Abstract
Grb2 is an SH2-SH3 protein adaptor responsible for linking growth factor receptors with intracellular signaling cascades. To study the role of Grb2 in cell growth, we have generated a new COS7 cell line (COS7shGrb2), based on RNAi technology, as null mutations in mammalian Grb2 genes are lethal in early development. This novel cell line continuously expresses a short hairpin RNA that targets endogenous Grb2. Stable COS7shGrb2 cells had the shGrb2 integrated into the genomic DNA and carried on <10% of normal levels of Grb2. Silencing Grb2 expression reduced, but did not eliminate, basal cell growth rate. This could be reversed, by either the addition of neomycin to the cell cultures or by rescuing with an Xpress-Grb2SiL construct (made refractory to the shRNA-mediated interference), but not with an SH2-deficient mutant (R86K). Thus, a viable knock-down and rescue protocol has demonstrated that Grb2 is crucial for cell proliferation.
Keywords: RNA silencing, Grb2, cell growth, signaling
INTRODUCTION
The Grb2/Sos complex serves to connect the EGFR with the activation of the MAPK signaling pathway [1]. The central SH2 domain of Grb2 binds tyrosine phosphorylated residues within the consensus sequence YXNX [2,3], whereas two flanking SH3 domains interact with proline-rich regions of other proteins, such as those found in the Ras guanine nucleotide exchange factor, Sos [1,4-6]. The Grb2/Sos complex promotes GTP loading of Ras, which leads to the activation of Ras effectors. As such, it has multiple functions in embryogenesis, cancer, regulation of the cytoskeleton, cell differentiation and DNA synthesis [1,7,8].
In spite of these known functions, demonstrating a direct role of Grb2 in cell growth has remained elusive. This is because null mutations in mammalian Grb2 genes are lethal in early development [8]. Depletion of Grb2 in cultured cells via RNAi is, in principle, a viable alternative, but this approach results in transient, partial or ineffective downregulation of the target protein, limiting such design to the study of the acute actions of gene silencing. Thus, permanent down-regulation becomes the only real alternative to study the long-term actions of gene silencing in cultured cells.
The answer to these problems is short-hairpin RNAs, which, under the transcriptional control of the U6 snRNA promoter and the first 27 bp of the human U6 snRNA (pU6+27-shRNA vectors), drive gene expression to very low or even undetectable levels [9]. In the present report, we describe a ‘deletion and rescue’ strategy in which COS-7 cells had stably integrated an U6+27-shRNA cassette against Grb2, making endogenous expression of Grb2 barely detectable. We demonstrate in this new experimental model that cell growth and DNA synthesis were vastly reduced, underscoring the vital role of Grb2 in cell growth. Further, two strategies have been devised to overcome this loss of function that will be of enormous usefulness to investigators who want to non-lethally silence a target cell signaling and study survival, growth, differentiation and oncogenesis.
MATERIALS AND METHODS
Materials and plasmids
The anti-Grb2 antibody was from Upstate (Updtate, NY), the anti-Xpress monoclonal antibody, DMEM, Lipofectamine and Plus reagents, Opti-MEM, DNase I were from Invitrogen (Carlsbad, CA); electrophoresis chemicals were from Bio-Rad Laboratories (Richmond, CA). Precast 4-20% polyacrylamide gels were from ISC BioExpress (Kaysville, UT). Taq polymerase and restriction enzymes were from New England Biolabs (Ipswich, MA). Total cellular RNA and genomic DNA isolation kits were from Qiagen, Inc. (Valencia, CA). The shRNA Grb2 encoding construct used for Grb2 knock-down (pU6+27-shGrb2) and its control (empty vector, pU6+27-shControl) were from Panomics, Inc (Fremont, CA). The human U6 snRNA promoter, the first 27 bp of the U6 snRNA (U6+27) [9] and the shRNA against human Grb2 were confirmed by direct sequencing. Site-directed mutagenesis was used to generate silent versions of Xpress-tagged Grb2 (XGrb2) encoded in the construct pcDNA-XGrb2 and renamed pcDNA-XGrb2SiL.
