Abstract
Apurinic endonuclease 1 / redox factor-1 (Ape1/Ref-1 or Ape1) is an essential protein with two distinct functions. It is a DNA repair enzyme in the base excision repair (BER) pathway and a reduction-oxidation (redox) signaling factor maintaining transcription factors in an active reduced state. Our laboratory previously demonstrated that Ape1 is overexpressed in ovarian cancer and potentially contributes to resistance. Therefore, we utilized siRNA technology to knockdown protein levels of Ape1 in ovarian cancer cell line, SKOV-3x. Knocking Ape1 down had dramatic effects on cell growth in vitro but was not due to an increase in apoptosis and at least partially due to an extension in transit time through S-phase. Similarly, human ovarian tumor xenografts with reduced levels of Ape1 protein demonstrated a dramatic reduction in tumor volume (p<0.01) and also statistically significant (p=0.02) differences in 18F-fluorodeoxyglucose (FDG) uptake indicating reduced glucose metabolism and cellular proliferation. Ape1’s role in DNA repair and redox signaling is important to our basic understanding of ovarian cancer cell growth and these findings strongly support Ape1 as a therapeutic target.
Keywords: base excision repair, Apurinic endocuclease1/Redox effector factor 1, tumor xenograft, ovarian cancer, cell cycle
1. INTRODUCTION
Ovarian cancer is the fourth most common cause of cancer death in women, and the leading cause of gynecologic cancer death in developed countries. In 2007, there are expected to be over 22,000 new cases of ovarian cancer diagnosed and over 15,000 deaths due to this deadly disease [1]. Approximately 90% of ovarian cancers are of the epithelial type (versus germ cell or stromal tumors), and 75–80% of patients present with metastatic disease beyond the ovaries. Our laboratory previously demonstrated that apurinic endonuclease 1 / redox factor-1 (Ape1/Ref-1; Ape1) is overexpressed in ovarian cancer and patterns of Ape1 expression change as the tumor grade increases [2]. Ape1 is an essential enzyme in the base excision repair (BER) pathway which is responsible for the repair of oxidative and alkylation damage in DNA and thus protects cells against the toxic effects of endogenous and exogenous agents [3]. Ape1 possesses DNA repair activity as an endonuclease that hydrolyzes the phosphodiester backbone immediately 5’ to an apurinic/apyrimidinic (AP) site. The DNA repair activity of Ape1 serves to protect the cell from the cytotoxic and mutagenic AP sites that can accumulate in DNA and may well be involved in ovarian cancer resistance to chemotherapy [2].
In addition to its role in DNA repair, Ape1 functions as a reduction-oxidation (redox) signaling factor maintaining transcription factors in an active reduced state (reviewed in [4] and [5]). Ape1 stimulates the DNA binding activity of numerous transcription factors that are involved in cancer promotion and progression such as AP-1 (fos/jun), NFκB, HIF-1α, CREB, p53 and others[4, 5].
The importance of Ape1 to cell viability is demonstrated by the lethality of Ape1 mouse knockouts at E3.5 to E9.5 and the lack of viable cell lines completely deficient for Ape1[6] and reviewed in [7]. In human colon and breast cancer cells as well as human lymphoblastoid cells, siRNA directed against Ape1 results in a decrease in proliferation, an increase in AP sites and increased levels of apoptosis. Knocking out Ape1 or using various dominant-negative forms of Ape1 (nuclease deficient ED mutant and redox deficient C65A mutant) lead to chemotherapeutic agent sensitization and emphasize the importance of both functions of Ape1 in both normal and tumor cells [8–10]. Targeted reduction of Ape1 protein by specific anti-sense oligonucleotides or siRNA renders mammalian cells hypersensitive to a variety of chemotherapeutic agents [11–15]. Studies using methoxyamine (MX) which can block the DNA repair function of Ape1, but not its redox function, lead to tumor cell sensitivity in combination with alkylating agents such as temozolomide (TMZ)[16–22].
Based on the above studies, we investigated the effect of siRNA knockdown of Ape1 in ovarian cancer cells and demonstrated that knocking Ape1 down in SKOV-3x cells results in a decrease in proliferation in vitro which appears to be due in part to an extension of S-phase. In addition, siRNA knockdown of Ape1 severely inhibited the growth of ovarian tumor xenografts. This data demonstrates that the Ape1 protein is crucial in the regulation of ovarian cancer cell growth and is a rational therapeutic target for drug development in the treatment of ovarian cancer.
2. MATERIALS & METHODS
Cell lines
SKOV-3x cells were kindly provided by Dr. Robert Bigsby (Department of Ob/Gyn, Indiana University School of Medicine) and were grown in McCoy’s 5a Media (Invitrogen; Carlsbad, CA) supplemented with 5% Cosmic Calf Serum (Hyclone; Logan, UT) and sodium pyruvate (final concentration 1mM) at 37°C and 5% CO2.
Transfection of SKOV-3x cells with Ape1 and Scrambled siRNA
Sequences of the double-stranded siRNAs are scrambled (5′ CCAUGAGGUCAGCAUGGUCUG 3′, 5′ GACCAUGCUGACCUCAUGGAA 3′) and Ape1 (5′ GUCUGGUACGACUGGAGUACC 3′, 5′ UACUCCAGUCGUACCAGACCU 3′) as previously described [8, 23]. The 21-base sequence was subjected to a BLAST-search (NCBI) database of EST libraries to ensure that only one gene was targeted. Approximately 2.5×105 SKOV-3x cells are plated in a 10 cm dish and allowed to attach overnight. The next day, oligofectamine reagent (Invitrogen, Carlsbad, CA) was used to transfect in the Ape1 and scrambled siRNA at concentrations between 25 and 100 nM following the manufacturer’s indicated protocol. Opti-MEM, siRNA, and oligofectamine was left on the cells for at least 6 h and then regular SKOV-3x media was added. Cells were assayed for Ape1 protein expression, apoptosis, and cell cycle from 1 to 10 days following transfection.
Western blot analysis
Exponentially growing controls, Ape1 siRNA- or scrambled siRNA- treated SKOV-3x cells were harvested, washed in cold PBS, and lysed in ~50 µl of RIPA buffer containing phosphatase and protease inhibitors (Santa Cruz Biotechnology, Santa Cruz, CA). Protein was quantified and electrophoresed (20 µg) in SDS gel-loading buffer on a 12% SDS–polyacrylamide gel. Mouse monoclonal anti-Ape1 Ab (1:1000) (Novus Biologicals, Littleton, CO) was used to confirm a reduction in the levels of Ape1 protein. Anti-actin antibody (1:1000; as a loading control, LabVision Corporation, NeoMarkers, Fremont, CA) was used to probe for protein levels as previously described [10].
