Abstract
Palmitoylation is a reversible, post-translational modification observed in a number of G-protein-coupled receptors. To gain a better understanding of its role in visual transduction, we produced transgenic knock-in mice that expressed a palmitoylation-deficient rhodopsin (Palm−/−). The mutant rhodopsin was expressed at wild-type levels and showed normal cellular localization to rod outer segments, indicating that neither rhodopsin stability nor its intracellular trafficking were compromised. But Palm−/− rods had briefer flash responses and reduced sensitivity to flashes and to steps of light. Upon exposure to light, rhodopsin became phosphorylated at a faster rate in mutant than in wild-type retinas. Since quench of rhodopsin begins with its phosphorylation, these results suggest that palmitoylation may modulate rod photoreceptor sensitivity by permitting rhodopsin to remain active for a longer period.
Palmitoylation is a covalent, post-translational modification occurring at the C terminus in many G-protein coupled receptors (GPCR)1 (1). Up to three cysteine residues located ~10–14 amino acids downstream of the seventh transmembrane domain may be palmitoylated. Although there is no definable consensus sequence (2), the evolutionary conservation of C-terminal cysteine palmitoylation implicates its importance in GPCR function. In vitro studies of several GPCRs in which the palmitoylated cysteine(s) was mutated have shown changes in G-protein coupling efficiency and selectivity, receptor phosphorylation and desensitization, and cell surface expression and trafficking (1). However, the effects were highly variable probably due to intrinsic receptor differences and to the utilization of non-native heterologous cell systems.
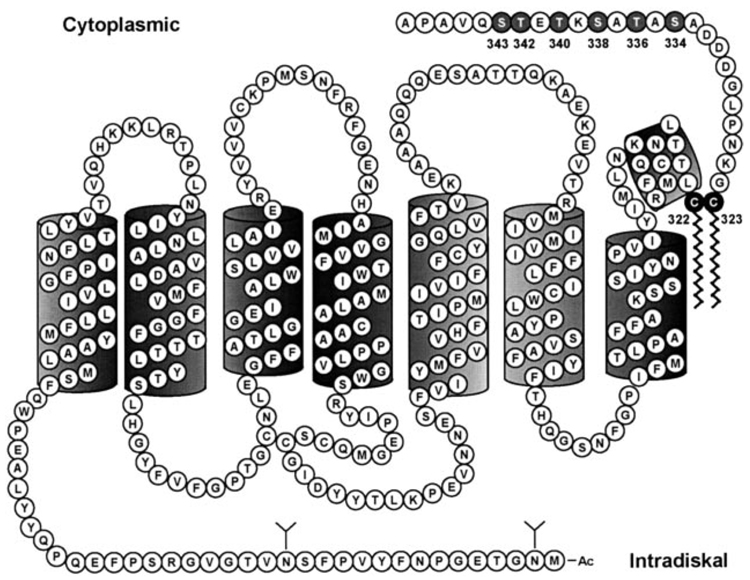
Rhodopsin was the first GPCR in which palmitoylation was reported (3). Palmitates, bound to cysteines 322 and 323 through a reversible thioester bond (4), insert into the membrane and anchor the C terminus of rhodopsin (5, 6), creating a fourth intracellular domain (Fig. 1). Residues in this domain are important for transducin activation (7, 8). Furthermore, serines and threonines in the C terminus downstream of the palmitoylation anchor are phosphorylated by rhodopsin kinase to terminate rhodopsin activity. Thus, it was logical to examine G-protein signaling activity and receptor phosphorylation in the palmitoylation-deficient rhodopsin mutant.
FIG. 1. Two-dimensional model of WT mouse rod opsin.
Cysteine residues at positions 322 and 323 were mutated to non-palmitoylatable threonine and serine residues, preventing the attachment of the membrane anchor at the C-terminal end of the eighth α-helix. Light-dependent phosphorylation occurs at C-terminal serine (334, 338, 343) and threonine (336, 340, 342) residues.
Here we examine the role of receptor palmitoylation in vivo using the rod-specific GPCR rhodopsin as the subject of study. To achieve this goal, we generated a transgenic mouse line that makes only palmitoylation-deficient rhodopsin. Our studies reveal palmitoylation-deficient (Palm−/−) rods to be less sensitive to light, due to increased rhodopsin phosphorylation.
EXPERIMENTAL PROCEDURES
Generation of Palm−/− Mice
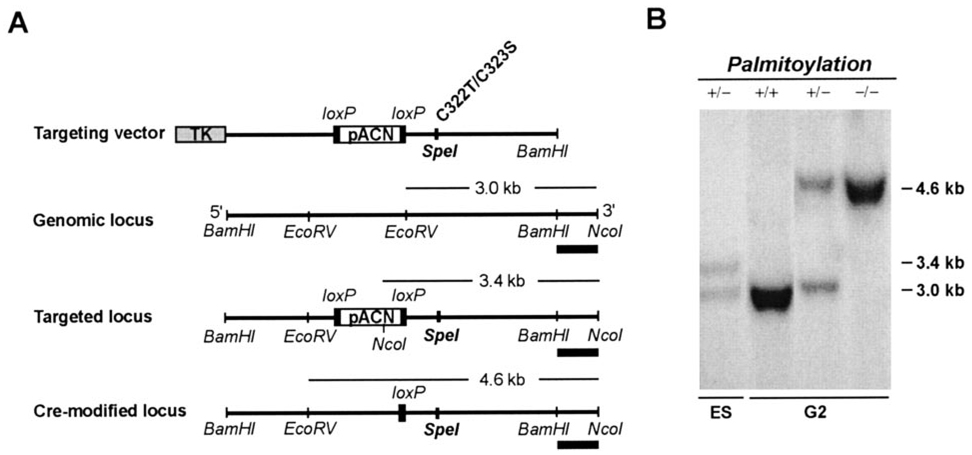
A mouse 129SV genomic library (Stratagene) was used to make a rhodopsin-targeting construct with two modifications, Cys322 → Thr and Cys323 → Ser, to eliminate both palmitoylation sites (Fig. 1). The modifications introduced a new SpeI restriction site in the gene, which we used diagnostically for genotypic identification. Presence of the desired mutations was verified by sequencing. The targeting strategy included a tACE-Cre-Neo cassette for self-excision of loxP-flanked cassettes in the male germ line of transgenic mice (9). J1 embryonic stem cells (10) were electroporated with the targeting construct (Fig. 2A) and selected with G418 and gancyclovir. The homologous recombinants were identified by Southern blot analysis using the targeting strategy shown in Fig. 2. We injected targeted ES clones into C57BL/6 blastocysts to generate chimeras. Male chimeras were crossed with C57BL/6 females, and offspring heterozygous for the mutant rhodopsin were interbred to produce mice that were homozygous for the palmitoylation deficiency. Animals were handled in accordance with the guidelines provided by the Association for Research in Vision and Ophthalmology and the Tufts-New England Medical Center Institutional Animal Care and Use Committee. Mice were reared on a 14 h light:10 h dark cycle and used at 4–6 weeks of age, except where noted otherwise.