Mutagenesis of pcDNA-XGrb2
pcDNA-XGrb2 plasmid (6315 bp) was used as a template for site directed mutagenesis following the QuickChange XL site-directed mutagenesis protocol (Stratagene, La Jolla, CA). pcDNA-XGrb2SiL was generated by introducing seven silent mutations without changing the amino acid sequence of human Grb2 to make XGrb2SiL resistant to the continuous presence of Grb2 shRNA (nt #310-330: GAT GTG CAG CAC TTC AAG GTT). The sense primer for the mutagenesis was: 5′-293CT GTC AAG TTT GGA AAC GAC GTC CAA CAT TTT AAA GTA CTC CGA GAT GGA GCC GGG348-3′ (in bold are silently-mutated, underlined are the restriction sites Aat II and Dra I). Introducing a point mutation a critical residue within the SH2 domain of Grb2 (R86K) generated the SH2-deficient version of pcDNA-XGrb2SiL. The mutant inserts were PCR-amplified and each point mutation checked by restriction analysis and full DNA sequencing. The following primers were used for PCR amplification: XGrb2SiL (850 bp), sense: 5′-−16T GTG GTG GAA TTC AGA A1TG GAA GCC ATC GCC15-3′, antisense: 5′-833ACT CGA GCG GCC GCC CCT CCC ACC CCC TAA803-3′; XGrb2SiL/Grb2 (438 bp), sense: 5′-196CCC AGA GCC AAG GCA GAA GAA A217-3′, antisense: 5′-633GGT GAC ATA ATT GCG GGG AAA CAT610-3′.
Generation of a stable COS7shGrb2 cell line
COS7 were initially seeded at 1×105 cells/well in 6-well tissue culture plate in 1.5 ml D-MEM containing 10 % FBS. Cells were grown at 37°C in a CO2 incubator until the cells were ∼70% confluent. Transient transfection was done as described in detail elsewhere. Stable transfection of COS7 cells with pU6+27 plasmids was done with 3 Ůg of linearized vectors. Two days after, cells were incubated in the presence of neomycin (0.5-1 mg/ml). Once all untransfected COS7 cells were dead in the presence of neomycin, COS7 cells resistant to neomycin were isolated, grown up and tested by PCR for pU6+27 plasmid integration into genomic DNA. The NeoR marker and the U6+27-shGrb2 or U6+27-shControl cassettes were PCR-amplified by using the following primers: sense, 5′-CAG GGG CGC CCG GTT CTT TTT GTC AAG-3′, or sense: 5′-GAC TTT CCA CAC CCT AAC TGA CAC A-3′, respectively. Double-positive clones were diluted 1:2000 and single cells re-plated. Endogenous Grb2 expression was followed by Western blot. After 8 additional passages, four COS7 clones expressing minimal endogenous Grb2 (named: COS7shGrb2) and four control clones expressing normal levels of Grb2 (named: COS7shControl) were selected, propagated and used for transient transfection with constructs expressing PLD2 (pcDNA-mycPLD2) and/or silent versions of human Grb2 (pcDNA-XGrb2SiL).
Proliferation kinetics of COS7shRNA cells and thymidine incorporation
COS7shGrb2 and COS7shControl cells were seeded in 6-well plates at a very low density (1-5 cells/well) and single colonies in 6 different wells counted every 24-30 hours until COS7shControl cells reached confluency in at least 50 % of the clones (∼300 cells/100X field). After this time, COS7shGrb2 cells were allowed to grow in the absence of neomycin. De novo DNA synthesis was assayed as described [15]. Briefly, cells were washed twice with sterile PBS and incubated for 3 hours in serum-free DMEM. COS-7 cells were treated with 1 ŮCi/ml [3H]-thymidine/well for 16 h. Cells were washed twice with ice-cold PBS and precipitated with 5 % TCA at room temperature for 5 minutes. Precipitable [3H]-DNA was measured by scintillation counting.
RESULTS
Integration of shGrb2 into the genomic DNA results in a 90% decrease of Grb2 expression
We have used a pU6+27-based plasmid encoding an shRNA against nt# 310-330 of Grb2 (pU6+27-shGrb2) (Suppl-A) to stably transfect COS7 cells. But first, the U6+27-shGrb2 and U6+27-shControl (Suppl-B) cassettes encoded in the pU6+27 vectors were amplified by PCR (Suppl-C) and sequenced (Supl-D). A predicted computer-generated stem-loop model of the shRNA, based on the confirmed sequence, is shown in Figure 1A, indicating the specific Grb2 region targeted for silencing. Transient transfection of COS7 cells with pU6+27-shGrb2 or pU6+27-shControl did not significantly change endogenous Grb2 expression between 12-48 hr (Fig. 1B) or at any of the DNA concentrations tested (2.5-10 Ůg) (Fig. 1C), based on that Grb2 expression could be readily detected in Western blots of COS-7 cell lysates. Next, COS-7 cells were transfected and at least 40 clones were analyzed by Western blot. Selected clones were grown in 12-well plates and analyzed again by immunoblotting. A few clones were selected with neomycin. After a total of 6 weeks of continued selection, three neomycin-resistant clones showed markedly (>90%) reduced expression of Grb2 as compared to shControl or WT COS-7 cells (Fig. 1D).