Immunocytochemistry (ICC)
The methods for ICC were conducted as previously described [2, 24]. Briefly, following cytospin, cells were coated with anti-Ape1 antibody (mouse anti-human Ape1/Ref-1 monoclonal; Novus Biologicals, Littleton, CO) and incubated overnight at 4°C at a 1:200 dilution in 10% goat serum in PBS. The following day, slides were washed three times for 5 minutes in PBS and incubated with biotinylated goat anti-mouse IgG (Vector Labs; Burlingame, CA) at 15 µg/ml in 10% goat serum for one hour. After two PBS washes for 5 minutes each, slides were incubated with avidin and biotinylated horseradish peroxidase complex (ABC elite kit; Vector Labs) for 45 minutes. Slides were then incubated with diaminobenzidine (Vector Labs). After the development of color signal the sections were briefly washed in dH2O, counterstained with eosin, dehydrated through a graded alcohol to xylene sequence, coverslipped, analyzed and photographed. As a negative control preimmune IgG (50 µg/ml) was used as the primary antibody in place of anti-Ape1. H & E (hematoxylin and eosion) staining was as previously described.
SRB assay for determination of growth rate of cells
SKOV-3x cells previously treated with mouse and Ape1 siRNA as described above were used in this assay as previously published.[18] Four thousand cells were aliquoted in quadruplicate three days after transfection for each sample in each of six 96-well plates and allowed to adhere overnight. One well of media alone was plated for each sample on each plate as a control. On days 3–8 after transfection, the cells were fixed with 50% Trichloracetic Acid for 1h at 4°C, rinsed eight times with water, air dried and stored until assayed. The fixed cells were then stained with SRB (sulforhodamine B) reagent for 30min, washed eight times with 10% acetic acid and air-dried. Once plates for each timepoint were fixed and air-dried, the incorporated SRB was dissolved in a volume of 10mM Tris equal to the starting culture volume. Lastly, the absorbance was read at 565nm and 690nm using a plate reader. The background 690nm reading was subtracted from the experimental reading at 565nm. The values were standardized to wells containing media alone.
Apoptosis Assays via Alexa Fluor 488-Conjugated Annexin-V (Annexin-V) / Propidium Iodide Staining
To analyze the cells for apoptosis, cells were plated and allowed to attach overnight. Cells were transfected as described above. Apoptosis was assayed at Day 2 and 3 following transfection. Cells were trypsinized, pelleted, washed in ice-cold PBS, and resuspended in 1X Binding Buffer (10 mM Hepes/NaOH pH7.4, 140 mM NaCl, 2.5 mM CaCl2). Apoptosis was analyzed using the Alexa Fluor® 488 Annexin-V from Vybrant® Apoptosis Assay Kit in combination with propidium iodide (PI) (Molecular Probes, Eugene, OR) as previously described [17]. Cells that were strongly Annexin-positive were considered positive for apoptosis. The samples were analyzed by flow cytometry in the Indiana University Cancer Center (IUCC) flow cytometry facility.
Cell cycle staining protocols
PI staining
To stain the cells for DNA content and analyze the percentage of cells in G0/G1, S, and G2/M, approximately 2.5×105 cells were plated in a 10 cm dish, allowed to attach overnight, and transfected as described above. Cells were harvested at Day 3 following Ape1 knockdown and then fixed in 70% ice-cold EtOH and stored at 4°C until PI staining. Fixed cells were pelleted, washed in PBS, and incubated with RNase (0.1 mg/mL) at 37°C for 30 minutes. Cells were pelleted again, washed in PBS to remove the RNase, and then resuspended in PI stain solution (0.1 mg/mL) with a final cell concentration 1×106 cell/mL. The cells were then incubated on ice for 30 min and analyzed by flow cytometry in the Indiana University Cancer Center (IUCC) flow cytometry facility.
BrdU staining
To stain the cells for DNA content and analyze the movement of cells through G0/G1, S, and G2/M, approximately 2.5×105 cells were plated in a 10 cm dish, allowed to attach overnight, and transfected as described above. A BrdU pulse (10µM) was added to the cells for 30 min and then the cells were washed twice with PBS. Cells were harvested at Day 3 following Ape1 knockdown and then processed following the BrdU Flow Kit protocol as indicated by the manufacturer (BD Pharmingen, San Diego, CA). Briefly, cells were fixed and permeablized with BD Cytofix/Cytoperm Buffer, then the nuclear membrane was permeabilized using BD Cytoperm Plus Buffer. From here, cells were treated with DNase (30µg per sample) to expose BrdU epitopes and then stained with the BrdU antibody (1:50 dilution) provided in the kit. Total DNA was stained with 7-AAD. The cells were then analyzed for DNA content (%G1, %S, %G2/M) and BrdU incorporation by flow cytometry utilizing ModFit (Topsham, ME) and CellQuest (BD Biosciences, San Jose, CA).
Xenograft tumors in nude mice
SKOV-3x cells were transfected with Ape1 and scrambled siRNA ex vivo as described above and 107 SKOV-3x cells in 0.2 ml of Hank's balanced salt solution were implanted subcutaneously (s.c.) into the right flanks of female nude mice (Harlan Inc, Indianapolis, IN). The tumors were measured two times per week beginning at Day 4 and followed for up to 3 weeks. Tumor volumes were monitored by caliper measurement [tumor volume = length × (perpendicular width)² × 0.5] and the average tumor volume in mm³ for each treatment group was plotted versus days post-injection for each treatment group. Average tumor volume ± SD for the scrambled and Ape1 siRNA-treated SKOV-3x ovarian cancer cells from three - five independent experiments at Days 4, 7, and 11 plus tumor volume doubling times were compared by a standard T-test.
PET Imaging Methods
Small Animal PET Imaging System
Studies were performed using the IndyPET-II scanner.[25] The IndyPET-II system has a 23 cm and 15cm transxial and axial field of view(FOV) respectively. Within an axial image plane the average full-width-at-half-maximum (FWHM) resolution is 2.5 mm at the center of the FOV. The NEMA-2001 sensitivity is 9030 cps/MBq at the center of the scanner and 4250 cps/MBq at a distance of 10cm from the center of the FOV. All data are acquired with a 350–650 keV energy window. The IndyPET-II scanner collects all data in list mode (the time and location of individual events is recorded). Following data acquisition events are combined to form event versus position histograms (sinogram) used for image reconstruction.
Small Animal CT Imaging System
Each animal imaging session included a high resolution x-ray CT scan using the EVS MS-9 scanner (Enhanced Vision Systems Corp, London, Ontario N6G 4X8) in the 100 micron voxel resolution mode. Images were reconstructed using a Feldcamp cone beam reconstruction algorithm. The CT image data serves as an anatomical reference to aid interpretation of the PET image data.
