Abstract
A universal base that is capable of substituting for any of the four natural bases in DNA would be of great utility in both mutagenesis and recombinant DNA experiments. This paper describes the properties of oligonucleotides incorporating two degenerate bases, the pyrimidine base 6H,8H-3,4-dihydropyrimido[4,5-c][1,2]oxazin-7-one and the purine base N6-methoxy-2,6-diaminopurine, designated P and K, respectively. An equimolar mixture of the analogues P and K (called M) acts, in primers, as a universal base. The thermal stability of oligonucleotide duplexes were only slightly reduced when natural bases were replaced by P or K. Templates containing the modified bases were copied by Taq polymerase; P behaved as thymine in 60% of copying events and as cytosine in 40%, whereas K behaved as if it were guanine (13%) or adenine (87%). The dUTPase gene of Caenorhabditis elegans, which we have found to contain three nonidentical homologous repeats, was used as a model system to test the use of these bases in primers for DNA synthesis. A pair of oligodeoxyribonucleotides, each 20 residues long and containing an equimolar mixture of P and K at six positions, primed with high specificity both T7 DNA polymerase in sequencing reactions and Taq polymerase in PCRs; no nonspecific amplification was obtained on genomic DNA of C. elegans. Use of P and K can significantly reduce the complexity of degenerate oligonucleotide mixtures, and when used together, P and K can act as a universal base.
Analogues of the naturally occurring nucleic acid bases that can prime DNA synthesis when incorporated into oligodeoxyribonucleotides have been used, inter alia, to decrease the chain multiplicity of primers and probes when incorporated at positions of degeneracy. This situation arises frequently when a DNA sequence is derived from a known protein sequence, because of the degenerate nature of the genetic code. A variety of analogues have been investigated. Many publications attest to the successful use of deoxyinosine (dI); the rationale for its use derives from observations relating to hypoxanthine as the wobble base in tRNAs (1, 2). Although hybridization probes and primers containing 2′-deoxyinosine have been widely used, the nucleoside suffers from some drawbacks. Its base pairs with the four normal bases have very different interaction energies (3), and indeed, the evidence overall suggests that stacking interactions mainly account for the stabilities of duplexes containing it (4, 5).
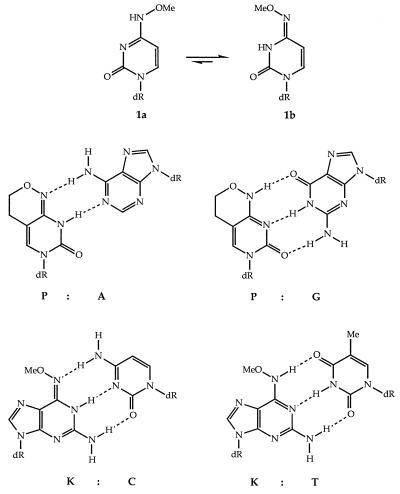
Earlier, we took the view that a pair of bispecific bases, one designed to mimic both of the natural pyrimidine bases and the other designed to mimic both purines, would provide a useful means of reducing degeneracy. The transition mutagenic bases (6) provided a starting point for such bispecificity. Thus the stable base pairing of N4-methoxycytosine (mo4C) (1) with both adenine (A) and guanine (G) in oligonucleotides established the principle (7). It showed that this correlated well with the effect of the electronegative oxygen atom in altering the tautomeric constant KT of cytosine from about 105 to 0.1–0.03, favoring the imino form (8, 9). Thus the small free energy difference between the tautomers permitted stable hydrogen bonding with both A and G. The preferred syn orientation of the methoxyl group (10, 11) with respect to N3 in mo4C (Fig. 1, compound 1b) gave some destabilization in base pairing. This was confirmed by the synthesis of the analogue nucleoside dP that when inserted into oligonucleotides, gave markedly more stable duplexes when paired with both A and G (12). The nucleoside, too, proved to be highly mutagenic to microorganisms (13, 14).
Figure 1.
Anti (1a) and syn (1b) conformation of N4-methoxycytidine (mo4C), base pairing of the analogue 6H,8H-3,4-dihydropyrimido[4,5-c][1,2]oxazin-7-one (P) with adenine and with guanine, and base pairing of the analogue N6-methoxy-2,6-diaminopurine (K) with cytosine and thymine.