FIGURE 1. A novel Grb2-deficient cell line.
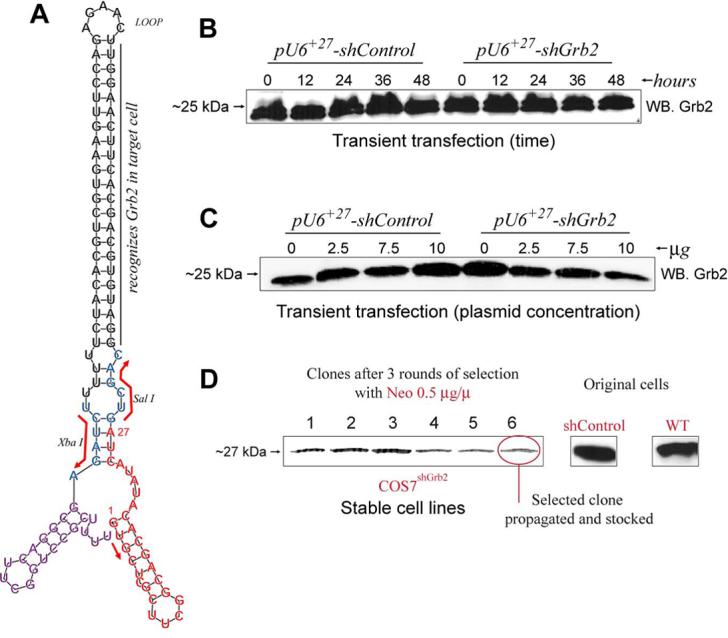
A, Predicted computer-generated stem-loop model using the RNAfold program (Vienna RNA Secondary Structure Prediction program web interface: http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) and the shRNA sequence from G+1 to the termination signal UUUU of the RNA polymerase III (position +113) as input. As reference, Sal I and Xba I restriction sites are shown. B, transient transfection of COS7WT cells with 2 mg of pU6+27-shGrb2 or pU6+27-shControl. COS7WT cells were transfected and, at the times indicated, protein samples were obtained in order to monitor endogenous Grb2 protein levels by Western Blot. C, COS7WT cells were transfected with the indicated amounts of pU6+27-shGrb2 or pU6+27-shControl and samples analyzed by Western Blot. D, Generation of stable cell lines. COS7 cells were stably transfected with pU6+27-shGrb2 or pU6+27-shControl as indicated in Methods. Forty clones resistant to neomycin and expressing pU6+27-shGrb2 were screened for Grb2 expression by Western blot. The process was repeated a total of three times. In the end, 3 subclones were stocked and used for further studies. Shown is the expression of Grb2 in comparison with that of original cells.
We next wanted to make sure that the shRNA had been permanently established and we had a true cell line in hand. After 8-10 passages, genomic DNA from stable transfectants was obtained and used as a template for PCR in order to detect the aminoglycoside phosphotransferase gene [neomycin resistance (NeoR)] and each of the U6+27-shRNA-encoding cassettes (as depicted in Fig. 2A). Genomic PCR analysis of parental COS7 cells stably transfected with pU6+27-shGrb2 or pU6+27-shControl plasmids detected the NeoR sequence (513 bp) and the respective U6+27-shRNA-encoding cassettes of the expected sizes (Figs. 2A,B). Thus, COS7 cells positive for genomic integration of the NeoR-U6+27-shGrb2 (that we named COS7shGrb2) and NeoR-pU6+27-shControl (that we named COS7shControl) sequences were further cloned and screened for Grb2 expression. In order to prevent phenotypic drift, the COS7shGrb2 cultures were used for only 3-5 passages before reverting to frozen stocks from an earlier passage.
FIGURE 2. Demonstration of stable integration of shGrb2 into the COS7 genome.
