Abstract
DBTBS, first released in 1999, is a reference database on transcriptional regulation in Bacillus subtilis, summarizing the experimentally characterized transcription factors, their recognition sequences and the genes they regulate. Since the previous release, the original content was extended by the addition of the data contained in 569 new publications, the total of which now reaches 947. The number of B. subtilis promoters annotated in the database was more than doubled to 1475. In addition, 463 experimentally validated B. subtilis operons and their terminators have been included. Given the increase in the number of fully sequenced bacterial genomes, we decided to extend the usability of DBTBS in comparative regulatory genomics. We therefore created a new section on the conservation of the upstream regulatory sequences between homologous genes in 40 Gram-positive bacterial species, as well as on the presence of overrepresented hexameric motifs that may have regulatory functions. DBTBS can be accessed at: http://dbtbs.hgc.jp.
INTRODUCTION
Bacillus subtilis is one of the best-studied Gram-positive bacteria, and hence serves as a model organism, much like Escherichia coli for Gram-negative bacteria. Bacillus subtilis databases, such as SubtiList (1,2) or the database of transcriptional regulation in B. subtilis (DBTBS) (3,4), are therefore used as a reference not only by B. subtilis researchers, but also by researchers focusing on more or less distant organisms. Indeed, it is assumed that at least part of the knowledge gained on the model organism can be extended to these other organisms; the amount of this extension being related to the distance between both organisms.
We previously presented DBTBS, which offers detailed information about the B. subtilis transcription system. Besides increasing the amount of B. subtilis data contained in DBTBS, we added a new section to the database aimed at helping wet-lab researchers assess the relevance of extending B. subtilis knowledge to the bacteria they study. This section presents upstream intergenic region conservation profiles for homologous proteins of a same Gram-positive genus, as well as hexameric motifs conserved between different profiles.
UPDATES AND NEW FEATURES
Since Release 3 of DBTBS in 2004 (4), a significant increase in the number of referenced publication, from 378 to 947, has occurred (Table 1). This increase resulted in the inclusion of six new transcription factors, bringing their number to 120. At the same time, the number of promoters rose from 633 to 1475 and the number of regulated operons went from 525 to 736. Indeed, the regulated genes were reorganized in regulated operons, and all the regulated genes are now reported, in contrast to only the first gene of the operon. In addition to the extension of the existing data, 463 experimentally validated B. subtilis operons (5) and their terminators have been included as well (6). As previously, researchers worldwide are encouraged to report outdated, incorrect or missing information in order to make DBTBS as complete and accurate as possible.
Table 1.
Summary of the updated data
| Category | Release 3 | Release 5 |
|---|---|---|
| Referenced publications | 378 | 947 |
| Transcription factors | 114 | 120 |
| Position-specific scoring matrices | 45 | 45 |
| Promoters | 633 | 1475 |
| Regulated operons | 525 | 736 |
| Terminators | 0 | 463 |
The number of items of each category present in release 3 (November 2004) and release 5 (September 2007) of DBTBS are given.
To facilitate the identification of regulatory elements, two new tools have been added. First, a matrix search function allows users to identify which transcription factors correspond to the position-specific weighted matrix they submitted by querying DBTBS for the top 10 weight matrices similar to it. Second, following a user request, a B. subtilis motif location search tool was added as a remedy for the disappearance of the GRASP-DNA tool developed by Schilling et al. (7). This tool allows a user to input a list of binding site sequences and returns the B. subtilis genome locations matched by the position-specific weighted matrix calculated from them. For each location, the two nearest upstream and downstream open reading frames are also reported. Furthermore, the generated position-specific weighted matrix can be directly used to search for the 10 most similar DBTBS weighted matrices matrices by using the provided link.
Upstream intergenic region conservation
In order to provide upstream intergenic region conservation information, groups of homologous proteins from 40 Gram-positive bacteria (Supplementary Table 1) were built. Homologies between the proteins were determined by all-against-all protein BLAST (8) searches, where a protein A was considered homologous to a protein B if an identity higher than 40% on more than 50% of the length of A was found. Each group was then divided into subgroups based on genus, and each subgroup further divided based on the lengths of the upstream intergenic region of its members. Although orthologous and paralogous genes are first grouped together, subdividing the genus-specific groups based on the length of the upstream intergenic regions is expected to separate paralogous genes that are differently regulated.
The upstream intergenic regions of each of the subgroups containing more than two members were aligned with ClustalW (9), and the last 300 positions of the alignment, representing the nucleotides directly upstream of the gene starts, were kept for further analysis.