The purine analogues N6-methoxyadenine (mo6A) (Z) and N6-methoxy-2,6-diaminopurine (K) correspondingly showed specific pairing with cytosine (C) and thymine (T) in duplexes (15–17). The analogue K gave greater duplex stabilities and so was chosen for further studies. Preliminary results using either P or K in PCR primers have been published (18); herein we develop this further and investigate the enzymatic recognition properties of the bases in oligodeoxyribonucleotides, used as both primers and templates.
MATERIALS AND METHODS
Syntheses of Monomers and Oligodeoxyribonucleotides.
The phosphoramidite monomers of dP and dK were synthesized as described (12, 16) or obtained from Glen Research (Sterling, VA); 2′-deoxyinosine monomer was supplied by Cruachem (Herndon, VA). Oligonucleotides were synthesized on an Applied Biosystem 380B synthesizer with the normal synthesis cycle. For several oligonucleotides, the P and K monomers were coupled as a premixed 50:50 solution. Oligomers containing P and K mixtures were isolated as trityl-containing compounds, purified by reverse-phase HPLC, and then detritylated. Further purification was carried out by HPLC on a Hichrom Partisil 10 SAX column. All other oligomers were purified by urea/PAGE in 20% polyacrylamide gels (except for the template oligonucleotides, for which 6% polyacrylamide was used), extraction using 0.5 M ammonium acetate/1 mM EDTA buffer, concentration, and then desalting using a Sephadex G-25 column (NAP-10, Pharmacia).
Hybridization Analyses.
Ultraviolet spectra were recorded on a Perkin–Elmer Lambda 2 spectrophotometer fitted with a Peltier cell. Melting transitions were measured at 260 nm in PCR buffer (10 mM Tris/50 mM KCl/1.5 mM MgCl2/0.001% gelatin, pH 8.3, at 25°C) or 6× SSC (0.9 M NaCl/0.09 M sodium citrate, pH 7), at an oligomer strand concentration of approximately 3 μM. Absorbance vs. temperature for each duplex was obtained at a heating and cooling rate of 0.5°C per min and melting temperatures (Tm) were determined as the maxima of the differential curves, with an error of ±1°C. Thermodynamic analysis of the melting curves was carried out with the “all or none” two-state model developed by Gralla and Crothers (19). From the enthalpy and free energy, the entropy of the transition was calculated. Enthalpy and entropy are assumed independent of temperature, and free energies are reported under standard conditions [310 K, 3 μM strand concentration, 1 M (6× SSC) or 0.1 M (PCR buffer) salt].
Templating Properties of P and K Bases.
Two closely related oligonucleotides, one 61 residues long containing six P residues and the other 64 residues long containing six K residues, were synthesized and used as templates in PCRs. For each oligonucleotide template, 48 identical reactions were performed. These were set up as two sets of half reactions in a 96-well plate; one half contained the enzyme Taq polymerase and the other half contained the oligonucleotide template. After equilibration to 72°C on a Techne PHC-3 PCR block, the half reactions were mixed with a multichannel pipettor and then incubated under the following conditions: 1 cycle at 96°C for 2 min, followed by 30 cycles of 96°C for 5 sec and 50°C for 5 sec, and finally 1 cycle at 72°C for 2 min. The completed reactions were pooled and the products were purified on a 20% polyacrylamide gel, before being ligated to a T vector prepared from pBluescript II SK− as described (20). After transformation, randomly picked plasmids were sequenced by using Sequenase version II (United States Biochemical).
Construction of Plasmid Templates for PCR and Sequencing.
Seven plasmids were constructed by using standard cloning methods and the vector pBluescriptII SK− (Stratagene). The first plasmid pdutR contains a 4.8-kb SalI–HindIII fragment from cosmid K07A1 (21) cloned between the same sites of the vector polylinker. This fragment contains the complete Caenorhabditis elegans dUTPase gene, which is composed of three homologous, but nonidentical, repeats (F.H., unpublished results). From this, three further plasmids, each containing only one of the repeats, were constructed. pTemplate 1 contains the first repeat as a 560-bp RsaI fragment in the SmaI site of the polylinker. pTemplate 2 contains the second repeat on a 583-bp RsaI fragment, also in the SmaI site. pTemplate 3, containing the third repeat, was constructed by deleting pdutR between the XhoI site in the polylinker (which was filled in with the Klenow fragment of DNA polymerase I and all four dNTPs) and the third Eco47III site, leaving an insert of 1.3 kb. Each of the remaining three plasmids contains one of the repeats amplified from a full-length cDNA clone cm06e3 (22). The first repeat was amplified with the primers ECX1 and ECX2, the second was amplified with ECX3 and ECX4, the third was amplified with ECX5 and ECX6 (ECX1, CCCATATGACTCTTACCGTTGCTGCT; ECX2, CCCATATGTTATTATGTAGATTCTCCAGTTGAGCC; ECX3, CCCATGGGTAACTCCGAAACGGCTAACCC; ECX4, CCCATGGTTATTACGTATTTTGTCCAGTCGATCC; ECX5, CCCATGGAAGCGGAACCAGCTTTG; ECX6, CCCATGGTTATTAAAGGGTTATATACTGCCC).The PCR products were treated with T4 DNA polymerase and all four dNTPs and cloned into the SmaI site of pBluescript II SK−. The orientation of the cDNA clones is opposite to that of the corresponding genomic clone.