Image Registration & Fusion
A standardized approach was used for registration of data from the IndyPET-II PET scanner and the EVS RS-9 micro-CT scanner. Both imaging systems have a common stage with mounting pins for precise localization of the animal bed relative to the scanner gantry. A 6-parameter rigid body registration transform of the PET image to the CT image space is calculated using PET and CT images of a registration phantom consisting of parallel and perpendicular tubes filled with a mixture of [18F]FDG and iodinated contrast agent. The overall fiducial registration error is less of this approach is less than 0.4 mm. The phantom derived registration transformation parameters were applied to the animal CT and PET data sets to register the anatomical and functional image data sets enabling the use of the CT anatomical data to help guide the localization of tumors in the PET image data.
Imaging Protocol
A series of PET imaging studies were performed using [18F]fluorodeoxyglucose ([18F]FDG) to measure glucose utilization. [18F]FDG was prepared by the Hamacher method using a commercial synthesis unit provided by PETNET/CTI (Knoxville, TN).[26] One mCi of [18F]FDG was administered intravenously to the mouse via tail vein.
Image Reconstruction
PET images were reconstructed into an 80 mm transaxial field of view, 0.625 mm pixels and 47 slices spaced 3.15 mm apart. Reconstructions were performed with a filtered back-projection algorithm using a Hanning filter with a cutoff frequency of 4.2/cm.
FDG Utilization Estimates
FDG uptake estimates were generated by placing ROIs on PET images from 45 – 60 minutes post tracer administration. An index of tumor FDG uptake was generated for each tumor by calculating the total FDG uptake in the tumor normalized by the mass of the animal and the injected dose of tracer. Equation 1 was used to generate FDG uptake indices (UIFDG).
| (1) |
Serial Imaging Studies
PET and CT imaging studies were performed at 2, 4, and 6 weeks following administration of tumor cells. The FDG uptake index was calculated for each study and compared for differences in total FDG uptake.
3. RESULTS
Ape1 siRNA effectively reduces Ape1 protein levels in ovarian cancer cells
SKOV-3x ovarian cancer cells were treated with a range of Ape1 siRNA concentrations from 25 to 100 nM resulting in a reduction in the amount of Ape1 protein by >85% versus scrambled siRNA controls. Figure 1A is a representative Western blot demonstrating the knockdown of Ape1 protein levels in whole cell extracts with increasing amounts of siRNA. Panel B is a representative Western blot demonstrating the expression of Ape1 at Day 2, 3, 5, 7 and 9 following transfection, and Panel C demonstrates the quantitation of the levels of Ape1 expression over time. Ape1 expression is normalized first to actin and then to SKOV-3x cells treated with oligofectamine. Ape1 expression is beginning to return over time and is reduced 34% at Day 9 compared to ~80% at Day 2 and 3 (Figure 1 B, C). Actin is included to demonstrate the presence of protein in all lanes and serves as a loading control. Panel D includes immunocytochemistry (ICC) utilizing Ape1 antibody and H & E (hematoxylin and eosion) staining. Figure 1 clearly demonstrates that significant knockdown of Ape1 protein in the ovarian cancer cell line, SKOV-3x following treatment with Ape1 siRNA is possible. A scrambled control siRNA is included in all panels to demonstrate the specificity of the Ape1 siRNA. However, as observed in panel 4 of Figure 1C, while Western blot demonstrates the total amount of Ape1 protein is significantly decreased in the sample, individually the amount of Ape1 that is knocked down varies per cell, but is still significantly decreased relative to the scrambled control.
Figure 1.
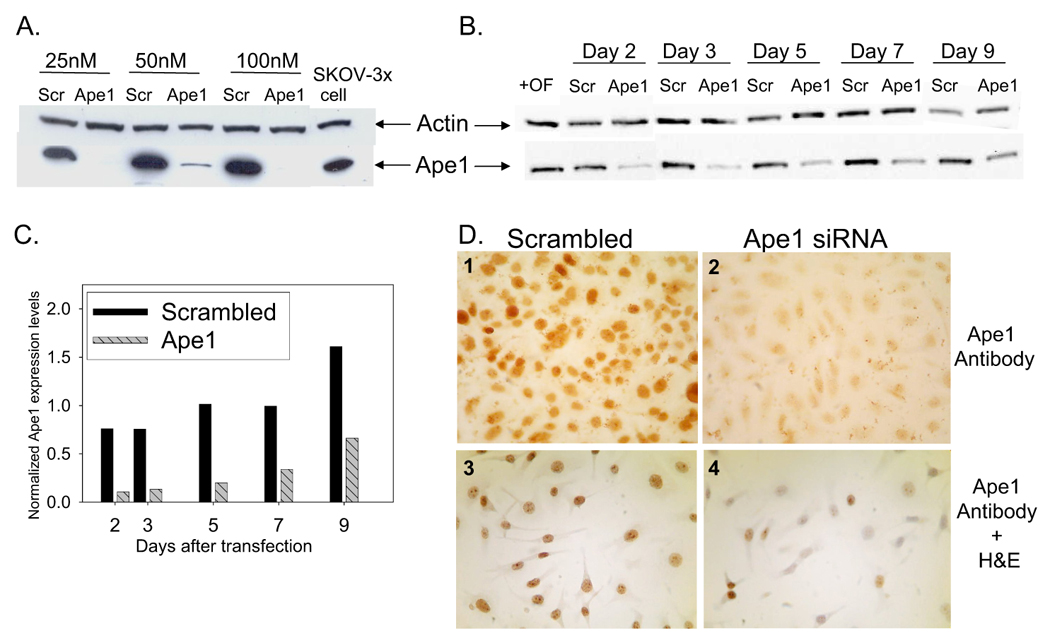
Ape1 protein expression following siRNA treatment. A. Representative western blot of Ape1 protein levels on Day 3 following transfection with scrambled or Ape1siRNA at 25, 50, and 100 nM. B. Time course of Ape1 protein levels via western blot following transfection with scrambled or Ape1siRNA at 50nM, with Day 1 referring to 24 h after the addition of oligofectamine. Actin was utilized as a loading control. C. Quantitation of Ape1 levels following Ape1 (grey striped bars) and scrambled (black bars) siRNA transfection at 50nM. Samples were normalized first to actin and then to SKOV-3x cells treated with oligofectamine alone. D. Immunocytochemical staining of SKOV-3x cells on Day 3 with monoclonal Ape1 antibody and H & E (#3, 4 only).
