight correlation between in vivo activation and specific in vitro RNA-binding affinities (3, 40, 41). All fusions strongly activated CAT expression, and the best fusions were 5–10-fold more active than Tat-Rev14 (Fig. 5B). Binding to the RRE IIB site was specific as judged by the inability of the fusions to activate through an RNA-binding mutant reporter in which G46:C74 was changed to C:G (3) (data not shown).
Glutamine-Mediated Binding Specificity.
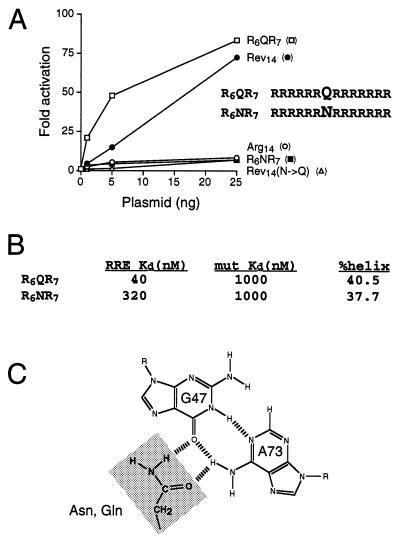
Most of the selected RRE binders contained at least one glutamine residue, most often located at a position corresponding to Asn-40 of Rev (Fig. 5A). This was especially surprising given the recent report of a change of specificity Rev mutant in which an Asn-40→Gln substitution allowed recognition of a mutant RRE IIB site (A73→G) but abolished binding to the wild-type site (27). We constructed a variant, R6QR7, in which glutamine was placed at the equivalent of position 40 in an otherwise all-arginine background and measured activation of the RRE IIB-CAT reporter (Fig. 6A). Remarkably, this variant was substantially more active than the Rev peptide, and equally remarkably, a variant containing asparagine at the same position, R6NR7, was inactive. The opposite result was obtained in the context of the Rev sequence; the wild-type peptide, which contains asparagine, was active whereas the glutamine mutant was inactive (Fig. 6A) (27).
Figure 6.
(A) Activities of Tat fusions containing R6QR7, R6NR7, Rev14, a Rev14(N→Q) mutant, and Arg14. Activities were determined as in Fig. 5B. (B) Apparent Kds and α-helical content of R6QR7 and R6NR7 peptides. Apparent Kds were measured by gel shift assays in the presence of tRNA by using wild-type RRE IIB RNA or a G46:C74→C:G mutant to assess nonspecific binding (affinities in the absence of tRNA competitor are substantially higher; ref. 3). Helical content was estimated from circular dichroism ellipticity at 222 nm (3). (C) Interaction of the carboxamide group of Asn-40 with the RRE G47:A73 bp, as determined by NMR (6, 7).
To confirm that the reporter assays accurately reflect peptide RNA-binding affinities, we synthesized R6QR7 and R6NR7 peptides and measured relative affinities and specificities for the RRE by using gel shift assays. R6QR7 bound RRE IIB with 8-fold higher affinity than R6NR7 whereas both peptides bound weakly to a mutant RNA containing a G46:C74→C:G substitution (Fig. 6B). Circular dichroism spectra indicated that both R6QR7 and R6NR7 were substantially α-helical (Fig. 6B), suggesting that the differences in binding affinity do not reflect different peptide conformations and that R6QR7 probably binds RRE IIB as an α-helix.
DISCUSSION
We have developed an HIV LTR-GFP reporter system based on transcriptional activation by Tat that can be used to screen expression libraries for RNA-binding molecules in mammalian cells. By screening a combinatorial library in which only four positions were randomized within a polyarginine context, we identified several peptides that bind with high affinity to the RRE IIB site and, in the simplest case, found that incorporating a single glutamine is sufficient for specific recognition. The results support the view that arginine-rich peptides provide an excellent framework for designing specific RNA-binding molecules, in part because they adopt different conformations and generally use few amino acids other than arginine for recognition (24). Polyarginine may be viewed as a primordial RNA-binding framework from which specificity may be evolved.