Sequencing.
Single-stranded DNA was prepared from each of the three cDNA clones (the noncoding strand was packaged in each case) and from the three genomic repeat clones (the sense strand being packaged in each case) with the helper phage M13K07 (23). Sequencing reactions were carried out according to the manufacturers instructions using a commercially available kit (Sequenase version 2; United States Biochemical) and 35S-labeled dATP. Either 1 or 10 pmol of oligonucleotide primer was used. For double-stranded sequencing, a Thermosequenase kit was used according to the manufacturers instructions with 33P-labeled dideoxynucleotide triphosphates (both from Amersham).
PCRs.
For plasmid templates, 100 ng of template DNA, linearized at the unique SacI site in the polylinker, was used in 50-μl reactions containing 10 mM Tris⋅HCl (pH 8.3 at 25°C), 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin, all four dNTPs (each at 200 μM), 50 pmol of each primer, and 2.5 units of AmpliTaq enzyme (Perkin–Elmer). The vector primers used were either T7 (5′-TAATACGACTCACTATAGGGC) or Reverse (5′-ACAGGAAACAGCTATGACCA). Reaction conditions (unless otherwise stated) were 1 cycle at 96°C for 3 min (during which Taq polymerase was added); then 30 cycles of (i) denaturation at 96°C for 5 sec, (ii) annealing at 44°C for 1 min, when primers containing some K were used, but otherwise annealing at 50°C for 5 sec, and (iii) extension at 72°C for 30 sec. A final cycle of 2 min at 72°C completed the reactions. For genomic DNA reactions, 100 ng of C. elegans genomic DNA was used as a template, and primer concentrations were increased to 200 pmol. After amplification, the PCR products were separated on 2% agarose gels containing ethidium bromide in 1× TAE buffer (23), and photographed with UV illumination. Propagation of C. elegans (strain N2) and preparation of genomic DNA were as described (24).
Southern Blotting.
The products amplified from genomic DNA were separated on a 3% agarose gel and Southern blotted by standard procedures (23). The blot was probed sequentially with three oligonucleotide probes labeled with 32P using T4 polynucleotide kinase and [γ-32P]ATP. Each probe was specific for one of the homologous repeats. Hybridization was performed at 42°C; washing was at 37°C in 6× SSC. The hybridization solution was 40 ml of 2× BSA, 800 μl of salmon sperm DNA (5 mg/ml; depurinated with acid and denatured with base before being neutralized), and 400 μl of heparin (120 mg/ml). The 2× BSA was made from 4 ml of 2× SSC, 2.5 ml of 1 M sodium phosphate (pH 7.0), 2 ml of 100 mM EDTA, 50 mg of sodium pyrophosphate, and 2 g of BSA, which was brought to 40 ml with water. The Southern blot was stripped between each hybridization by washing in 0.5% SDS at 75°C for 1 h and checked by overnight exposure. Exposure time for detecting hybridization was 2 h.
RESULTS
The bases P and K were incorporated into oligonucleotide duplexes and their melting temperatures were established in high salt (1 M; 6× SSC) and in low salt (0.1 M; PCR buffer). The former are shown in Table 1; the trend is the same in low salt buffer but the Tm values are depressed by approximately 10°C. From these measurements, it is clear that when substituted for T, P had no noticeable effect on the Tm but was mildly destabilizing when substituted for C. For K, there was a decrease in Tm when it was substituted for A (ΔTm = 3°C) that was more pronounced when it was substituted for G (ΔTm = 7°C). It is of interest that P⋅K base pairs too are quite stable, as can be seen by the Tm values and high stacking enthalpies.
Table 1.