FIG. 2. Generation of the palmitoylation-defective rhodopsin gene.
A, gene targeting strategy for rhodopsin palmitoylation mutant knock-in. The targeting vector (top) carries a negative selection marker, the thymidine kinase (TK) gene, attached to the 5′-end of the targeting construct. The positive selection marker, encoding the neomycin gene under the control of the PolII promoter in the pACN cassette, is flanked by loxP sites. It also includes the angiotensin-converting enzyme promoter, which drives expression of Cre-recombinase in sperm of the first generation of transgenic mice (9). Introduction of the Cys322 → Thr, Cys323 → Ser mutations created a SpeI restriction site. EcoRV and NcoI double digestions distinguished the targeted locus and the Cre-recombinase modified locus from the genomic locus on Southern blots. The DNA probe, shown as a solid rectangle, lies exterior to the targeting vector. B, Southern blot of EcoRV- and NcoI-digested DNA from ES cells (ES) and from mouse tails of transgenic second-generation mice (G2). WT (+/+) mice yielded a 3.0-kb fragment. A 4.6-kb fragment was present in homozygous mutant mice (−/−), while both the 3.0- and 4.6-kb fragments were present in heterozygotes (+/−). The recombinant ES cell clone shows the 3.0-kb WT band and a 3.4-kb band that reflects homologous recombination prior to Cre-recombinase-mediated excision of the pACN drug selection cassette.
Determination of RNA and Protein Expression in Mutant Mice
Total RNA isolation and RT-PCR procedures were performed using an RNeasy mini kit (Qiagen) and Ready-to-go RT-PCR beads (Amersham Biosciences) following the manufacturer’s protocols. Rhodopsin cDNA was amplified with an exon 4 forward primer (5′-CATCTTCTTCCTGATCTGCT-3′) and an exon 5 reverse primer (5′-CTTTCAAGCCACAGTCTCTG-3′). The PCR products were digested by SpeI restriction enzyme to identify the palmitoylation mutation.
Protein immunoblot analysis was performed to assess the expression levels of rhodopsin and other phototransduction proteins. Extracts were prepared by homogenizing retinas in radioimmunoprecipitation buffer containing 1% Nonidet P-40, 1% sodium deoxycholate, 150 mm NaCl, 2 mm EGTA, and a protease inhibitor mixture (Complete Mini Protease Inhibitor Mixture, Roche Applied Science, Mannheim, Germany). Total protein concentration was determined using the bicinchoninic acid assay (Pierce). Equivalent amounts of proteins were electrophoresed in 10 or 12% SDS-polyacrylamide gels and electrotransferred onto nitrocellulose membranes. Membranes were stained with anti-rhodopsin (1D4, 1:5000) (11), transducin α-subunit (Tα1A, 1:1000, a gift from M. Simon, California Institute of Technology, Pasadena, CA), rhodopsin kinase (G8, 1:1000, a gift from K. Palczewski, University of Washington, Seattle, WA), arrestin (C10C10, 1:5000) (12), recoverin (P26, 1:50,000) (13), RGS9 (R4433, 1:5000, a gift from T. Wensel, Baylor College of Medicine, Houston, TX), and β-actin (1:1000, Sigma) antibodies and subsequently with goat anti-mouse or anti-rabbit IgG conjugated to horseradish peroxidase (BD Biosciences). Immunodetection was performed using enhanced chemiluminescent substrate (Pierce).
Retinal Histology and Immunohistochemistry
Mouse eyes were marked for orientation, enucleated, perforated at the cornea, and immersed in 4% paraformaldehyde, 0.1% glutaraldehyde in 0.1 m phosphate buffer (pH 7.4) at 4 °C overnight. Tissues were then dehydrated in a graded series of alcohols and embedded in paraffin. Eyes were bisected along the vertical meridian through the optic nerve head and stained with hematoxylin and eosin for light microscopy. Morphometric analysis was performed by counting rows of photoreceptor cells in the outer nuclear layer (ONL) from four different areas in the central and midperipheral retina across both the superior and inferior hemispheres. Regional values were averaged for each retina. The counts were taken from three to five animals for each genotype at each age. Results from age-matched WT and mutant mice were compared by an analysis of variance.
To analyze rhodopsin localization, retinal sections were incubated in blocking buffer (3% bovine serum albumin, 5% goat serum, and 0.2% Triton X-100 in 0.1 m phosphate buffer solution) for 1 h and then with anti-rhodopsin antibody (1D4, 1:500) at 4 °C overnight, followed by the rhodamine-conjugated secondary antibody for 1 h at room temperature.
Single-cell Suction Electrode Recording
Eighteen Palm−/− mice and nine WT mice, aged 5–9 weeks, were used for rod photoresponse recordings. A mouse was dark-adapted for at least 12 h the night before the experiment. All tissue preparations were performed under infrared light. Retinas were dissected, put into oxygenated Leibovitz’s L-15 medium (Invitrogen) supplemented with 0.1 mg ml−1 bovine serum albumin (Fraction V, Sigma) and 10 mm glucose and stored on ice until use. A small piece of tissue was chopped finely in L-15 medium containing DNase I (Type IV-S, Sigma) and transferred into an experimental chamber. The tissue was perfused with Locke’s medium (mm): 139 Na+, 3.6 K+, 2.4 Mg2+, 1.2 Ca2+, 123.3 Cl−, 20 HCO3−, 10 HEPES, 3 succinate, 0.5 l-glutamate, 0.02 EDTA, 10 glucose enriched with minimal essential medium amino acids (Invitrogen) and basal medium Eagle vitamins (Sigma), equilibrated with 95% O2/5% CO2, and heated to 36.5–37.5 °C (pH 7.4). The outer segment of a rod was pulled into a silanized glass pipette filled with (mm): 140 Na+, 3.6 K+, 2.4 Mg2+, 1.2 Ca2+, 145.8 Cl−, 10 HEPES, 0.02 EDTA, 10 glucose. Photocurrents were recorded with a current-to-voltage converter (Axopatch 200A, Axon Instruments, Union City, CA), filtered at 30 Hz (−3 dB, 8-pole Bessel, Frequency Devices, Haverhill, MA), and digitized at 400 Hz (Pulse Version 8.07, HEKA Elektronik, Lambrecht/Pfalz). No corrections were made for the delay introduced by filtering. Light provided by a xenon arc source passing through a six-cavity interference filter (500 nm, Omega Optical, Brattleboro, VT) was used to stimulate the rod. The duration and intensity of the exposure were controlled by an electronic shutter and neutral density filters, respectively.
In Vitro Rhodopsin Phosphorylation
Mice were dark-adapted overnight and retinas were collected under infrared illumination. Rod outer segments (ROS) were purified by flotation on a layered 10%/20% stepped density gradient according to a previously published method (14). Rhodopsin content was determined spectrophotometrically. ROS preparations were phosphorylated by incubation with (mm): 25 Tris-HCl (pH 7.5), 6 MgCl2, 1 EDTA, 0.0005 okadaic acid, and 2 ATP under 2000 lux white light at room temperature for various times. Reactions were quenched with SDS-PAGE sample buffer and then the samples were analyzed by 12% SDS-PAGE. The blots were probed with anti-phospho-Ser338 and -Ser334 rhodopsin antibodies, kindly provided by Dr. D. Williams (University of California, San Diego, CA) (15). After stripping, the membranes were reprobed with anti-rhodopsin antibody (1D4).