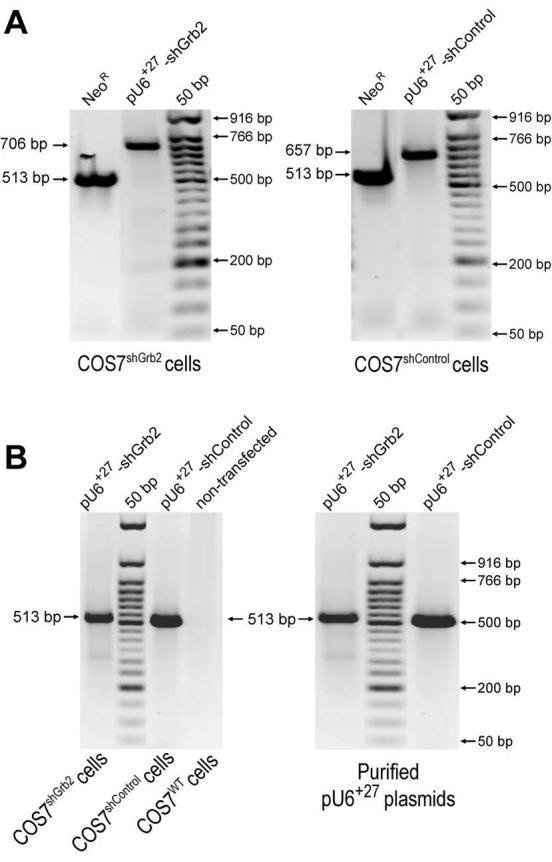
A, PCR amplification of pU6+27-shGrb2 and pU6+27-shControl markers (NeoR and shRNA insert) in COS7shGrb2 and COS7shControl cells, respectively. After eight cellular passages in the continuous presence of 0.5 mg/ml neomycin, genomic DNA from COS7shGrb2 and COS7shControl cells were obtained and amplified by PCR in order to detect the neomycin resistance gene (NeoR: 513 bp) or each of the inserts encoded in the shRNA inserts (shGrb2: 706 bp or shControl: 657 bp). The PCR fragments of 706 bp (the shGrb2 insert) and 657 bp (the shControl insert) obtained from COS7shGrb2 and COS7shControl cells, respectively, were sequenced in both directions using the same PCR primers used for PCR amplification (not shown). B, As controls, the NeoR gene encoded exclusively in the pU6+27 plasmids was also PCR-amplified from COS7shGrb2, COS7shControl and COS7WT genomic DNA, as well as directly from the purified plasmids pU6+27-shGrb2 or pU6+27-shControl.
A ‘rescue’ plasmid was able to restore cellular Grb2 levels
We next designed a way to ‘add back’ the lost protein/functionality to the cell. A rescuing reagent would also serve to demonstrate specificity of the shRNA-mediated silencing in the first place [10]. We introduced seven ‘silent’ mutations that rendered the Xpress-Grb2 unrecognizable by shRNA (in the process, we also introduced two new restriction sites, Aat II and Dra I for faster clone screening). This new plasmid was named ‘pcDNA-Xrb2SiL-WT’ (Fig. 3A). In addition to this, a mutant version (Grb2 SH2-deficient ) of it was also produced (pcDNA-Xrb2 SiL -R86K) (Fig. 3B). Figure 3C shows comparable expression levels of the RT-PCR fragments (850 bp and 438 bp) with respect to COS7shControl cells, suggesting that XGrb2SiL mRNA can be ectopically and transiently expressed in these cells. The identity of XGrb2SiL-derived expression was further demonstrated by restriction digestion of the RT-PCR products with Aat II or Dra I (Figure 3D). The minimal levels of endogenous Grb2 mRNA detected by RT-PCR in COS7shGrb2 cells after Aat II or Dra I digestion correlated with very low levels of endogenous Grb2 protein.
FIGURE 3. Restoration of Grb2 expression with a silent Grb2 plasmid.