Melting temperatures and stacking enthalpies of oligonucleotide duplexes containing dP and dK
| Sequences | Tm, °C | ΔH, kJ/mol |
|---|---|---|
| 5′-ACTTGGCCACCATTTTG | 72°C | −514 |
| 3′-TGAACCGGTGGTAAAAC | ||
| 5′-ACTTKGCCACCATTTTG | 65°C | −442 |
| 3′-TGAACCGGTGGTAAAAC | ||
| 5′-ACTTGGCCACCATPTTG | 72°C | −500 |
| 3′-TGAACCGGTGGTAAAAC | ||
| 5′-ACTTGGCCACCATTTTG | 69°C | −482 |
| 3′-TGAACCGGTGGTAKAAC | ||
| 5′-ACTTGGCCACCATTTTG | 67°C | −488 |
| 3′-TGAAPCGGTGGTAAAAC | ||
| 5′-ACTTKGCCACCATPTTG | 64°C | −502 |
| 3′-TGAACCGGTGGTAAAAC | ||
| 5′-ACTTGGCCACCATTTTG | 62°C | −403 |
| 3′-TGAAPCGGTGGTAKAAC | ||
| 5′-ACTTKGCCACCATTTTG | 65°C | −546 |
| 3′-TGAAPCGGTGGTAAAAC | ||
| 5′-ACTTGGCCACCATPTTG | 69°C | −435 |
| 3′-TGAACCGGTGGTAKAAC | ||
| 5′-ACTTKGCCACCATPTTG | 61°C | −433 |
| 3′-TGAAPCGGTGGTAKAAC |
To examine the specificity of the degenerate bases as template bases for Thermus aquaticus (Taq) DNA polymerase, two oligonucleotide templates, 61 or 64 residues long, and containing modifications at six positions, were synthesized (Fig. 2); one template contained six dP residues and the other contained six dK residues. The templates were designed to include modified bases both 5′ and 3′ to each of the four natural bases. For each template, 48 identical PCRs were assembled at 72°C to ensure multiple copying events, and the template was amplified by PCR with two flanking oligonucleotide primers; the PCR products were subsequently cloned and sequenced. The resulting sequences are tabulated in Fig. 2. In 96 copying events, the analogue P was copied as C and as T in a ratio of 1:1.5. The K base showed a pronounced bias toward behaving as A rather than G (ratio of 7:1).
Figure 2.
Template properties of the degenerate bases. An oligonucleotide template containing unnatural bases, represented by X, and the two primers used to amplify, by PCR, the template are shown at the top; two columns of sequences showing only the regions between the two primers are shown below. The top row of each column of sequences shows the relevant portion of the oligonucleotide template (underlined sequence), on the left, are the sequences of 16 clones copied from a template containing six P residues, and on the right, are those copied from a template containing six K residues.
Previously, we have shown that oligomers containing either P or K could be used to prime in PCR. We were interested to determine whether oligomers containing a mixture of P and K (designated M) and, therefore, equivalent to having all four natural bases (N) at designated positions could be used for PCR and sequencing and to compare the results with dI. As a model system in which to test primers, we chose the dUTPase gene of C. elegans, which is composed of three homologous but not identical repeats (unpublished data and Fig. 3A). Six plasmids were constructed; three contained each of the repeats subcloned separately from genomic DNA and the other three contained each of the repeats subcloned individually from a full-length cDNA clone cm06e3 (22). Each repeat encodes five motifs characteristic of dUTPase genes (25); the third and fifth motifs, respectively, encode the seven amino acid peptide sequences IDSDYRG and GGFGSTG.
Figure 3.
(A) Diagram of the full-length cDNA encoding the deoxyuridine triphosphatase of C. elegans (GenBank accession no. U96695). The positions of 10 bp derived from the 22-bp spliced leader (SL1), the poly(A) tail (represented by AAA), and the start (ATG) and stop (TAA) codons are shown. The positions of five motifs characteristic of dUTPase genes (25), each present three times, are represented by solid boxes. (B) The nucleotide sequences encoding the amino acid motifs 3 and 5 in each of the homologous repeats of the dUTPase gene, beneath the amino acids that they encode. Third codon positions are shown in boldface type. Beneath these nucleotide sequences are shown the two series of oligonucleotides containing base analogues that were examined for their ability to prime at these positions.