Analyses of Rhodopsin Palmitoylation and Phosphorylation by Mass Spectrometry
To verify the absence of palmitoylation in Palm−/− mice, dark-adapted ROS were prepared from Palm−/− and WT mice. Cyanogen bromide fragmentation, high performance liquid chromatography (HPLC) separation, and mass spectrometric mapping of the Palm−/− and WT mouse rhodopsin followed methodology published elsewhere (16, 17).
To quantify the phosphorylation of rhodopsin in vivo, Palm−/− and WT mice were dark-adapted overnight, given 0.5% tropicamide and 5% phenylephrine hydrochloride to dilate their pupils and then exposed to white light for 10 min at 500, 1000, or 2000 lux. After illumination, the retinas were isolated and immediately put into 8 m urea. Asp-N digestion yielded C-terminal rhodopsin peptides, which were separated by HPLC and analyzed by mass spectrometry as described elsewhere (17). Tandem mass spectral data were acquired for the most abundant ion in each mass spectrum for sequence analysis, but for phosphorylation studies tandem mass spectral data were only acquired for rhodopsin phosphopeptides. The mass spectrometer bias against the negatively charged phosphopeptides was corrected (18), and the average number of phosphates incorporated was calculated with the following formula,
| (Eq. 1) |
where is the corrected intensity for n, the number of phosphorylations per opsin. The results were subjected to an analysis of variance (analysis of variance, Stata Version 6, StataCorp, College Station, TX), treating light as a continuous variable.
RESULTS
Abrogation of Rhodopsin Palmitoylation in Palm−/− Mice
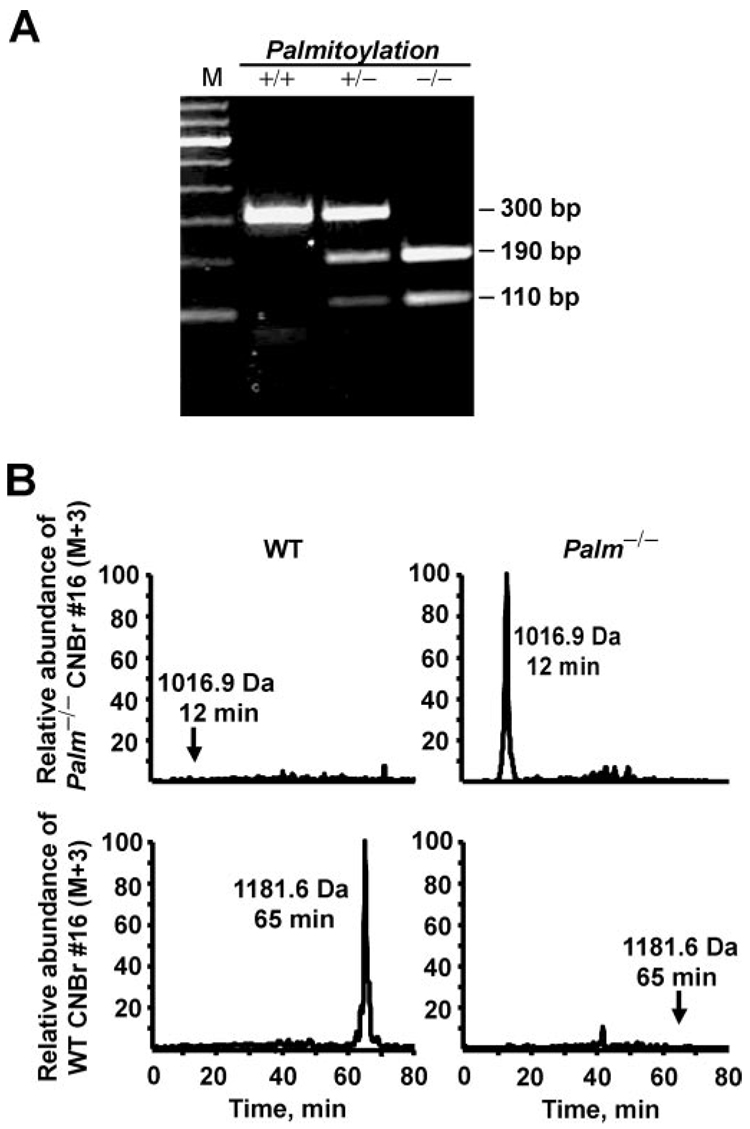
Palmitoylation-deficient rhodopsin animals were produced by mutation of the cysteine residues at positions 322 and 323 of rhodopsin to non-palmitoylatable threonine and serine sites, respectively. The mutation introduced a unique SpeI restriction site, which facilitated the identification of germ line transmission of the mutated gene in Palm−/− animals by Southern blot analysis (Fig. 2). Expression of the palmitoylation-deficient rhodopsin mRNA was verified by RT-PCR (Fig. 3A). Retinas from WT mice yielded a 300-bp band, while Palm−/− mice yielded bands at 110 and 190 bp following SpeI restriction digestion. All three bands were present in Palm+/− mice. Finally, the absence of palmitates on rhodopsin protein in Palm−/− mice was confirmed by mass spectrometry (Fig. 3B).
FIG. 3. Disruption of rhodopsin palmitoylation in Palm−/− mice.
A, RT-PCR and SpeI digestion to verify the presence of the palmitoylation-deficient rhodopsin mRNA. RNA prepared from WT retinas (+/+) yielded a 300-bp fragment, while homozygous mutant mice (−/−) yielded 190- and 110-bp fragments, due to the unique SpeI site in the mutant rhodopsin gene. Heterozygotes (+/−) have 300-, 190-, and 110-bp fragments, reflecting the presence of both WT and mutant mRNAs. M, DNA size marker. B, confirmation that rhodopsin from transgenic retinas lacked palmitoylation by mass spectrometry. The mutant C-terminal peptide fragment showed a reduction in molecular mass to 1016.9 Da (+3 charge state; upper right panel) from the palmitoylated WT C-terminal peptide fragment at 1181.6 Da (lower left panel) consistent with the mutation of palmitoylated cysteine residues 322 and 323 to non-palmitoylatable threonine and serine residues. In addition, HPLC run time decreased from 65 min (lower left panel) to 12 min (upper right panel), due to the substantial decrease in hydrophobicity. The identities of the peaks were confirmed by sequencing the peptides.