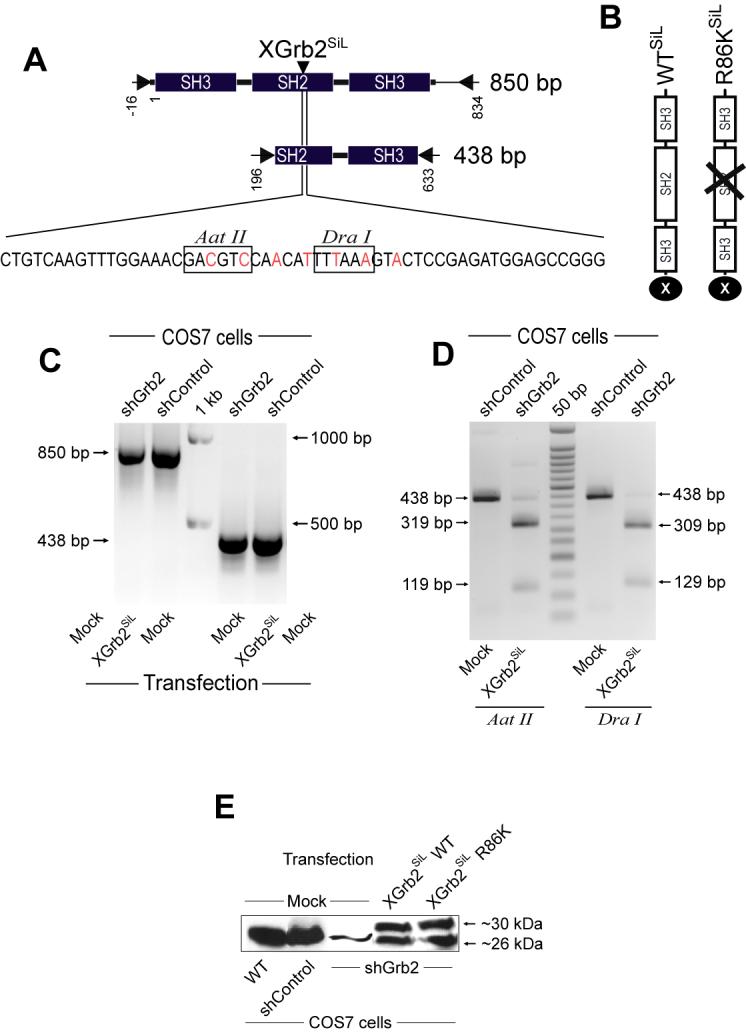
A, Representation of the silent version of the Xpress-tagged Grb2 (XGrb2SiL) encoded in pcDNA-XGrb2SiL. Boxes show the unique restriction sites created simultaneously for Aat II and Dra I that facilitated quick identity screening. B, Representation of the typical SH3-SH2-SH3 domain structure of the protein XGrb2SiL WT and an R86K (with impaired SH2 motif) with the Xpress tag in the C-end. C, COS7shGrb2 cells were transfected with 2 μg of pcDNA-XGrb2SiL. After 48 hours, total RNA was extracted, DNAse I-digested and the expression of XGrb2SiL mRNA monitored by RT-PCR using the specific sets of primers indicated in A. Total RNA from COS7shControl cells were also analyzed. D, Restriction digestion of RT-PCR products. The 438 bp RT-PCR products obtained from transfection were digested. The predicted restriction fragments of the XGrb2SiL (438 bp) segment are detected (Aat II=319 bp + 119 bp and Dra I=309 bp + 129 bp). E, Expression of two different XGrb2SiL constructs (WT and the SH2 deficient version R86K) at the protein level, analyzed simultaneously by Western blot using an anti-Grb2 antibody. Shown is a representative experiment in which XGrb2SiL is detected as a ∼30 kDa band, whereas endogenous Grb2 is ∼26 kDa. F, Direct immunofluorescence detection of XGrb2SiL WT in COS7shGrb2 cells. COS7shGrb2 cells transiently expressing pcDNA-XGrb2SiL for 48 hours were serum-starved overnight and analyzed for XGrb2SiL localization by direct immunofluorescence using a FITC-conjugated antibody against the Xpress-tag of XGrb2SiL. Nucleus is DAPI-stained.
Western blot analysis using antibodies against human Grb2 indicated that the XGrb2SiL (29.7 kDa) and the endogenous Grb2 (25.2 kDa) proteins are expressed in COS7shGrb2 cells transfected with two different pcDNA-XGrb2SiL constructs, WT or the SH2-deficient mutant R86K (Figure 3E). This panel also indicates that the endogenous expression of Grb2 in COS7shGrb2 cells was reduced by ∼90%. Normal XGrb2SiL WT expression and localization in COS7shGrb2 cells were demonstrated by direct immunofluorescence using a FITC-conjugated anti-Xpress antibody. XGrb2SiL transiently expressed in COS7shGrb2 cells localizes diffusely and exclusively in the cytoplasm, as demonstrated by others for different Grb2 constructs in COSWT cells [11]. Taken together, these results indicate that endogenous Grb2 mRNA and protein expression can be both specifically knocked-down and rescued in COS7shGrb2 cells.