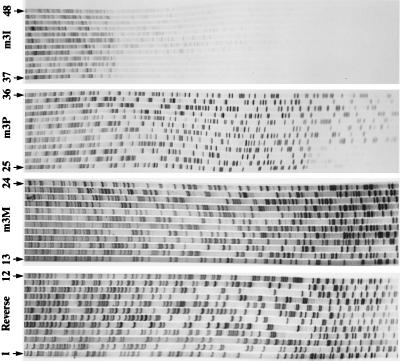
Two sets of oligonucleotides were synthesized (Fig. 3B); the motif 3 (m3X) series corresponds to the sense strand of the motif IDSDYRG, and the motif 5 (m5X) series of oligonucleotides corresponds to the antisense strand of the motif GGFGSTG. Each set of oligonucleotides contains four sequences that differ only in the modified base used in the six positions corresponding to the third position of each codon. Each primer was tested for its ability to prime a modified T7 DNA polymerase (Sequenase version II) in Sanger dideoxynucleotide sequencing reactions, on each of the three templates. The primer sequences and the priming sites are shown in Fig. 3. The K primers (m3K and m5K) produced only a single sequence ladder: m5K primed on template 2; in this case there are two mismatches (K versus a purine) 12 and 15 bases from the 3′ end of the primer. Failure to prime on the other templates is to be expected if K hybridizes only to T or C. Of the P primers, the motif 5 primer (m5P) failed to prime on templates 1 or 2 but primed on template 3 when there were mismatches (P versus a pyrimidine) 9 and 15 residues from the 3′ end of the primer (data not shown), but the motif 3 primer (m3P) primed efficiently on all three motif 3 templates (Fig. 4, lanes 25–36). This was expected because all priming sites for this oligonucleotide contain a pyrimidine at all the variable third codon positions (Fig. 3). The primers containing mixed P and K residues (m3M and m5M) worked approximately equally well on all six templates (Fig. 4, lanes 13–24 for m3M), but only when the concentration of primer used was 10 pmol per reaction, rather than 1 or 0.5 pmol usually used. The inosine oligomers primed on some of the templates but with variable efficiencies; the m3I primer produced only weak sequence ladders on all three templates (Fig. 4, lanes 37–48), whereas the m5I primer failed to sequence on template 3. Thus, in sequencing reactions the P/K primers proved to be more efficient than those containing inosine.
Figure 4.
Autoradiographs of sequencing reactions, using T7 DNA polymerase, on cDNA templates 1, 2, and 3 and each of four primers: a vector primer, Reverse (lanes 1–12), primer m3M (lanes 13–24), primer m3P (lanes 25–36), and primer m3I (lanes 37–48). Each reaction is run in the order TGCA; 10 pmol of each primer was used in the labeling reactions, except for Reverse, where 0.5 pmol was used.
Each oligonucleotide in both motif series was tested individually in a PCR with an unmodified vector primer that could prime at a site flanking the polylinker. Neither of the K primers, m3K and m5K, produced any products from any of the templates when used individually with the vector primer. Of particular note, the m5K primer failed to work despite priming in the sequencing reaction. The primer m3P amplified the expected product from all three templates when used in combination with a vector primer. The primer m5P primed only on template 3, mirroring the results in sequencing. The combination of the two primers, m3P and m5P, amplified only from the third template.
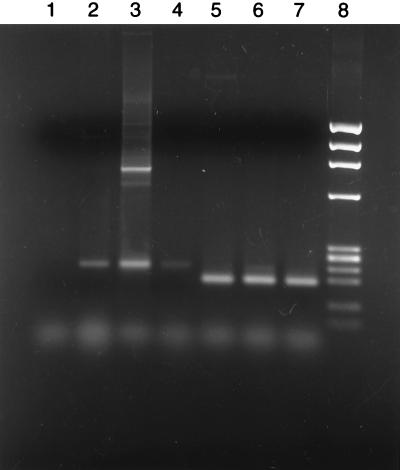
Unlike the sequencing experiments, each inosine primer amplified DNA in PCRs from all three templates with an efficiency sufficient to produce visible products on gel electrophoresis when used in combination with a vector primer, but the yields varied depending on the template and paralleled their efficiency in priming T7 DNA polymerase sequencing reactions. However, when used in combination, the two primers m3I and m5I produced PCR products from only two of the plasmid templates and only at high primer concentrations (200 pmol of each). Primers containing P/K mixtures of both the motif 3 and motif 5 series amplified the correct products from all three templates in reactions with the vector primer. Importantly, both P/K primers (m3M and m5M) worked in combination with each other, yielding approximately equal amounts of PCR product, whether 50 or 200 pmol of each primer was used (Fig. 5, lanes 5–7).