Minimal Retinal Degeneration in Palm−/− Mice
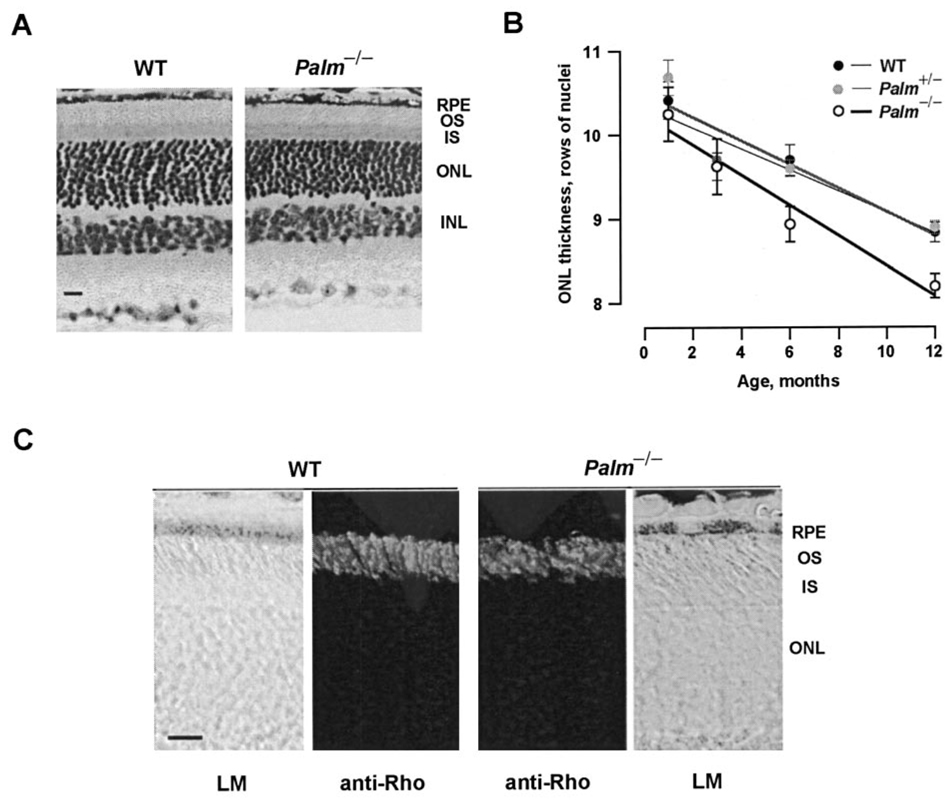
We examined retinal morphology by light microscopy to ascertain whether rhodopsin lacking palmitoylation gave rise to a retinal degeneration. The numbers of photoreceptor nuclei in normal and Palm−/− mice gradually declined with age, as judged by ONL thickness (p < 0.0001), but in Palm−/− mice there were fewer nuclei (p < 0.007). By 12 months of age, Palm−/− rod outer segment lengths were shortened slightly, and ONL thickness had decreased by nearly one row of nuclei relative to WT, indicating a loss of about 10% of the rods (Fig. 4, A and B). Thus, the gross morphology of the retina was minimally affected by the deficiency in rhodopsin palmitoylation.
FIG. 4. Minimal retinal degeneration and normal rhodopsin distribution in Palm−/− mice.
A, light micrographs of WT and Palm−/− retinas at 1 year of age. Palm−/− retinas have fewer nuclei in the ONL and slightly shortened outer segments (OS), indicating a mild retinal degeneration at older ages. INL, inner nuclear layer; IS, inner segment; RPE, retinal pigment epithelium. Scale bar = 20 µm. B, mild thinning of ONL in Palm−/− retinas. From the slopes of the regression lines, photoreceptors were lost at a rate of 0.122, 0.134, and 0.178 rows of nuclei per month in WT, Palm+/−, and Palm−/− retinas, respectively. Although the linear regressions were carried out on collected results, means and standard errors are plotted due to the large numbers of coincident points. C, rhodopsin immunohistochemistry. In both WT and Palm−/− retinas, rhodopsin labeling was restricted to the outer segments (second and third panel from the left). Outer panels show corresponding bright field images (LM). Scale bar = 20 µm.
Palmitoylation-deficient Rhodopsin Showed Normal Expression Levels and Localization
In 4–8-week-old mice, the quantity of mutant rhodopsin in Palm−/− ROS (0.17 ± 0.04 nmol/ retina, n = 22), measured spectroscopically, matched that of WT rhodopsin in control retinas (0.19 ± 0.06 nmol/retina, n = 32). The immunohistochemistry of retinas showed that rhodopsin in Palm−/− mice localized to outer segments of rod cells in a staining pattern indistinguishable from that of WT retinas (Fig. 4C). Therefore the absence of palmitoylation did not substantially alter rhodopsin expression, stability, or transport to the rod outer segment. Other key phototransduction proteins were probed by Western analysis to find out whether their expression had changed. The levels of transducin α-subunit, rhodopsin kinase, recoverin, arrestin, and RGS9 in Palm−/− retinas were no different from those of age-matched controls (data not shown).
Faster Photoresponse Recovery in Palm−/− Rods
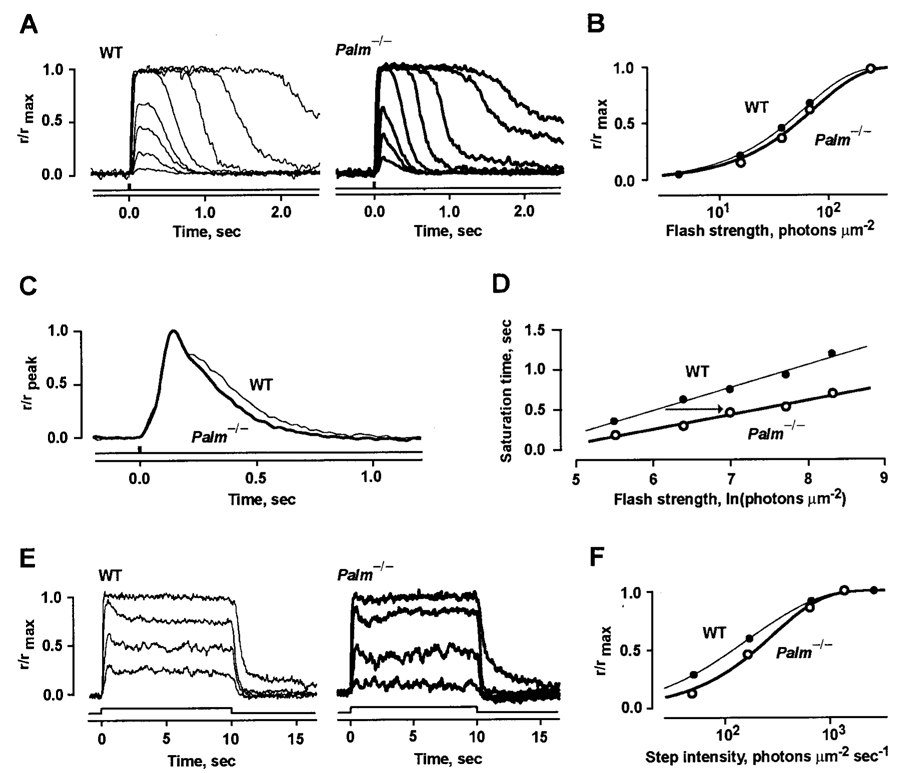
Non-palmitoylated rhodopsin proved capable of G-protein signaling in single cell recordings although there were subtle differences between flash responses of Palm−/− and WT rods (Fig. 5 and Table I). The rising phase and time to peak of the Palm−/− rod response were normal, but recovery was accelerated (Fig. 5C), which shortened integration time. Palm−/− rods were somewhat less sensitive to flashes than WT rods as evidenced by their higher i½. Reduced sensitivity could have arisen from reductions in quantal response amplitude, rod collecting area, or both. Any reductions in quantal response or in outer segment size were too small to resolve, however. Palm−/− rods recovered from bright flashes sooner than did WT rods (Fig. 5, A and D). Since the time in saturation increases with flash strength, the briefer saturation period in Palm−/− rods reflected a decrease in phototransduction gain at late times after the flash. The average magnitude of the change assessed by the increase in flash strength required to achieve a criterion saturation time of 0.5 s was 1.9-fold, Palm−/− rods having the lower gain (Fig. 5D). Palm−/− rods were also 2-fold less sensitive than wild type rods to steps of light (e.g. Fig. 5, E and F). For the cells recorded, the change in sensitivity to steps followed from their 1.5-fold reduction in integration time and 1.2-fold increase in i½. Overall, the lowered sensitivity and accelerated recovery kinetics of the responses to dim and bright flashes in Palm−/− rods suggests that palmitoylation normally hinders the shutoff of photoexcited rhodopsin activity.