Cell growth and DNA synthesis are reversibly impaired with shGrb2
Once the needed molecular reagents were in hand, we investigated whether or not Grb2 was important for overall cell growth. First, we analyzed the proliferation kinetics of Grb2-deficient cells. As shown in Figure 4A, COS7shControl cells reached confluency faster than COS7shGrb2 cells in the continuous presence of neomycin. Importantly, we discovered that when neomycin was withdrawn from the culture media, the cellular growth kinetics of COS7shGrb2 became comparable to that of COS7shControl cells. We designed an experiment to show that COS7shGrb2 cells are deficient in de novo DNA synthesis. To this end, cells were serum-starved for 24 hs and incubated in the presence of [3H]-thymidine overnight. As shown in Figure 4B, [3H]-thymidine incorporation into de novo synthesized DNA was comparable in COS7WT and COS7shControl (Figure 4B, left bars), whereas COS7shGrb2 cells showed a dramatic reduction in [3H]-thymidine incorporation (Figure 4B, central bar). However, when COS7shGrb2 cells were either incubated in the absence of neomycin or transfected with pcDNA-XGrb2SiL, COS7shGrb2 de novo DNA synthesis was quickly restored (Figure 4B, right bars). Lastly, in cells transfected with the SH2-deficient mutant the R86K, the rescue was not possible (not shown). These experiments suggest that depletion of endogenous Grb2 expression in COS7 cells correlates with impaired de novo DNA synthesis and demonstrates that Grb2 is a key factor for normal cell proliferation.
FIGURE 4. Acute effect of Grb2 knock-down on basal cell growth rate.
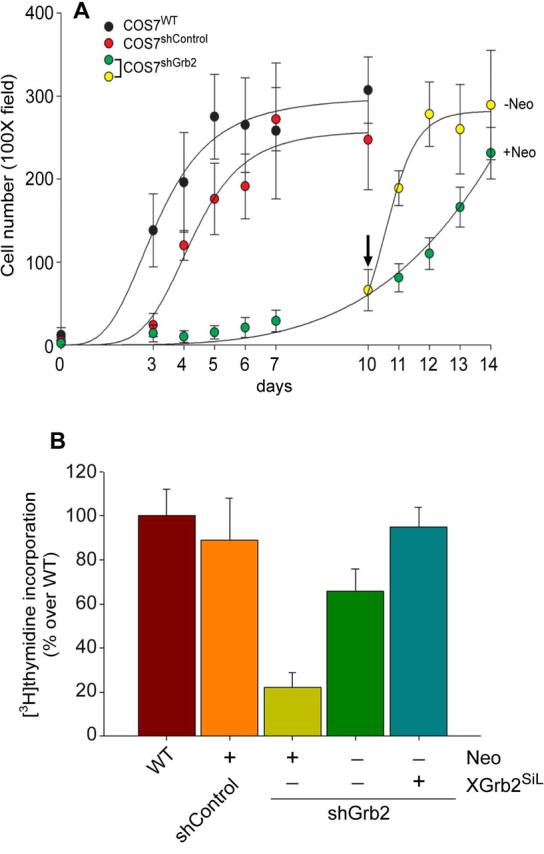
COS7WT and COS7shControl versus COS7shGrb2, proliferation curve and response to EGF. A, COS7shControl and COS7shGrb2 were plated in the presence of 1 mg/ml neomycin at a density of 1-5 cells/well. Cell number was monitored during the indicated times by counting at least 3 different colonies in 6 different wells in the 100X field. When COS7shControl cells reached confluency, neomycin was withdrawn from three of the six wells of COS7shGrb2 (indicated by an upside down vertical arrow). Results are expressed as cell number directly counted in 3-6 100X fields ± SEM. B, [3H] thymidine incorporation. De novo DNA synthesis in COS7WT, COS7shControl and COS7shGrb2 (in the presence or absence of neomycin, or transfected with pcDNA-XGrb2SiL) was estimated under basal conditions. Cells were incubated with 1 μCi/ml overnight. TCA-precipitable [3H]-DNA was counted (%controls)
DISCUSSION
Using RNA interference, a COS7 cell line was generated expressing ∼10% of normal levels of Grb2. Transient expression of a plasmid encoding an shRNAs against endogenous Grb2 in COS7 was ineffective suggesting that Grb2 protein has a very slow turn over. However, we were successful in creating a stable cell line permanently expressing very low levels of Grb2, vital for investigating the effects of Grb2 depletion in cell growth.
Silencing endogenous Grb2 in COS7 cells appears to be specific since COS7shGrb2 cells showed pcDNA-XGrb2SiL, an Xpress-tagged Grb2 version made silent to the shGrb2 actions. Overexpression of XGrb2SiL did not result in acute upregulation of endogenous Grb2 mRNA due to transfection artifacts, demonstrating that endogenous Grb2 silencing can be rescued at the protein level. Our laboratory has previously shown that Grb2, as part of a Shc-Grb2-SOS complex is essential for the signaling of the phospholipase D2 (PLD2) isoform through the SOS-RAS-RAF-MAPKK-MAPK signaling cascade and eventual effect on DNA synthesis and cell growth [12,13]. Efficient signaling requires association of the lipase with Grb2 through an identified residue, (as the Src homology ‘SH’ region-2 binds to phosphorylated tyrosine residues). Given the importance of PLD as a regulator of cell proliferation, the Grb2-cell line is potentially a useful tool to investigate the role of SH2 binding domains in a target protein in the regulation of human tumoral cell growth.