Ape knockdown reduces cell growth and alters cell cycle profile of ovarian cancer cells
Following treatment with Ape1 siRNA, the growth rate of ovarian cancer cell line, SKOV-3x was monitored. As observed in Figure 2, the reduction of Ape1 protein in the SKOV-3x cells dramatically slowed their growth rate following transfection. The cell population doubling time for the control siRNA-treated cells between four and six days after transfection is 0.5 days while the Ape1 siRNA-treated cells still have not doubled eight days after transfection. We next examined whether this reduced growth rate was due to an increase in apoptotic cell death. However, a reduction in Ape1 did not coincide with an increase in apoptosis which was clearly demonstrated in siRNA-control cells both by Annexin-V staining at Day 2 (data not shown) and Day 3 and sub-G1 DNA content using flow cytometry analysis (Figure 3A, B). Dramatic reduction of Ape1 protein from SKOV-3x cells does not induce an apoptotic response in the absence of exogenous DNA damage. SKOV-3x cells are not defective in an apoptotic response as several laboratories, including ours, have demonstrated that this cell line can undergo apoptosis when treated with DNA damaging agents such as paclitaxel, platinum compounds, temozolomide, and others [17, 27, 28].
Figure 2.
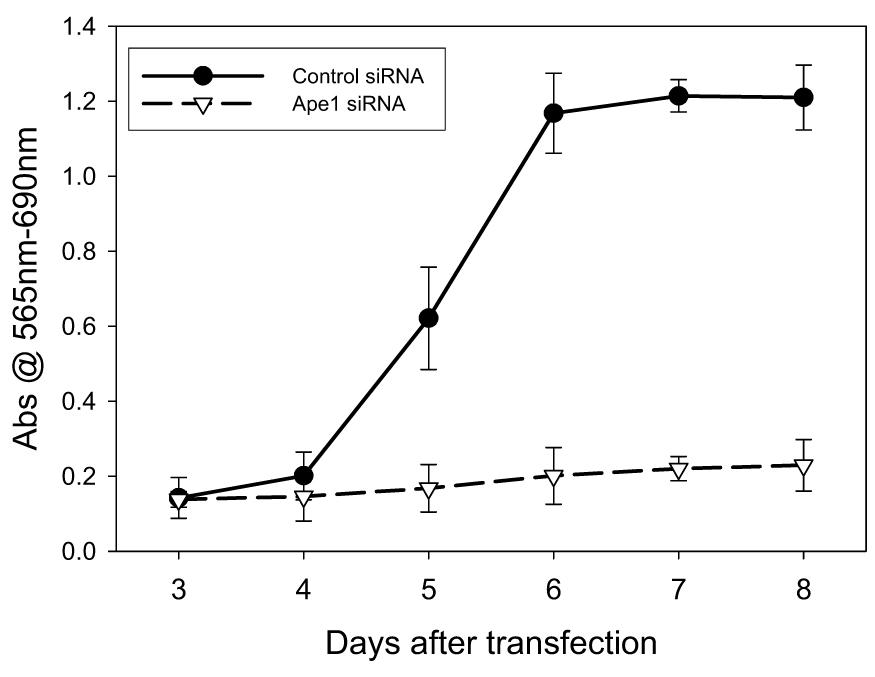
Growth rate of SKOV-3x cells following 25 nM siRNA treatment. SRB assay was used to determine the growth rate.
Figure 3.
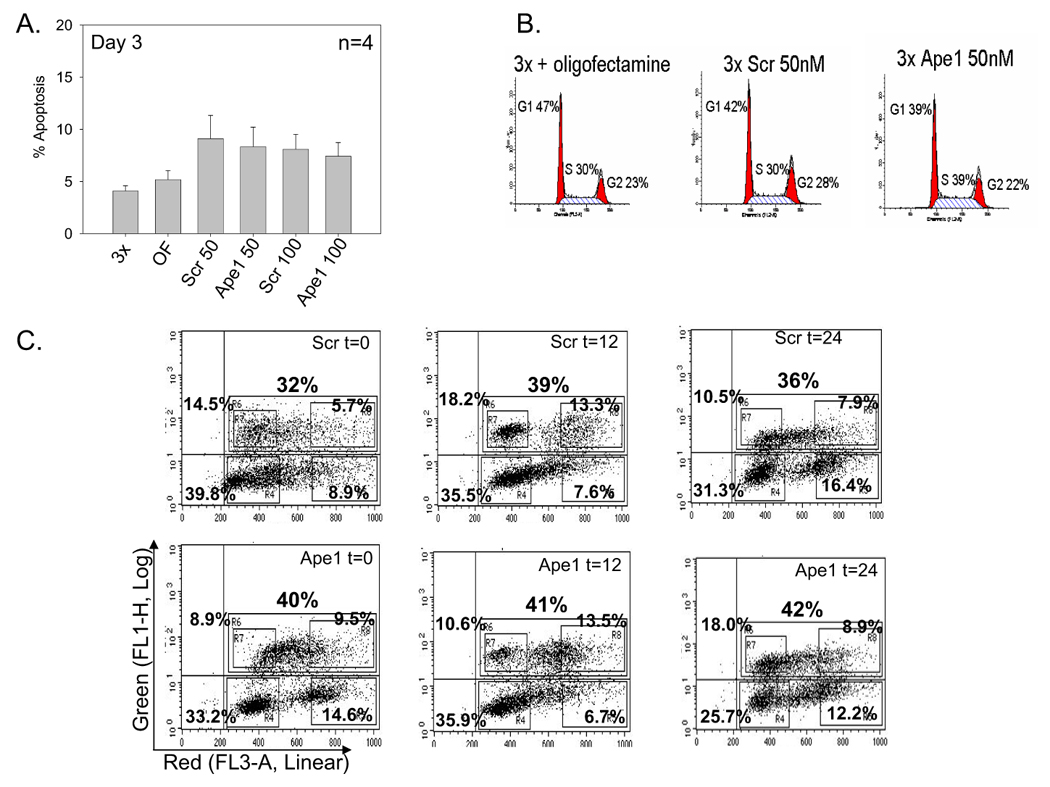
Apoptosis and cell cycle analysis of SKOV-3x cells following siRNA treatment at Day 3. A. Bar graph showing the average % apoptosis in 4 independent experiments ± SE; B Representative histogram plots of SKOV-3x cells treated with oligofectamine, scrambled (scr) siRNA 50nM, and Ape1 siRNA 50nM stained with PI; C. BrdU incorporation assays at t=0, 12, & 24 hr. Representative dot plots of SKOV-3x cells treated with scrambled (scr) siRNA 50nM (top panel), and Ape1 siRNA 50nM (bottom panel) stained with BrdU-FITC antibody and 7-AAD.