FIG. 5. Lower sensitivity and faster flash response recovery in Palm−/− rods.
A, normalized averaged responses of a wild-type rod (thin traces) and a Palm−/− rod (thick traces) to flashes at 500 nm. The rmax was 8.2 pA for the WT rod and 10.3 pA for the Palm−/− rod. The flash monitor is displayed below the traces. B, stimulus response relations for the cells shown in A. Results were fit with the saturating exponential equation: r/rmax = 1 − exp(−ki), where i is the flash strength, and k is a constant. C, normalized dim flash responses of 75 Palm−/− (thick trace) and 43 WT (thin trace) rods. Flashes eliciting responses with mean amplitudes <0.2 rmax were considered to be dim. The time to peak was 129 ms for both types of rods, and the integration times were 335 and 252 ms for the WT and Palm−/− rods, respectively. D, reduced saturation time after bright flashes for the Palm−/− rod shown in A. Traces corresponding to some points were omitted in A for clarity. Saturation time was taken as the period from midflash to 0.9 rmax. The slopes obtained by linear regression were 290 and 182 ms for WT (thin line) and Palm−/− (thick line) rods, respectively. The reduced slope reflects a faster shutoff of phototransduction in the Palm−/− rod (49, 50). The flash strength required to produce a saturation time of 0.5 s was 1.3 natural log units greater (arrow) for the Palm−/− rod due to its lower sensitivity. E, averaged, normalized responses of a WT rod (thin traces) and a Palm−/− rod (thick traces) to steps of light. The rmax values were 7.2 and 11.7 pA for WT and Palm−/−, respectively. F, intensity response relations for the cells shown in E. Continuous lines show fits of the rods with a modified saturating exponential equation: r/rmax = 1 − exp (−(k1 + k2 exp(−k3I))*I), where k1–3 are constants, and I is light intensity (51). I½, the step intensity producing a half-maximal response, was 120 photons µm−2 s−1 for the WT rod and 206 for the Palm−/− rod.
TABLE I. Light response parameters of WT and Palm−/−.
Values given as mean ± S.E. (n). The current circulating in darkness was given by the amplitude of the response to a saturating flash. Single photon response amplitude was estimated by dividing the ensemble variance by the mean response amplitude. Kinetics were determined from dim flash responses whose amplitudes were a fifth of the maximum. Time to peak was measured from mid-flash to the response peak. Integration time was calculated as the integral of the response divided by response amplitude. i½ and I½ are the light strengths giving rise to half-maximal flash and step responses, respectively. Comparisons were made using a t test. ns, not significant.
| WT | Palm−/− | p | |
|---|---|---|---|
| Circulating current, pA | 8.8 ± 0.3 (59) | 8.8 ± 0.2 (95) | ns |
| Single photon response | |||
| Amplitude, pA | 0.53 ± 0.08 (5) | 0.43 ± 0.04 (10) | ns |
| Time to peak, ms | 128 ± 3 (43) | 128 ± 3 (75) | ns |
| Integration time, ms | 335 ± 17 (43) | 252 ± 9 (75) | <0.0001 |
| Sensitivity to flashes, i½, photons µm−2 | 46 ± 2 (58) | 52 ± 2 (95) | <0.01 |
| Sensitivity to steps, I½, photons µm−2 s−1 | 120 ± 11 (8) | 238 ± 30 (14) | <0.01 |
Increased Phosphorylation of Palmitoylation-deficient Rhodopsin
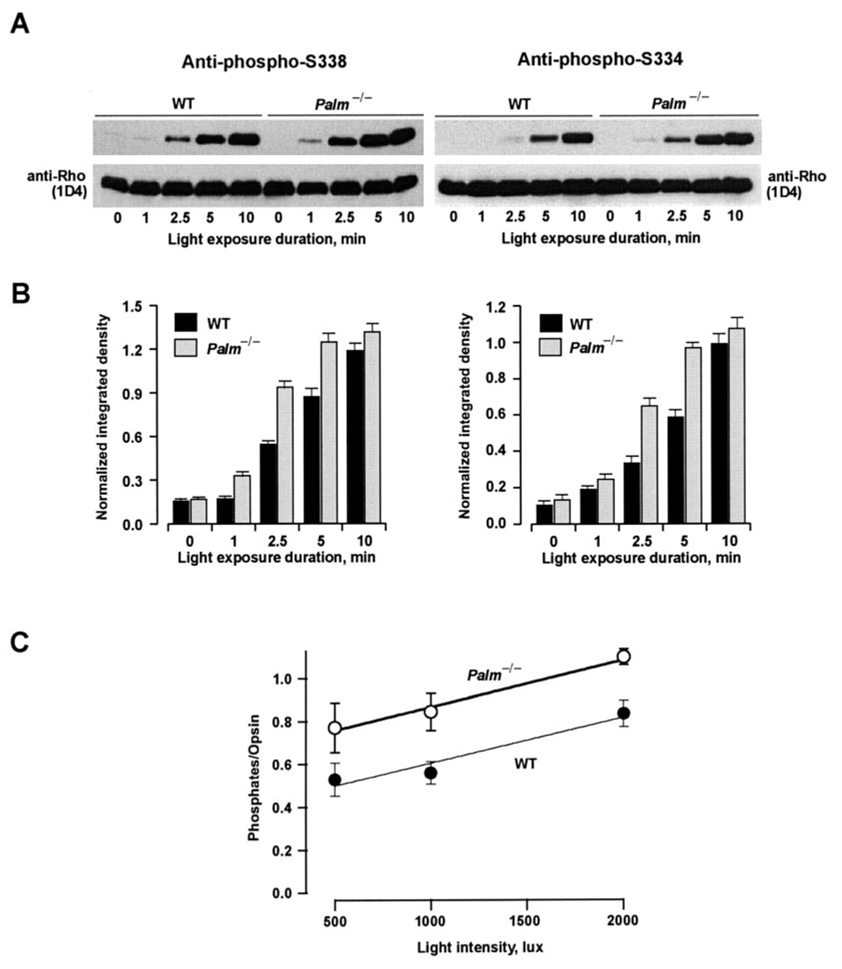
Shutoff of photoexcited rhodopsin begins with its phosphorylation at C-terminal serine and threonine residues by rhodopsin kinase. We assessed phosphorylation in rod outer segment preparations by immunoblotting with two antibodies that selectively recognized phosphoserine residues at positions 334 or 338 of rhodopsin (15). As shown in Fig. 6, A and B, little phosphorylated rhodopsin was detected by the two antibodies in dark-adapted ROS from either type of mouse. With increasing time after exposure to bright light, mutant and WT ROS showed increases in Ser334 and Ser338 phosphorhodopsin, but phosphates accumulated at both sites at a faster rate in Palm−/− than in WT ROS.