In the Rev peptide, Asn-40 makes a specific contact to a G47:A73 bp, with its carboxamide group donating a hydrogen bond to the O6 group of G47 and accepting a hydrogen bond from the N6 group of A73 (Fig. 6C) (6, 7). We propose that the carboxamide group of glutamine makes a similar contact to the G-A pair in the selected peptides. While the Rev-RRE arrangement accommodates a coplanar orientation between Asn-40 and the G-A pair, the extra methylene group of glutamine cannot be accommodated, yet the converse is true in the polyarginine context. The orientation and depth of penetration of the Rev α-helix in the major groove is determined by a set of contacts to functional groups on the bases and to backbone groups (6, 7), and we imagine that additional arginines in the polyarginine peptide orient the helix less deeply in the groove, thereby allowing glutamine to contact the G-A pair. The observation that the glutamine-containing peptides bind more tightly than the Rev peptide suggests that the presumed new orientation may be energetically more favorable, though detailed structural information is clearly needed to establish the basis for improved binding. In DNA-protein interactions, coplanar amino acid-base arrangements are commonly seen in which arginines form two hydrogen bonds to the guanines of G-C bp and glutamine or asparagine form two hydrogen bonds to the adenines of A-T pairs (42). It appears that glutamine or asparagine also are well-suited to form coplanar hydrogen-bonded interactions with G-A bp (Fig. 6C), which may be common in RNA structures (43).
The mammalian GFP reporter system described here should complement bacterial and yeast screening systems for identifying RNA-binding proteins (23–28). Because plasmid delivery to mammalian cells is nearly clonal and plasmids can be efficiently recovered, it is possible to screen libraries of ≈106–107 complexity with reasonable confidence, a level sufficient to clone low-abundance cDNAs. In the experiments described, GFP reporters were stably integrated into mammalian cells, however additional experiments indicate that both the reporter and Tat-expression plasmids can be delivered simultaneously from the same protoplast if compatible origins are used (A. J. Lynn and A.D.F., unpublished results), eliminating the need to establish stable cell lines and allowing RNA libraries to be screened in the reporter plasmid.
In the cDNA expression cloning strategy developed by Seed and Aruffo (29), plasmids containing a SV40 origin were used because they replicate episomally to high levels in COS cells expressing large T-antigen. Others have shown that vectors containing an Epstein–Barr virus origin and expressing the Epstein–Barr virus-encoded nuclear antigen 1 protein can replicate in a variety of cell types but copy numbers are lower than with SV40-based plasmids (32). Infection with retroviral vectors can overcome some cell type restrictions but can be limited by low viral titers, infection efficiencies, or vector recovery (31). Using the conditions described here, plasmids can be delivered by protoplast fusion and recovered directly without replication, in principle eliminating cell type restrictions. Though our experiments have focused on RNA-protein interactions, other GFP reporters might be devised to screen for protein-protein or protein-DNA interactions.
Acknowledgments
We thank Nigel Kileen for CD4 reagents and use of the FACScan, Steve Landt for the Tat-U1A plasmid, Cynthia Honchell for DNA and peptide synthesis, Paul Dazin (Howard Hughes Medical Institute) for FACS sorting, Nigel Kileen, Lily Chen, Mark Feinberg, and John Young for helpful discussions, and Nigel Kileen, David Julius, and Robert Edwards for comments on the manuscript. This work was supported by an American Foundation for AIDS Research scholarship (R.T.) and by the National Institutes of Health.
ABBREVIATIONS
- BIV
bovine immunodeficiency virus
- RRE
Rev response element
- LTR
long terminal repeat
- FACS
fluorescence-activated cell sorting
- SV40
simian virus 40
- GFP
green fluorescent protein
- CAT
chloramphenicol acetyltransferase
- TAR
transactivating response element
- CMV
cytomegalovirus
References
- 1.Weeks K M, Crothers D M. Cell. 1991;66:577–588. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- 2.Calnan B J, Biancalana S, Hudson D, Frankel A D. Genes Dev. 1991;5:201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- 3.Tan R, Chen L, Buettner J A, Hudson D, Frankel A D. Cell. 1993;73:1031–1040. doi: 10.1016/0092-8674(93)90280-4. [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Frankel A D. Biochemistry. 1994;33:2708–2715. doi: 10.1021/bi00175a046. [DOI] [PubMed] [Google Scholar]