We then investigated whether the reduction of Ape1 may slow cell cycle time or the length of the G1, S, or G2/M phases of the SKOV-3X cells using DNA cell cycle analysis and BrdU incorporation assays. The percentage of cells in a particular phase of the cell cycle is proportional to the average length in time for that particular cell cycle phase to occur [29, 30]. In Table 1 and in Figure 3B, we demonstrate that at Day 3 following transfection, the percentage of cells in S-phase is statistically significantly increased in the Ape1 siRNA-treated cells versus the scrambled control (p<0.05). This is confirmed in the BrdU studies in Figure 3C with an increase of ~25% more cells in S-phase in the Ape1-deficient cells. Furthermore, following the BrdU labeled cells versus time post-labeling as they progress around the cell cycle demonstrates that the progression out of S-phase is slower and the return of these labeled cells to G1-phase is delayed in the Ape1 knockdown cells versus scrambled control cells. For example, in scrambled control cells, we observe a 42% increase in cells that have returned to G1 compared to the Ape1 deficient cells at t=12. The transit time through S-phase is extended at least 25% in cells treated with Ape1 siRNA, however no apoptosis was observed either by Annexin-V staining or a sub-G1 peak. These data suggest that knockdown of Ape1 slows cell cycle progression and increases the length of S-phase in ovarian cancer cells.
Table 1.
Cell cycle percentages for the SKOV-3x cells treated with oligofectamine, Scrambled siRNA (50 & 100nM) and Ape siRNA (50 & 100nM). Experiment was conducted three separate times.
| Treatment | %G1 ± SE | %G2/M ± SE | %S ± SE |
|---|---|---|---|
| Oligofectamine control | 45.0 + 1.5 | 22.7 + 0.3 | 32.3 + 1.9 |
| Scrambled (50nM) | 43.7 + 2.2 | 26.3 + 0.9 | 30.3 + 1.5 |
| Ape (50nM) | 40.3 + 1.3 | 23.7 + 1.2 | 35.7 + 1.8* |
| Scrambled (100nM) | 43.0 + 2.1 | 26.0 + 1.2 | 30.7 + 2.3 |
| Ape (100nM) | 39.0 + 1.2 | 24.3 + 0.9 | 37.0 + 0.0* |
p<0.05 using student’s t test, comparing Scrambled siRNA control vs Ape siRNA at corresponding dose.
Ape1 knockdown inhibits ovarian cancer tumor xenograft growth
Based on the in vitro data demonstrating that knocking Ape1 down caused a dramatic effect on the growth rate of SKOV-3x ovarian cancer cells and a slower cell cycle, we turned to ovarian tumor xenografts to determine if a similar result would be observed in vivo following Ape1 knockdown. Tumor xenografts from SKOV-3x cells that have reduced levels of Ape1 protein demonstrate a dramatic reduction in growth rate compared to tumors that have wild type levels of Ape1 protein (Figure 4A, B). Quantitation of the tumor doubling time showed an increase in tumor doubling time by 3.2-fold from 5 days to over 15 days in the tumors which have Ape1 protein knocked down and statistically significant smaller tumor volumes at timepoints when Ape1 levels are reduced (Figure 4B, C).
Figure 4.
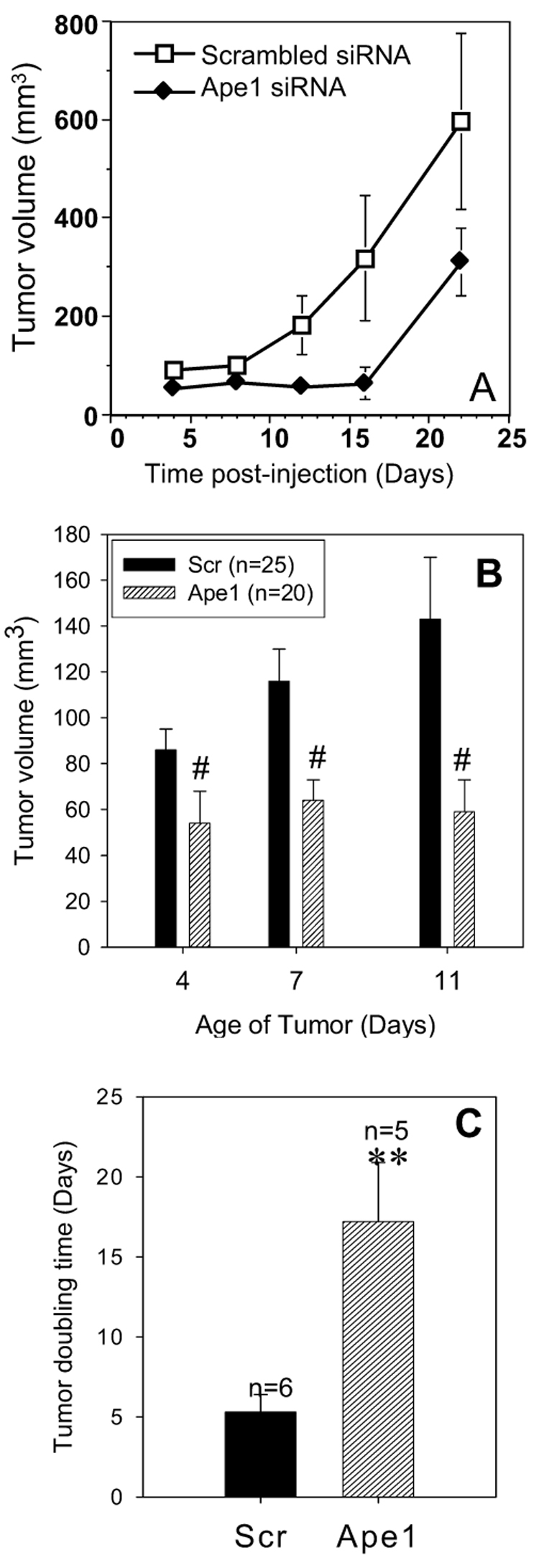
A. Representative tumor growth curve from one experiment with 4 mice per treatment group. B. Summary graph representing all the animals that were injected with cells that were transfected with Scrambled control or Ape1 siRNA as described in Materials & Methods, # p<0.001. C. Average tumor doubling time for tumors that express Ape1 and tumors that do not express Ape1, ** p<0.01.
Ape1 protein expression in cells from the xenografted tumor
To gain a better understanding of the relationship between the growth rate of the ovarian tumors and the Ape1 protein expression after siRNA treatment ex vivo, we collected tumor tissue at various timepoints following implantation of SKOV-3x cells transfected with either scrambled or Ape1 siRNA. After 7 days in the flank of the mouse, Ape1 protein expression is nearly completely suppressed. It is only after 14 days that Ape1 protein levels begin to recover but remain reduced compared to scrambled controls (Figure 5). We therefore observe a strong correlation between Ape1 protein expression and tumor growth rate. As the Ape1 siRNA is diluted out of the SKOV-3x tumor cells, the xenografted tumors slowly recover and begin to grow at a rate that is similar to SKOV-3x cells that have been transfected with scrambled control (Figure 4A and 5).