FIG. 6. Increased phosphorylation of rhodopsin in Palm−/− mice.
A, rhodopsin phosphorylation levels assessed by Western blot analysis using antibodies specific for phospho-Ser338 (upper left panel) or phospho-Ser334 (upper right panel). ROS prepared from dark-adapted WT or Palm−/− mice were kept in the dark or exposed to 2000 lux for 1, 2.5, 5, or 10 min. Blots probed with anti-phosphorhodopsin antibodies were stripped and reprobed with anti-rhodopsin 1D4 antibody (lower panels), which recognizes both phosphorylated and non-phosphorylated rhodopsin. As the relative affinity of the two phosphorhodopsin antibodies have not been measured, the phosphorylation at Ser338 and Ser334 was not compared. B, histograms showing the densitometric analyses of phospho-Ser338 and -Ser334, normalized to the density for total rhodopsin as probed with 1D4, from three independent experiments. Error bars show S.E. C, increased numbers of phosphates per opsin molecule in Palm−/− mice compared with WT mice after exposure to light for 10 min as determined by mass spectrometry. There were six determinations for each point except for Palm−/− at 2000 lux, where there were three. Error bars show S.E.
Phosphorylation of rhodopsin at these and other positions was probed further by mass spectrometry. The introduced Cys322 → Thr and Cys323 → Ser mutation sites were not phosphorylated in the light or in the dark (data not shown). Rhodopsin phosphorylation was not detected at any C-terminal sites in dark-adapted Palm−/− or WT retinas. Upon illumination, however, both the Palm−/− and WT animals showed mono to triple phosphorylation with involvement of all six C-terminal sites. There was no distinguishing pattern of phosphorylation between the two sets of animals, although the Palm−/− animals consistently showed a higher level of phosphorylation. In both WT and Palm−/− mice, phosphorylation increased with light intensity (p < 0.0007). In addition, Palm−/− mice had a greater mean number of phosphates per opsin compared with WT mice (p < 0.0004) (see Fig. 6C). The increased phosphate incorporation in Palm−/− mice arose from a lower proportion of unphosphorylated rhodopsin (p < 0.0015) and a higher ratio of di- and triphosphorylated rhodopsins relative to the monophosphorylated form (p < 0.0012). Thus, increased rhodopsin phosphorylation could account for the faster flash response recovery in Palm−/− rods.
DISCUSSION
The aim of the present study was to explore the physiological function of rhodopsin palmitoylation in vivo. Toward that end, we created a transgenic mouse that made only palmitoylation-deficient rhodopsin. To avoid phenotypic changes produced by alterations in levels of rhodopsin transgene expression (19, 20), the palmitoylation sites in the endogenous rhodopsin gene were mutated by homologous recombination. The pair of palmitates on rhodopsin normally insert into the membrane and create a fourth cytoplasmic loop. Although it has been suggested that the absence of palmitates untethers the rhodopsin C terminus, spin labeling studies indicate that the mobility of the rhodopsin C terminus is only very slightly affected (21). Nevertheless, the palmitates could dictate the folding and arrangement of the cytoplasmic surface. Since this surface possesses an intracellular localization sequence and regions that interact with transducin, rhodopsin kinase, and arrestin (8), absence of palmitates could potentially interfere with rhodopsin translocation and/or G-protein signaling.
Defective receptor palmitoylation has been reported to affect the proper membrane localization of several GPCRs. Palmitoylation-deficient mutants of the luteinizing hormone receptor (22), the H2 histamine receptor (23), the vasopressin V2 receptor (24), the CCR5 chemokine receptor (25, 26), and the thyrotropin receptor (27) exhibited reduced cell surface expression in heterologous systems. The underlying mechanisms varied. In the case of luteinizing hormone receptor, the absence of palmitates increased internalization, while for the thyrotropin receptor, delivery to the plasma membrane was delayed. Palmitoylation-deficient rhodopsins did not accumulate intracellularly when expressed in COS cells (28). In rods of mutant mice, we saw no evidence of rhodopsin accumulation in intracellular compartments other than the outer segment (Fig. 4C) nor were there differences in the levels of rhodopsin in Palm−/− retinas at 1 month of age, which might occur if fewer rhodopsin molecules were transported to the outer segment. Therefore, rhodopsin palmitoylation does not appear to be required for rhodopsin synthesis or for trafficking to the outer segment in intact rods.
Palmitoylation has been shown to affect coupling of receptors to G-proteins. For example, loss of all three palmitates on the human endothelin B receptor reduces Gq and Gi signaling (29). Activation of transducin increases in retinas treated with 1 m hydroxylamine to chemically depalmitoylate rhodopsin (30) but is not different for rod outer segments treated in a different way to remove the palmitates (7) or for mutant C322T, C323S rhodopsin expressed in COS cells (28, 31). Perhaps high concentrations of hydroxylamine have nonspecific effects, which impinge on the results. In intact Palm−/− rods, the rising phase of the flash response and the time to peak were normal (Fig. 5C), consistent with normal transducin activation (32). Opsin with all-trans-retinal bound non-covalently shows reduced signaling activity in the absence of palmitates, suggesting that palmitoylation may impact adaptation after exposures to very bright light (7). This issue was not addressed in our study and warrants further investigation.