Although there are several reports taking advantage of an overexpressed Grb2 and its effect on signaling or physiological functions, like endothelial cell migration and angiogenesis [14], or the oncogenic protein v-ErbB and its association to Grb2 through Shc [15], very few studies exist on the effect of knocking down its presence. Microinjecting cells with an anti-Ash/Grb2 antibody results in the inhibition of ruffle induction and stress fiber formation but had no effect on S phase entry in the cell cycle [16]. Dominant-negative mutants of Grb2 are known to induce reversal of the transformed fibroblast phenotypes [17]. Depletion of Grb2 by RNA interference increases EGF-induced Stat3 tyrosine phosphorylation. Grb2 inhibits the interaction between Stat3 and EGFR by competitive binding to the receptor, demonstrating a negative regulation [18]. Using liposomes to deliver deoxioligonucleotides specific for the Grb2 mRNA lead to inhibition of breast cancer cell growth, but mainly on cells expressing high levels of the tyrosine kinase ErbB2 [19].
As demonstrated in this study, cell proliferation was greatly diminished in COS7shGrb2 cells. Still, the cells managed to still grow, at albeit a slow pace, and stably, making this system extremely useful for future studies, which is compounded by the results obtained with neomycin and presented in Figure 4. Addition of media without neomycin to the stable clones resulted in the quick restoration of nearly normal cell growth. Thus, cells with the Grb2-deficient phenotype can be practically reversed by either transfection of an ‘add back’ Grb2-refractory plasmid or by the removal of the antibiotic.
In conclusion, we have described here in detail a ‘silencing and rescue’ approach in which COS7 cells stably integrated a NeoR-U6+27-shRNA cassette directed against Grb2, making endogenous expression of Grb2 extremely low. This new cell line will undoubtedly help future research of the key roles of Grb2 in cell survival, growth, differentiation and oncogenesis, as it was clear that Grb2 is important overall for cell proliferation.
Supplementary Material
pU6+27-shGrb2 and -shControl vectors. A, Representation of the pU6+27-shGrb2 plasmid used in this study. The short hairpin RNA (shRNA) against Grb2 (shGrb2, 5′-GAU GUG CAG CAC UUC AAG GUU-3′) is shown as part of the 54 bp insert between Sal I/Xba I sites of the U6+27 cassette. B, Representation of the pU6+27-shControl region of the plasmid used in this study. In red is the sequence corresponding to the U6+27 promoter, the control sequence GGG is denoted between the two cloning restriction sites, Sal I and Xba I, followed by the U6 termination sequence in purple. C, PCR amplification of the U6-shRNA cassette. pU6+27 plasmids were PCR-amplified by using the primers indicated as arrows in A or as boxed sequences in D. U6+27 cassette sequence. Plasmids were amplified by PCR using primers that anneal the boxed sequences. The relative location of the insert (shGrb2, 54 bp; or shControl, 3 bp) is also indicated.
ACKNOWLEDGEMENTS
This work has been supported by grants from the National Institutes of Health (HL056653) to J.G.-C. We thank Kathleen Frondorf for maintaining the cultures of COS7shControl and COS7shGrb2 cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Hu P, Margolis B, Skolnik EY, Lammers R, Ullrich A, Schlessinger J. Interaction of phosphatidylinositol 3-kinase-associated p85 with epidermal growth factor and platelet-derived growth factor receptors. Mol Cell Biol. 1992;12:981–90. doi: 10.1128/mcb.12.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–78. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 3.Songyang Z, Shoelson SE, McGlade J, Olivier P, Pawson T, Bustelo XR, Barbacid M, Sabe H, Hanafusa H, Yi T. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol Cell Biol. 1994;14:2777–85. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rozakis-Adcock M, Fernley R, Wade J, Pawson T, Bowtell D. The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature. 1993;363:83–5. doi: 10.1038/363083a0. [DOI] [PubMed] [Google Scholar]
- 5.Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature. 1993;363:85–8. doi: 10.1038/363085a0. [DOI] [PubMed] [Google Scholar]