For each subgroup, a conservation profile was calculated based on information content, thus giving the degree of conservation of each position and allowing the determination of conserved regions. In our analysis, conserved regions were determined by setting the threshold for the degree of conservation to 75%, while allowing at most three consecutive positions to have lower values. All the possible 6-bases-long position-specific weighted matrices were then created from the determined conserved regions and clustered using the quality cluster algorithm (10) and a Kullback–Leibler divergence (11) of 0.3 as the maximum cluster diameter. Matrices clustering together were merged to yield the hexameric motif matrices available from DBTBS.
Through this process, 29 520 hexameric motif matrices were created; 5652 of them were specific to Bacillus, 1516 to Staphylococcus and 184 to Streptococcus (Table 2 and Supplementary Table 2). These numbers are largely influenced by the grouping method, which results in only few groups for genus with few members, and hence lowers the potential number of motifs specific to that genus.
Table 2.
Repartition of the clusters and motifs
| Genus | Bacteria | Genes | Specific sub- groups | % | Specific motifs | % | |
|---|---|---|---|---|---|---|---|
| Bacillus | BAC | 7 | 7450 | 1581 | 79.0 | 5652 | 22.5 |
| Carboxydothermus | CAB | 1 | 7 | 2 | 100.0 | 0 | 0.0 |
| Clostridium | CLO | 3 | 271 | 46 | 53.5 | 11 | 0.4 |
| Enterococcus | ENT | 1 | 11 | 1 | 33.3 | 0 | 0.0 |
| Geobacillus | GEO | 1 | 19 | 4 | 100.0 | 0 | 0.0 |
| Lactobacillus | LAB | 3 | 299 | 24 | 25.5 | 13 | 0.4 |
| Lactococcus | LAC | 1 | 45 | 7 | 100.0 | 0 | 0.0 |
| Listeria | LIS | 2 | 51 | 9 | 64.3 | 0 | 0.0 |
| Mycoplasma | MYC | 9 | 298 | 40 | 58.0 | 24 | 0.8 |
| Oceanobacillus | OCB | 1 | 6 | 1 | 50.0 | 0 | 0.0 |
| Phytoplasma | OYP | 1 | 92 | 18 | 85.7 | 0 | 0.0 |
| Staphylococcus | STA | 4 | 3435 | 555 | 60.5 | 1516 | 8.3 |
| Streptococcus | STR | 5 | 1573 | 196 | 46.3 | 184 | 1.9 |
| Thermoanaerobacter | TAB | 1 | 59 | 10 | 100.0 | 0 | 0.0 |
Genuses are listed followed by the three-letter abbreviation used for cluster names. Bacteria: number of bacteria species used for the given genus; Genes: total number of genes reported in the bacteria of the given genus; Specific subgroups: number of subgroups for which no homologous subgroup is found in other genuses; Specific motifs: number of hexameric motif matrices found to bind exclusively in a given genus. Percentages are given based on the total number of subgroups, respectively motifs, found in a given genus.
The Gram-positive bacteria upstream intergenic conservation information can be accessed from the ‘Motif conservation’ link on the main page of DBTBS. Users can then search the data by submitting a gene name, a genus-subgroup name or a motif number. Also, because of the large number of hexameric motif matrices available, the desired ranges for the information content and the number of occurrences of a motif can be selected in order to filter the displayed motifs.
Submitting a gene name will return a table indicating which organisms contain a gene labeled with the given name, as well as in which genus-subgroup this gene is included and which motifs are found in that subgroup. Genus-subgroups and motifs are linked to the same pages that those obtained by directly searching with a genus-subgroup name or a motif number.
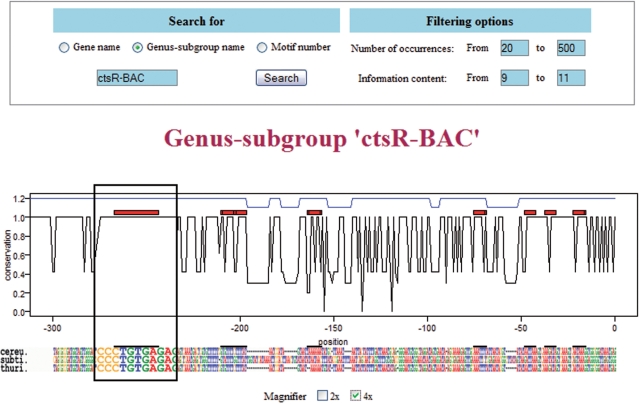
The result of a genus-subgroup name search is a page presenting the conservation profile of the subgroup, with the conserved regions and motifs positions. The upstream intergenic sequence alignment used to calculate the conservation profile is shown under it (Figure 1). Following the graphical display of the subgroup is a list of the genes included in the subgroup, and a list of the motifs present. This last list shows the motif logo (12) and indicates in which other groups the same motif is found. Again each genus-subgroup name and motif number in this list is linked to the same page as the one obtained by a direct search.
Figure 1.