A number of GPCRs undergo changes in phosphorylation upon loss of palmitoylation. Basal and agonist-stimulated phosphorylation increase for the A3 adenosine receptor palmitoylation mutant (33). With the β2-adrenergic receptor palmitoylation mutant, basal (34–36) and agonist-stimulated (37) phosphorylation levels are much higher, while its ability to couple to Gs is lower (34, 35, 37). The latter effect may be due to hyperphosphorylation because further mutation of two phosphorylation sites restores basal phosphorylation levels and proper coupling of the receptor to Gs (36). A steric regulatory role for palmitoylation is supported by reports that palmitoylation of the rat bradykinin β2 receptor at Cys356 and phosphorylation of Tyr352 are mutually exclusive, suggesting that phosphorylation can only occur upon depalmitoylation (38). In a heterologous system, light-dependent phosphorylation of rhodopsin lacking palmitate at position 323 was slightly lower than for wild-type rhodopsin, while rhodopsin lacking both palmitates behaved normally (28). Our immunoblots and mass spectrometric assays indicate that in outer segments, absence of palmitoylation did not lead to phosphorylated rhodopsin in darkness but did promote light-dependent phosphorylation (Fig. 6). Phosphorylation and dephosphorylation occur on similar time scales (39), so our results may include an impaired interaction between Palm−/− rhodopsin and phosphatase. Greater phosphate incorporation can explain the faster recovery of the photon response in Palm−/− rods. In this regard, there is a striking similarity between the responses of Palm−/− rods and those of rods lacking recoverin (40), a Ca2+-dependent regulator of rhodopsin kinase (41–44). In recoverin knock-out rods, the flash response recovers more rapidly probably because a restraint on rhodopsin phosphorylation was removed. But unlike recoverin’s action, the impact of palmitoylation does not fade in the presence of background light (45). Therefore, we propose that by organizing the cytoplasmic face of rhodopsin, palmitoylation prolongs the activity of rhodopsin by attenuating phosphorylation by rhodopsin kinase. Sensitivity to flashes improves slightly, but the main effect is a greater response to step changes in illumination. In contrast to rhodopsin, short wavelength-sensitive opsins bear only a single palmitoylation site on the C terminus, while middle/long wavelength-sensitive opsins have none (46, 47). The lack of palmitoylation may contribute to the faster phosphorylation in middle/long wavelength- sensitive cones than in rods (48). Palmitoylation may lose its importance in cones because they operate with fast response kinetics at high light intensities.
Acknowledgments
We thank F. Celestin of the Specialized Center of Research in Ischemic Heart Disease Histology Core for assistance with histological sections and M. Solovyeva for technical support. We also thank Drs. R. Molday, M. Simon, L. Donoso, A. Dizhoor, T. Wensel, and D. Williams for antibodies and F. Sangiorgi for helpful comments.
Footnotes
This work was supported by Research to Prevent Blindness (Medical University of South Carolina) and by National Institutes of Health (NIH) Grants F2EY13912A (to Z. W.), EY12008 (to J. L.), EY11358 (to C. L. M.), EY04939 (to R. K. C.), EY014799 (to Z. A.), and NIH P30 EY13078 Core Grant for Vision Research awarded to Tufts-New England Medical Center and NIH P30 EY14104 Core Grant for Vision Research awarded to Massachusetts Eye and Ear Infirmary.
This article was selected as a Paper of the Week.
The abbreviations used are: GPCR, G-protein coupled receptor; RT, reverse transcription; ONL, outer nuclear layer; WT, wild-type; ROS, rod outer segment(s); HPLC, high performance liquid chromatography.
REFERENCES
- 1.Qanbar R, Bouvier M. Pharmacol. Ther. 2003;97:1–33. doi: 10.1016/s0163-7258(02)00300-5. [DOI] [PubMed] [Google Scholar]
- 2.Probst WC, Snyder LA, Schuster DI, Brosius J, Sealfon SC. DNA Cell Biol. 1992;11:1–20. doi: 10.1089/dna.1992.11.1. [DOI] [PubMed] [Google Scholar]
- 3.O’Brien PJ, Zatz M. J. Biol. Chem. 1984;259:5054–5057. [PubMed] [Google Scholar]
- 4.Ovchinnikov Yu A, Abdulaev NG, Bogachuk AS. FEBS Lett. 1988;230:1–5. doi: 10.1016/0014-5793(88)80628-8. [DOI] [PubMed] [Google Scholar]
- 5.Moench SJ, Moreland J, Stewart DH, Dewey TG. Biochemistry. 1994;33:5791–5796. doi: 10.1021/bi00185a017. [DOI] [PubMed] [Google Scholar]
- 6.Krishna AG, Menon ST, Terry TJ, Sakmar TP. Biochemistry. 2002;41:8298–8309. doi: 10.1021/bi025534m. [DOI] [PubMed] [Google Scholar]
- 7.Sachs K, Maretzki D, Meyer CK, Hofmann KP. J. Biol. Chem. 2000;275:6189–6194. doi: 10.1074/jbc.275.9.6189. [DOI] [PubMed] [Google Scholar]
- 8.Menon ST, Han M, Sakmar TP. Physiol. Rev. 2001;81:1659–1688. doi: 10.1152/physrev.2001.81.4.1659. [DOI] [PubMed] [Google Scholar]
- 9.Bunting M, Bernstein KE, Greer JM, Capecchi MR, Thomas KR. Genes. Dev. 1999;13:1524–1528. doi: 10.1101/gad.13.12.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li E, Bestor TH, Jaenisch R. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 11.Molday RS, MacKenzie D. Biochemistry. 1983;22:653–660. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- 12.Donoso LA, Merryman CF, Edelberg KE, Naids R, Kalsow C. Investig. Ophthalmol. Vis. Sci. 1985;26:561–567. [PubMed] [Google Scholar]
- 13.Dizhoor AM, Ray S, Kumar S, Niemi G, Spencer M, Brolley D, Walsh KA, Philipov PP, Hurley JB, Stryer L. Science. 1991;251:915–918. doi: 10.1126/science.1672047. [DOI] [PubMed] [Google Scholar]
- 14.Tsang SH, Burns ME, Calvert PD, Gouras P, Baylor DA, Goff SP, Arshavsky VY. Science. 1998;282:117–121. doi: 10.1126/science.282.5386.117. [DOI] [PubMed] [Google Scholar]