- 6.Chardin P, Camonis JH, Gale NW, van Aelst L, Schlessinger J, Wigler MH, Bar-Sagi D. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science. 1993;260:1338–43. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 7.Egan SE, Giddings BW, Brooks MW, Buday L, Sizeland AM, Weinberg RA. Association of Sos Ras exchange protein with Grb2 is implicated in tyrosine kinase signal transduction and transformation. Nature. 1993;363:45–51. doi: 10.1038/363045a0. [DOI] [PubMed] [Google Scholar]
- 8.Lowenstein EJ, Daly RJ, Batzer AG, Li W, Margolis B, Lammers R, Ullrich A, Skolnik EY, Bar-Sagi D, Schlessinger J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70:431–42. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 9.Cheng AM, Saxton TM, Sakai R, Kulkarni S, Mbamalu G, Vogel W, Tortorice CG, Cardiff RD, Cross JC, Muller WJ, Pawson T. Mammalian Grb2 regulates multiple steps in embryonic development and malignant transformation. Cell. 1998;95:793–803. doi: 10.1016/s0092-8674(00)81702-x. [DOI] [PubMed] [Google Scholar]
- 10.Paul CP, Good PD, Winer I, Engelke DR. Effective expression of small interfering RNA in human cells. Nat Biotechnol. 2002;20:505–8. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- 11.Meyer S, LaBudda K, McGlade J, Hayman MJ. Analysis of the role of the Shc and Grb2 proteins in signal transduction by the v-ErbB protein. Mol Cell Biol. 1994;14:3253–62. doi: 10.1128/mcb.14.5.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kittler R, Pelletier L, Ma C, Poser I, Fischer S, Hyman AA, Buchholz F. RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture cells. Proc Natl Acad Sci U S A. 2005;102:2396–401. doi: 10.1073/pnas.0409861102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horn J, Lopez I, Miller MW, Gomez-Cambronero J. The uncovering of a novel regulatory mechanism for PLD2: Formation of a ternary complex with protein tyrosine phosphatase PTP1B and growth factor receptor-bound protein GRB2. Biochem Biophys Res Commun. 2005;332:58–67. doi: 10.1016/j.bbrc.2005.04.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Fulvio M, Lehman N, Lin X, Lopez I, Gomez-Cambronero J. The elucidation of novel SH2 binding sites on PLD2. Oncogene. 2006;25:3032–40. doi: 10.1038/sj.onc.1209340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laramee M, Chabot C, Cloutier M, Stenne R, Holgado-Madruga M, Wong AJ, Royal I. The scaffolding adapter gab1 mediates vascular endothelial growth factor signaling and is required for endothelial cell migration and capillary formation. J Biol Chem. 2007;282:7758–69. doi: 10.1074/jbc.M611327200. [DOI] [PubMed] [Google Scholar]
- 16.Matuoka K, Shibasaki F, Shibata M, Takenawa T. Ash/Grb-2, a SH2/SH3-containing protein, couples to signaling for mitogenesis and cytoskeletal reorganization by EGF and PDGF. Embo J. 1993;12:3467–73. doi: 10.1002/j.1460-2075.1993.tb06021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie Y, Pendergast AM, Hung MC. Dominant-negative mutants of Grb2 induced reversal of the transformed phenotypes caused by the point mutation-activated rat HER-2/Neu. J Biol Chem. 1995;270:30717–24. doi: 10.1074/jbc.270.51.30717. [DOI] [PubMed] [Google Scholar]
- 18.Zhang T, Ma J, Cao X. Grb2 regulates Stat3 activation negatively in epidermal growth factor signalling. Biochem J. 2003;376:457–64. doi: 10.1042/BJ20030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tari AM, Hung MC, Li K, Lopez-Berestein G. Growth inhibition of breast cancer cells by Grb2 downregulation is correlated with inactivation of mitogen-activated protein kinase in EGFR, but not in ErbB2, cells. Oncogene. 1999;18:1325–32. doi: 10.1038/sj.onc.1202422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
pU6+27-shGrb2 and -shControl vectors. A, Representation of the pU6+27-shGrb2 plasmid used in this study. The short hairpin RNA (shRNA) against Grb2 (shGrb2, 5′-GAU GUG CAG CAC UUC AAG GUU-3′) is shown as part of the 54 bp insert between Sal I/Xba I sites of the U6+27 cassette. B, Representation of the pU6+27-shControl region of the plasmid used in this study. In red is the sequence corresponding to the U6+27 promoter, the control sequence GGG is denoted between the two cloning restriction sites, Sal I and Xba I, followed by the U6 termination sequence in purple. C, PCR amplification of the U6-shRNA cassette. pU6+27 plasmids were PCR-amplified by using the primers indicated as arrows in A or as boxed sequences in D. U6+27 cassette sequence. Plasmids were amplified by PCR using primers that anneal the boxed sequences. The relative location of the insert (shGrb2, 54 bp; or shControl, 3 bp) is also indicated.