Hexameric motif conservation in an upstream intergenic region. The upper part shows the search entry box, with the criteria selected for filtering of the displayed hexameric motifs. The lower part is the resulting figure, showing the conservation profile as a black line and the alignment used to obtain it. The blue line represents the calculated conserved region and the red boxes the found hexameric motifs. A 2×/4× magnifier is available in order to conveniently scan the sequence alignment.
A motif number search shows first the motif logo, and then a list of genus-subgroups where the motif is found. In this list, the position of the motif in each subgroup is shown graphically, and for each subgroup, the list of the included genes and of the other motifs found in it is given, once again linked to the relevant pages.
CONCLUSIONS
Because of the high number of hexameric motif matrices, a careful picking of the filtering ranges is necessary to avoid the presence of too many motifs in the genus-subgroup pages. Although this might be seen as a drawback, it allows a higher flexibility in the type of searches users can perform. Indeed, by changing the range of the number of occurrences of a motif, one could search for conserved regions potentially indicating binding sites of minor or specialized transcription factors, which typically only bind at a few places in a genome, or target-binding sites of global regulators, which occur a lot more frequently.
Additionally, the provided conservation profiles and sequence alignments can by themselves offer valuable information that could not be captured by the matrix conservation analysis. For instance, the shift by a few bases of a binding site might result in sub-optimal sequence alignment, and hence in an apparent absence of conservation which will result in the lack of matrix motif at this position. However, a visual inspection of the alignment will nevertheless allow the identification of the conserved region, despite the failure of the automatic recognition. In fact, many factors influence the quality of the motifs obtained using the method presented here, such as the quality of the homologous genes grouping, the accuracy of the sequence alignment, the distance threshold used in the motif clustering, or the chosen length of the motifs. The exact effects of these parameters have not been investigated so far, and future work in that direction should be carried out to be able to decrease the number of biologically meaningless motifs more efficiently. The prediction of the operon structures for each strain used should also improve our results and will therefore be considered in the future. In addition, an analysis of the co-occurrence of motifs might provide interesting information about co-regulation by several transcription factors. Although the current version of this tool already offers a significant amount of information to interested scientists, it should consequently only be considered as a first step in the analysis of the conservation of upstream intergenic regions, setting ground for finer investigations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Y. Fujita for his constructive feature request. This work has been supported by BIRD of Japan Science and Technology Agency (JST). N.S. was also supported by the Japan Society for the Promotion of Science. Computation time was provided by the Super Computer System, Human Genome Center, Institute of Medical Science, University of Tokyo. Funding to pay the Open Access publication charges for this article was provided by provided by JST-BIRD.
Conflict of interest statement. None declared.
REFERENCES
- 1.Moszer I, Glaser P, Danchin A. SubtiList: a relational database for the Bacillus subtilis genome. Microbiology. 1995;141(Pt 2):261–268. doi: 10.1099/13500872-141-2-261. [DOI] [PubMed] [Google Scholar]
- 2.Moszer I, Jones LM, Moreira S, Fabry C, Danchin A. SubtiList: the reference database for the Bacillus subtilis genome. Nucleic Acids Res. 2002;30:62–65. doi: 10.1093/nar/30.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishii T, Yoshida K, Terai G, Fujita Y, Nakai K. DBTBS: a database of Bacillus subtilis promoters and transcription factors. Nucleic Acids Res. 2001;29:278–280. doi: 10.1093/nar/29.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makita Y, Nakao M, Ogasawara N, Nakai K. DBTBS: database of transcriptional regulation in Bacillus subtilis and its contribution to comparative genomics. Nucleic Acids Res. 2004;32:D75–D77. doi: 10.1093/nar/gkh074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Hoon MJL, Imoto S, Kobayashi K, Ogasawara N, Miyano S. Predicting the operon structure of Bacillus subtilis using operon length, intergene distance, and gene expression information. Pac. Symp. Biocomput. 2004:276–287. doi: 10.1142/9789812704856_0027. [DOI] [PubMed] [Google Scholar]
- 6.De Hoon MJL, Makita Y, Nakai K, Miyano S. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput. Biol. 2005;1:e25. doi: 10.1371/journal.pcbi.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schilling CH, Held L, Torre M, Saier M.H., Jr GRASP-DNA: a web application to screen prokaryotic genomes for specific DNA-binding sites and repeat motifs. J. Mol. Microbiol. Biotechnol. 2000;2:495–500. [PubMed] [Google Scholar]
- 8.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heyer LJ, Kruglyak S, Yooseph S. Exploring expression data: identification and analysis of coexpressed genes. Genome Res. 1999;9:1106–1115. doi: 10.1101/gr.9.11.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kullback S, and Leibler RA. On information and sufficiency. Ann. Math. Stat. 1951;22:79–86. [Google Scholar]
- 12.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.