Figure 5.
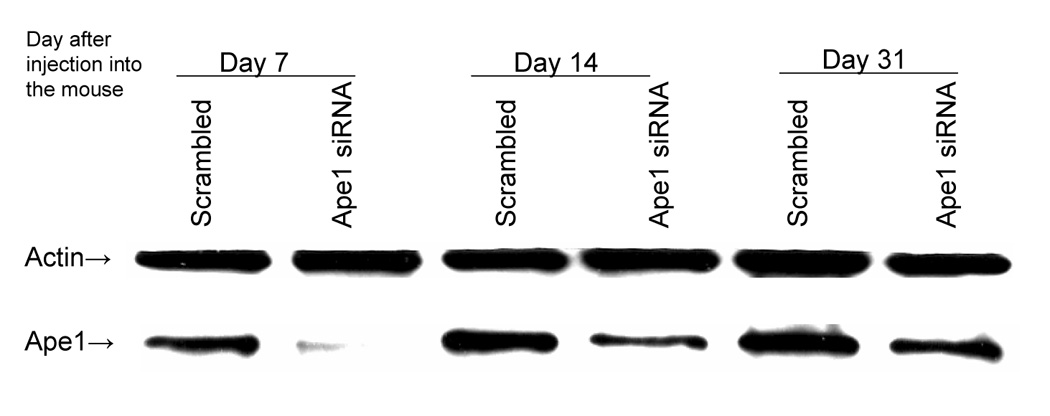
Western blot demonstrating the Ape1 protein levels in samples from tumor xenografts. Lanes 1 and 2 are samples collected 7 days after injection. Lanes 3 and 4 are samples collected at 14 days after injection, and Lanes 5 and 6 are samples collected 31 days after injection. Actin is utilized as a loading control.
Xenografted human ovarian tumors that are not expressing Ape1 have a reduced capacity for uptake of 18F-fluorodeoxyglucose (FDG)
To demonstrate whether the reduction of Ape 1 influenced ovarian tumor cell metabolism and could help explain the slowed cell cycle progression, SKOV-3x tumor xenografts, were allowed to grow for 4 weeks and 2-[18F]fluoro-2-deoxy-D-glucose (FDG) microPET and microCT studies were performed. PET imaging data was acquired continuously for 1 hour following a tail vein injection of FDG. Following the FDG study, the animal holder was transferred to the EVS RS-9 microCT scanner and a 93 mm resolution scan was performed. Figure 6A shows the fusion of a PET FDG image with the corresponding anatomy from a CT image. This fused image clearly shows the increased FDG uptake in the untreated xenograft ovarian tumor. The uptake of FDG in the SKOV-3x tumor can be readily identified as well as the predicted uptake of FDG in the bladder and liver (Figure 6A). The area of high FDG accumulation below the acquired CT image is the injection site in the tail vein. A plot of the FDG accumulation in the SKOV-3x tumor as a function of time after tracer administration is shown in Figure 6B. Figure 6C demonstrates the reduced FDG uptake in the tumor xenografts that are not expressing Ape1 protein. After 4 weeks, there is statistically significant (p=0.02) differences in FDG uptake between tumors that have endogenous Ape1 protein and tumors that have reduced levels of Ape1 (Table 1).
Figure 6.
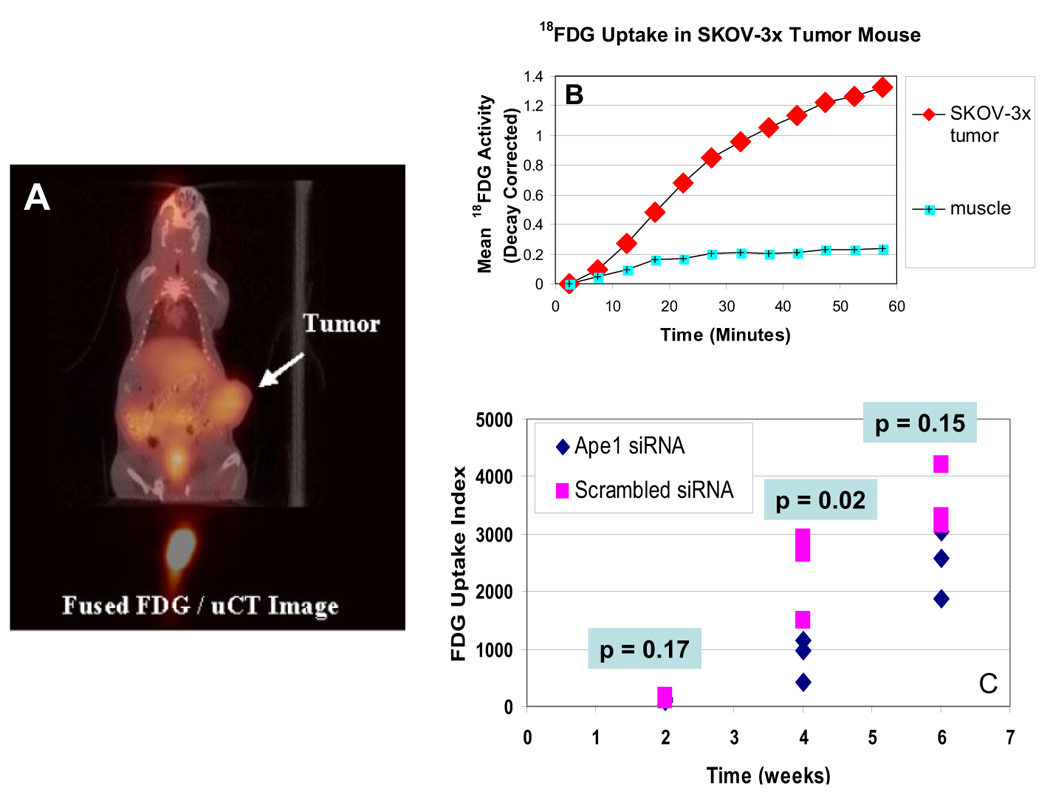
A. Fusion Image of FDG Uptake with x-ray CT anatomy. The uptake of FDG in the SKOV-3x tumor can be readily identified in the tumor as well as the bladder and liver. The area of high FDG accumulation below the acquired CT image is the injection site in the tail vein. B. Time activity curves for FDG uptake in SKOV-3x tumor xenograft. A region of interest was placed within the tumor volume shown and a second region of interest was placed over a skeletal muscle in the thigh of the mouse. The curve demonstrates a tumor/muscle contrast ratio of ~5 at 60 minutes post tracer injection. C. Graph showing the FDG uptake for individual mice at 2, 4, and 6 weeks following injection. An index of tumor FDG uptake was generated for each tumor by calculating the total FDG uptake in the tumor normalized by the mass of the animal and the injected dose of tracer. A paired two-tailed t test was used to determine significance.