- 5.Tan R, Frankel A D. Proc Natl Acad Sci USA. 1995;92:5282–5286. doi: 10.1073/pnas.92.12.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battiste J L, Mao H, Rao N S, Tan R, Muhandiram D R, Kay L E, Frankel A D, Williamson J R. Science. 1996;273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 7.Ye X, Gorin A, Ellington A D, Patel D J. Nat Struct Biol. 1996;3:1026–1033. doi: 10.1038/nsb1296-1026. [DOI] [PubMed] [Google Scholar]
- 8.Puglisi J D, Chen L, Blanchard S, Frankel A D. Science. 1995;270:1200–1203. doi: 10.1126/science.270.5239.1200. [DOI] [PubMed] [Google Scholar]
- 9.Ye X, Kumar R A, Patel D J. Chem Biol. 1995;2:827–840. doi: 10.1016/1074-5521(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 10.Kao S Y, Calman A F, Luciw P A, Peterlin B M. Nature (London) 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 11.Feinberg M B, Baltimore D, Frankel A D. Proc Natl Acad Sci USA. 1991;88:4045–4049. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marciniak R A, Sharp P A. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato H, Sumimoto H, Pognonec P, Chen C H, Rosen C A, Roeder R G. Genes Dev. 1992;6:655–666. doi: 10.1101/gad.6.4.655. [DOI] [PubMed] [Google Scholar]
- 14.Laspia M F, Wendel P, Mathews M B. J Mol Biol. 1993;232:732–746. doi: 10.1006/jmbi.1993.1427. [DOI] [PubMed] [Google Scholar]
- 15.Rosen C A, Sodroski J G, Haseltine W A. Cell. 1985;41:813–823. doi: 10.1016/s0092-8674(85)80062-3. [DOI] [PubMed] [Google Scholar]
- 16.Roy S, Delling U, Chen C H, Rosen C A, Sonenberg N. Genes Dev. 1990;4:1365–1373. doi: 10.1101/gad.4.8.1365. [DOI] [PubMed] [Google Scholar]
- 17.Ruben S, Perkins A, Purcell R, Joung K, Sia R, Burghoff R, Haseltine W A, Rosen C A. J Virol. 1989;63:1–8. doi: 10.1128/jvi.63.1.1-8.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauber J, Malim M H, Cullen B R. J Virol. 1989;63:1181–1187. doi: 10.1128/jvi.63.3.1181-1187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang C V, Lee W M. J Biol Chem. 1989;264:18019–18023. [PubMed] [Google Scholar]
- 20.Southgate C, Zapp M L, Green M R. Nature (London) 1990;345:640–642. doi: 10.1038/345640a0. [DOI] [PubMed] [Google Scholar]
- 21.Selby M J, Peterlin B M. Cell. 1990;62:769–776. doi: 10.1016/0092-8674(90)90121-t. [DOI] [PubMed] [Google Scholar]
- 22.Southgate C D, Green M R. Genes Dev. 1991;5:2496–2507. doi: 10.1101/gad.5.12b.2496. [DOI] [PubMed] [Google Scholar]
- 23.Laird-Offringa I A, Belasco J G. Proc Natl Acad Sci USA. 1995;92:11859–11863. doi: 10.1073/pnas.92.25.11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harada K, Martin S S, Frankel A D. Nature (London) 1996;380:175–179. doi: 10.1038/380175a0. [DOI] [PubMed] [Google Scholar]
- 25.Wilhelm J E, Vale R D. Genes Cells. 1996;1:317–323. doi: 10.1046/j.1365-2443.1996.25026.x. [DOI] [PubMed] [Google Scholar]
- 26.Fouts D E, Celander D W. Nucleic Acids Res. 1996;24:1582–1584. doi: 10.1093/nar/24.8.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain C, Belasco J G. Cell. 1996;87:115–125. doi: 10.1016/s0092-8674(00)81328-8. [DOI] [PubMed] [Google Scholar]
- 28.SenGupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seed B, Aruffo A. Proc Natl Acad Sci USA. 1987;84:3365–3369. doi: 10.1073/pnas.84.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yates J J, Warren N, Sugden B. Nature (London) 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 31.Kinsella T M, Nolan G P. Hum Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 32.Seed B. Curr Op Biotechnol. 1995;6:567–573. doi: 10.1016/0958-1669(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 33.Schaffner W. Proc Natl Acad Sci USA. 1980;77:2163–2167. doi: 10.1073/pnas.77.4.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandri-Goldin R M, Goldin A L, Levine M, Glorioso J C. Mol Cell Biol. 1981;1:743–752. doi: 10.1128/mcb.1.8.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rassoulzadegan M, Binetruy B, Cuzin F. Nature (London) 1982;295:257–259. doi: 10.1038/295257a0. [DOI] [PubMed] [Google Scholar]
- 36.Oubridge C, Ito N, Evans P R, Teo C H, Nagai K. Nature (London) 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 37.Frankel A D, Pabo C O. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 38.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 39.Hirt B. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 40.Tan R, Frankel A D. Biochemistry. 1994;33:14579–14585. doi: 10.1021/bi00252a025. [DOI] [PubMed] [Google Scholar]
- 41.Symensma T L, Giver L, Zapp M, Takle G B, Ellington A D. J Virol. 1996;70:179–187. doi: 10.1128/jvi.70.1.179-187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seeman N C, Rosenberg J M, Rich A. Proc Natl Acad Sci USA. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wyatt J R, Tinoco I., Jr . In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 465–496. [Google Scholar]