- 15.Adams RA, L X, Williams DS, Newton AC. Biochem. J. 2003;374:537–543. doi: 10.1042/BJ20030408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ablonczy Z, Kono M, Crouch RK, Knapp DR. Anal. Chem. 2001;73:4774–4779. doi: 10.1021/ac015563n. [DOI] [PubMed] [Google Scholar]
- 17.Ablonczy Z, Crouch RK, Goletz PW, Redmond TM, Knapp DR, Ma JX, Rohrer B. J. Biol. Chem. 2002;277:40491–40498. doi: 10.1074/jbc.M205507200. [DOI] [PubMed] [Google Scholar]
- 18.Lee KA, Craven KB, Niemi GA, Hurley JB. Protein Sci. 2002;11:862–874. doi: 10.1110/ps.3870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan E, Wang Q, Quiambao AB, Xu X, Qtaishat NM, Peachey NS, Lem J, Fliesler SJ, Pepperberg DR, Naash MI, Al-Ubaidi MR. Investig. Ophthalmol. Vis. Sci. 2001;42:589–600. [PubMed] [Google Scholar]
- 20.Lem J, Krasnoperova NV, Calvert PD, Kosaras B, Cameron DA, Nicolo M, Makino CL, Sidman RL. Proc. Natl. Acad. Sci. U. S. A. 1999;96:736–741. doi: 10.1073/pnas.96.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langen R, Cai K, Altenbach C, Khorana HG, Hubbell WL. Biochemistry. 1999;38:7918–7924. doi: 10.1021/bi990010g. [DOI] [PubMed] [Google Scholar]
- 22.Zhu H, Wang H, Ascoli M. Mol. Endocrinol. 1995;9:141–150. doi: 10.1210/mend.9.2.7776964. [DOI] [PubMed] [Google Scholar]
- 23.Fukushima Y, Saitoh T, Anai M, Ogihara T, Inukai K, Funaki M, Sakoda H, Onishi Y, Ono H, Fujishiro M, Ishikawa T, Takata K, Nagai R, Omata M, Asano T. Biochim. Biophys. Acta. 2001;1539:181–191. doi: 10.1016/s0167-4889(01)00104-5. [DOI] [PubMed] [Google Scholar]
- 24.Schulein R, Liebenhoff U, Muller H, Birnbaumer M, Rosenthal W. Biochem. J. 1996;313:611–616. doi: 10.1042/bj3130611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanpain C, Wittamer V, Vanderwinden JM, Boom A, Renneboog B, Lee B, Le Poul E, El Asmar L, Govaerts C, Vassart G, Doms RW, Parmentier M. J. Biol. Chem. 2001;276:23795–23804. doi: 10.1074/jbc.M100583200. [DOI] [PubMed] [Google Scholar]
- 26.Percherancier Y, Planchenault T, Valenzuela-Fernandez A, Virelizier JL, Arenzana-Seisdedos F, Bachelerie F. J. Biol. Chem. 2001;276:31936–31944. doi: 10.1074/jbc.M104013200. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka K, Nagayama Y, Nishihara E, Namba H, Yamashita S, Niwa M. Endocrinology. 1998;139:803–806. doi: 10.1210/endo.139.2.5911. [DOI] [PubMed] [Google Scholar]
- 28.Karnik SS, Ridge KD, Bhattacharya S, Khorana HG. Proc. Natl. Acad. Sci. U. S. A. 1993;90:40–44. doi: 10.1073/pnas.90.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okamoto Y, Ninomiya H, Tanioka M, Sakamoto A, Miwa S, Masaki T. J. Biol. Chem. 1997;272:21589–21596. doi: 10.1074/jbc.272.34.21589. [DOI] [PubMed] [Google Scholar]
- 30.Morrison DF, O’Brien PJ, Pepperberg DR. J. Biol. Chem. 1991;266:20118–20123. [PubMed] [Google Scholar]
- 31.Marin EP, Krishna AG, Zvyaga TA, Isele J, Siebert F, Sakmar TP. J. Biol. Chem. 2000;275:1930–1936. doi: 10.1074/jbc.275.3.1930. [DOI] [PubMed] [Google Scholar]
- 32.Lamb TD, Pugh EN., Jr. J. Physiol. (Lond.) 1992;449:719–758. doi: 10.1113/jphysiol.1992.sp019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer TM, Stiles GL. Mol. Pharmacol. 2000;57:539–545. [PubMed] [Google Scholar]
- 34.O’Dowd BF, Hnatowich M, Caron MG, Lefkowitz RJ, Bouvier M. J. Biol. Chem. 1989;264:7564–7569. [PubMed] [Google Scholar]
- 35.Moffett S, Mouillac B, Bonin H, Bouvier M. EMBO J. 1993;12:349–356. doi: 10.1002/j.1460-2075.1993.tb05663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moffett S, Adam L, Bonin H, Loisel TP, Bouvier M, Mouillac B. J. Biol. Chem. 1996;271:21490–21497. doi: 10.1074/jbc.271.35.21490. [DOI] [PubMed] [Google Scholar]
- 37.Moffett S, Rousseau G, Lagace M, Bouvier M. J. Neurochem. 2001;76:269–279. doi: 10.1046/j.1471-4159.2001.00005.x. [DOI] [PubMed] [Google Scholar]
- 38.Soskic V, Nyakatura E, Roos M, Muller-Esterl W, Godovac-Zimmermann J. J. Biol. Chem. 1999;274:8539–8545. doi: 10.1074/jbc.274.13.8539. [DOI] [PubMed] [Google Scholar]
- 39.Kennedy MJ, Lee KA, Niemi GA, Craven KB, Garwin GG, Saari JC, Hurley JB. Neuron. 2001;31:87–101. doi: 10.1016/s0896-6273(01)00340-3. [DOI] [PubMed] [Google Scholar]
- 40.Makino CL, Dodd RL, Chen J, Burns ME, Roca A, Simon MI, Baylor DA. J. Gen. Physiol. 2004;123:729–741. doi: 10.1085/jgp.200308994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawamura S. Nature. 1993;362:855–857. doi: 10.1038/362855a0. [DOI] [PubMed] [Google Scholar]
- 42.Gorodovikova EN, Philippov PP. FEBS Lett. 1993;335:277–279. doi: 10.1016/0014-5793(93)80746-h. [DOI] [PubMed] [Google Scholar]
- 43.Chen J, Makino CL, Peachey NS, Baylor DA, Simon MI. Science. 1995;267:374–377. doi: 10.1126/science.7824934. [DOI] [PubMed] [Google Scholar]
- 44.Klenchin VA, Calvert PD, Bownds MD. J. Biol. Chem. 1995;270:16147–16152. doi: 10.1074/jbc.270.27.16147. [DOI] [PubMed] [Google Scholar]
- 45.O’Brien PJ, St. Jules RS, Reddy TS, Bazan NG, Zatz M. J. Biol. Chem. 1987;262:5210–5215. [PubMed] [Google Scholar]
- 46.Stenkamp RE, Filipek S, Driessen CA, Teller DC, Palczewski K. Biochim. Biophys. Acta. 2002;1565:168–182. doi: 10.1016/s0005-2736(02)00567-9. [DOI] [PubMed] [Google Scholar]
- 47.Ebrey T, Koutalos Y. Prog. Retin. Eye Res. 2001;20:49–94. doi: 10.1016/s1350-9462(00)00014-8. [DOI] [PubMed] [Google Scholar]
- 48.Tachibanaki S, Tsushima S, Kawamura S. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14044–14049. doi: 10.1073/pnas.241396898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pepperberg DR, Okajima TI. Exp. Eye Res. 1992;54:369–376. doi: 10.1016/0014-4835(92)90049-x. [DOI] [PubMed] [Google Scholar]
- 50.Nikonov S, Engheta N, Pugh EN., Jr. J. Gen. Physiol. 1998;111:7–37. doi: 10.1085/jgp.111.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma J, Znoiko S, Othersen KL, Ryan JC, Das J, Isayama T, Kono M, Oprian DD, Corson DW, Cornwall MC, Cameron DA, Harosi FI, Makino CL, Crouch RK. Neuron. 2001;32:451–461. doi: 10.1016/s0896-6273(01)00482-2. [DOI] [PubMed] [Google Scholar]