4. DISCUSSION
Ovarian cancer is the fourth-leading cause of cancer death in women. There are currently no available screening tests for the early detection of ovarian cancer. Consequently, three-fourths of women with ovarian cancer present with advanced disease, and the majority of these women will ultimately succumb. Improved surgical techniques and the identification of new chemotherapy agents such as cisplatin and its derivatives, and more recently paclitaxel, have yielded modest improvements in survival. Future reductions in ovarian cancer mortality can only be obtained through the identification and implementation of effective screening tests for apparently-healthy women, and through the identification and implementation of better treatments [1, 31]. To this end, here we investigate the effects of knocking out Ape1, an important protein in DNA repair and in redox signaling. In this study, siRNA treatment effectively knocked out ~85 – 90% of the Ape1 protein in ovarian cancer cells. Our findings here demonstrate that reduced levels of Ape1 dramatically slow the growth of ovarian cancer cells, both in vitro and in vivo and is at least partially due to an extended time in S and perhaps G1 phase of the cell cycle. Unlike other reports[32, 33], the induction of cell death by apoptosis does not appear to play a role in the slower growth induced by siRNA reduction of Ape1. This is the first study in which the effects of reduced levels of Ape1 on tumor cell growth have been investigated in vivo. The tumors that did not express Ape1 protein were smaller, took longer to double in size, and had a statistically significant reduction of FDG uptake indicating reduced glucose metabolism and cellular proliferation. Along with caliper measurement of tumor growth, sophisticated imaging techniques verified the decrease in the volume of tumors that were not expressing Ape1 (data not shown).
Our studies confirmed published observations that Ape1 is important to the function and propagation of cells, both normal and cancer cells [11–15, 32, 33]. There are some conflicting reports regarding whether knocking Ape1 down results in apoptotic cell death. While we did not observe an increase in apoptosis in SKOV-3x ovarian cancer cells, other laboratories utilizing different cellular systems have observed that a dramatic reduction in Ape1 protein results in apoptosis even in the absence of DNA damage [32, 33]. Using siRNA specific for the Ape1 protein in JB6+ mouse epidermal cell lines and in JB6P+ cells which are resistant to H2O2 and Cadmium, a model system utilized to study transformation and tumor promotion, a reduction in colony formation of ~50% is observed when the Ape1 protein is not present. An increase in apoptosis is also observed in the transformed cells when Ape1 protein is knocked down to ~90% of wild type cells [33]. However, while hemangioblast development and primitive and definitive hematopoiesis from ES cells was reduced when Ape1 expression was knocked down by Ape1-specific siRNA, this reduction in hematopoiesis was not associated with increased apoptosis [34]. We at least partially attribute the differential apoptotic response to Ape1 knockdown to be cell-type dependent as different cell types would possess separate signaling and growth pathways and the role of Ape1 could vary between cell types. Different and distinct methods of knocking down Ape1 all result in slowed growth rates and recovery of growth rate as Ape1 expression is restored demonstrating that the Ape1 protein is vital to propagation of growth signaling and in certain cell types results in apoptotic cell death [32–34]. Our laboratory is continuing to investigate the contribution of redox and DNA repair activities of Ape1 to the growth rate of ovarian cancer cells.
A better understanding of Ape1’s involvement, either redox or DNA repair, in ovarian cancer growth and resistance to treatment is important as we seek to treat ovarian cancer more effectively. Other laboratories have demonstrated that knocking out Ape1 or using a dominant-negative form of Ape1 does lead to chemotherapeutic agent sensitization [8–10]. Small molecule, methoxyamine (MX) that blocks Ape1 DNA repair function, but not its redox function leads to ovarian, breast, and colon tumor cell sensitivity when combined with alkylating agents, TMZ and BCNU[16–22]. Regardless of the mechanism of slowed growth of the ovarian cancer xenografts, our data demonstrates the importance of Ape1 to ovarian tumor growth. In order to investigate the importance of redox and DNA repair, we plan to express Ape1 redox mutants or DNA repair mutants in ovarian cancer cells. Both of the activities of Ape1 have potential in sensitizing ovarian cancer cells to chemotherapeutic agents. Additionally, we have a small molecule inhibitor of Ape1’s redox function[34] and have recently selected Ape1 repair inhibitors in a chemical genomics screen. These small molecule inhibitors will be used in conjunction with the transgene approach to delineate each of Ape1’s main functions, redox and AP endonuclease activity, in tumor growth. Ape1’s role in DNA repair and redox signaling is important to our basic understanding of ovarian cancer cell growth. Some of the downstream transcription factors that Ape1 targets are involved in general cellular stress response which could also account for the effects observed in our studies. These studies all support Ape1 as a therapeutic target for enhancing currently used chemotherapy as well as the development of new agents as both combination or single-acting agents for this same purpose.
Table 2.
Mean FDG uptake corresponding to the graphical representation in Figure 6C.
| Week |
Ape1 siRNA |
Scrambled siRNA |
|---|---|---|
| 2 | 109 | 151 |
| 4 | 838 | 2366 |
| 6 | 2499 | 3558 |
Acknowledgements
This work was supported by grants CA094025, CA106298, CA114574 and Riley Children's Foundation (M.R. Kelley), CA122298 (M.L. Fishel) and a Marsha Rivkin Center for Ovarian Cancer Research grant (M.L. Fishel). Both authors were also supported on this work by the Ovar’coming Together foundation.
Abbreviations used
- BER
Base Excision Repair
- Ape1/Ref-1
Apurinic endocuclease1/Redox effector factor 1
- AP
Apurinc/Apyrinidinic
- MX
methoxyamine
- FDG
18F-fluorodeoxyglucose
- BrdU
Bromodeoxyuridine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.American Cancer Society. Cancer Facts and Figures. 2007 [Google Scholar]
- 2.Moore DH, et al. Alterations in the expression of the DNA repair/redox enzyme APE/ref-1 in epithelial ovarian cancers. Clin Cancer Res. 2000;6(2):602–609. [PubMed] [Google Scholar]
- 3.Fleck O, Nielsen O. DNA repair. J Cell Sci. 2004;117(Pt 4):515–517. doi: 10.1242/jcs.00952. [DOI] [PubMed] [Google Scholar]
- 4.Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat Res. 2000;461(2):83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 5.Tell G, et al. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid Redox Signal. 2005;7(3–4):367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 6.Xanthoudakis S, et al. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci U S A. 1996;93(17):8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsen E, et al. Organ and cell specificity of base excision repair mutants in mice. Mutat Res. 2007;614(1–2):56–68. doi: 10.1016/j.mrfmmm.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, Luo M, Kelley MR. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol Cancer Ther. 2004;3(6):679–686. [PubMed] [Google Scholar]
- 9.McNeill DR, Wilson DM., 3rd A Dominant-Negative Form of the Major Human Abasic Endonuclease Enhances Cellular Sensitivity to Laboratory and Clinical DNA-Damaging Agents. Mol Cancer Res. 2007;5(1):61–70. doi: 10.1158/1541-7786.MCR-06-0329. [DOI] [PubMed] [Google Scholar]
- 10.Vasko MR, Guo C, Kelley MR. The multifunctional DNA repair/redox enzyme Ape1/Ref-1 promotes survival of neurons after oxidative stress. DNA Repair (Amst) 2005;4(3):367–379. doi: 10.1016/j.dnarep.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Ono Y, et al. Stable expression in rat glioma cells of sense and antisense nucleic acids to a human multifunctional DNA repair enzyme, APEX nuclease. Mutat Res. 1994;315(1):55–63. doi: 10.1016/0921-8777(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 12.Walker LJ, et al. A role for the human DNA repair enzyme HAP1 in cellular protection against DNA damaging agents and hypoxic stress. Nucleic Acids Res. 1994;22(23):4884–4889. doi: 10.1093/nar/22.23.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bobola MS, et al. Apurinic/apyrimidinic endonuclease activity is associated with response to radiation and chemotherapy in medulloblastoma and primitive neuroectodermal tumors. Clin Cancer Res. 2005;11(20):7405–7414. doi: 10.1158/1078-0432.CCR-05-1068. [DOI] [PubMed] [Google Scholar]
- 14.Yang ZZ, Chen XH, Wang D. Experimental study enhancing the chemosensitivity of multiple myeloma to melphalan by using a tissue-specific APE1-silencing RNA expression vector. Clin Lymphoma Myeloma. 2007;7(4):296–304. doi: 10.3816/CLM.2007.n.006. [DOI] [PubMed] [Google Scholar]
- 15.Lau JP, et al. Effects of gemcitabine on APE/ref-1 endonuclease activity in pancreatic cancer cells, and the therapeutic potential of antisense oligonucleotides. Br J Cancer. 2004;91(6):1166–1173. doi: 10.1038/sj.bjc.6602080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fishel ML, et al. Imbalancing the DNA base excision repair pathway in the mitochondria; targeting and overexpressing N-methylpurine DNA glycosylase in mitochondria leads to enhanced cell killing. Cancer Res. 2003;63(3):608–615. [PubMed] [Google Scholar]
- 17.Fishel ML, et al. Manipulation of base excision repair to sensitize ovarian cancer cells to alkylating agent temozolomide. Clin Cancer Res. 2007;13(1):260–267. doi: 10.1158/1078-0432.CCR-06-1920. [DOI] [PubMed] [Google Scholar]
- 18.Rinne M, Caldwell D, Kelley MR. Transient adenoviral N-methylpurine DNA glycosylase overexpression imparts chemotherapeutic sensitivity to human breast cancer cells. Mol Cancer Ther. 2004;3(8):955–967. [PubMed] [Google Scholar]
- 19.Liu L, Gerson SL. Therapeutic impact of methoxyamine: blocking repair of abasic sites in the base excision repair pathway. Curr Opin Investig Drugs. 2004;5(6):623–627. [PubMed] [Google Scholar]
- 20.Liu L, et al. Inhibition of AP site repair coupled with action of topoisomerase II poison: A potential strategy to ehnance efficacy of chemotherapeutic alkylating agents. Proc Amer Assoc Ca Research; 2003. [Google Scholar]
- 21.Liu L, Nakatsuru Y, Gerson SL. Base excision repair as a therapeutic target in colon cancer. Clin Cancer Res. 2002;8(9):2985–2991. [PubMed] [Google Scholar]
- 22.Taverna P, et al. Methoxyamine potentiates DNA single strand breaks and double strand breaks induced by temozolomide in colon cancer cells. Mutat Res. 2001;485(4):269–281. doi: 10.1016/s0921-8777(01)00076-3. [DOI] [PubMed] [Google Scholar]
- 23.Fan Z, et al. Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nat Immunol. 2003;4(2):145–153. doi: 10.1038/ni885. [DOI] [PubMed] [Google Scholar]
- 24.Robertson KA, et al. Down-regulation of apurinic/apyrimidinic endonuclease expression is associated with the induction of apoptosis in differentiating myeloid leukemia cells. Cell Growth Differ. 1997;8(4):443–449. [PubMed] [Google Scholar]
- 25.Rouze N, Hutchings G. Design and Characterization of IndyPET-II: A High Resolution, High-Sensitivity Dedicated Research Scanner. IEEE Trans Nucl Sci. 2003;50:1491–1497. [Google Scholar]
- 26.Hamacher K, Coenen HH, Stocklin G. Efficient stereospecific synthesis of no-carrier-added 2-[18F]-fluoro-2-deoxy-D-glucose using aminopolyether supported nucleophilic substitution. J Nucl Med. 1986;27(2):235–238. [PubMed] [Google Scholar]
- 27.Liu GH, et al. Inhibition of nuclear factor-kappaB by an antioxidant enhances paclitaxel sensitivity in ovarian carcinoma cell line. Int J Gynecol Cancer. 2006;16(5):1777–1782. doi: 10.1111/j.1525-1438.2006.00652.x. [DOI] [PubMed] [Google Scholar]
- 28.Boccarelli A, et al. Differential processing of antitumour-active and antitumour-inactive trans platinum compounds by SKOV-3 ovarian cancer cells. Biochem Pharmacol. 2006;72(3):280–292. doi: 10.1016/j.bcp.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 29.Steel GG. Basic Clinical Radiobiology. 3rd ed. Arnold Press; 2002. [Google Scholar]
- 30.Hall EJ, Giaccia A. 6th ed. Lippincott Williams & Wilkins Press; 2006. Radiobiology for the Radiologist. [Google Scholar]
- 31.Eltabbakh GH. Recent advances in the management of women with ovarian cancer. Minerva Ginecol. 2004;56(1):81–89. [PubMed] [Google Scholar]
- 32.Fung H, Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol Cell. 2005;17(3):463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 33.Yang S, et al. Redox Effector Factor-1, Combined with Reactive Oxygen Species, Plays an Important Role in the Transformation of JB6 Cells. Carcinogenesis. 2007 doi: 10.1093/carcin/bgm128. [DOI] [PubMed] [Google Scholar]
- 34.Zou GM, et al. Ape1 regulates hematopoietic differentiation of embryonic stem cells through its redox functional domain. Blood. 2007;109(5):1917–1922. doi: 10.1182/blood-2006-08-044172. [DOI] [PubMed] [Google Scholar]