Figure 5.
Electrophoresis on a 2% agarose gel of PCR products amplified with 200 pmol of both m3M and m5M on various templates as follows: Lanes: 1, no template; 2, C. elegans genomic DNA; 3, genomic clone pdutR with a 30-sec extension at 72°C in each cycle; 4, genomic clone pdutR with no pause for extension at 72°C; 5, cDNA of repeat 1; 6, cDNA of repeat 2; 7, cDNA of repeat 3; 8, φX HaeIII markers.
As a more rigorous test of the P/K primer pair m3M and m5M, both a DNA clone, pdutR, containing a fragment of genomic DNA from C. elegans containing all three repeats, and genomic DNA itself were used as templates. Because the target products from each repeat were very similar in size, the PCR products were Southern blotted and probed with oligonucleotides specific for each repeat to determine which repeats had been amplified. The PCR products amplified from genomic DNA (Fig. 5, lane 2) were identical in length to the shortest products amplified from the genomic clone (Fig. 5, lanes 3 and 4). Due to the presence of an intron in each genomic repeat, the products from the cDNA clones are 45 bp shorter (Fig. 5, lanes 5–7). Longer PCR products amplified from the genomic clone (Fig. 5, lane 3) are amplicons starting in one repeat and extending into the neighboring repeat. These longer products were not present when the extension time in each cycle (at 72°C) was eliminated (Fig. 5, lane 4). Despite the presence of a complex mixture of nontarget sequences in genomic DNA, the three target sequences were amplified exclusively (Fig. 5, lane 2).
A Southern blot of the gel shown in Fig. 6 was hybridized sequentially with 32P-labeled probes specific for each repeat; the blot was stripped between each hybridization. This demonstrated that all three target sequences were amplified simultaneously, with approximately equal efficiency, from the genomic clone (Fig. 6, lanes 3 and 4) and from genomic DNA (Fig. 6, lane 2).
Figure 6.
Southern blotting of PCR products, shown in Fig. 5, with probes specific for each of the three amplified repeats. The sequence of the probes were as follows (5′ to 3′): Repeat 1 (probe 1), AGGTTAAGAGTTTGCCATCC; Repeat 2 (probe 2), CCGCAAGTGAATTGGAGAAT; Repeat 3 (probe 3), AAGTGGAGAGCCTTGAAGTC. Numbering of the lanes is as in Fig. 5.
DISCUSSION
A universal base that could substitute with equal efficiency for any of the four natural bases in DNA would be of great utility in recombinant DNA experiments. No compound has been found that can mimic all the bases in all circumstances. Hypoxanthine, the base that has been used most widely, forms stable duplexes when incorporated into oligonucleotides and has been used both in hybridization probes for screening libraries and in PCR primers, when, for example, the precise target sequence for the primer is not known (26, 27). However, as a template residue, dI behaves exclusively as deoxyguanosine (28).
It is necessary to distinguish a number of separate specificities for any base, namely, as a nucleoside triphosphate substrate, in a template, in a primer and, more generally, in hybridization. The four natural bases, of course, perform with consistent unique specificities in all these functions and it is only when nonnatural DNA bases are used that we can detect differences between the properties of a base in a primer and in a template. Many candidates for universal bases have been tested in hybridization but to a lesser extent for the other functions (refs. 26 and 29 and references therein). In the absence of a consistently successful universal base, we have examined the combination of two bases (Fig. 1) as a universal base equivalent. Of the two bases, P can substitute for either of the two natural pyrimidine bases, T or C, whereas K can replace either of the two natural purine bases, G or A.
It is worth noting that even if a very efficient universal base were available there are still many occasions in which a degenerate pyrimidine or purine base would be used in preference. As one example, the design of primers based only on knowledge of an amino acid sequence is complicated by the degeneracy of the genetic code. Only two amino acids, Met and Trp, have unique codons; but a further nine amino acids (Cys, Asp, Glu, Phe, His, Lys, Asn, Gln, and Tyr) require only a degenerate purine or pyrimidine in the third position of their codons to have “unique” codons. The remainder, including stop codons, are coded for without ambiguity with both a purine and a pyrimidine in the third position. In a related approach, 5-fluorouracil has been proposed as a pyrimidine analogue (30). The use of degenerate pyrimidine/purine bases at the first and second codon positions allows many amino acids to be encoded by oligonucleotides of low complexity, which is advantageous in, for example, site-directed mutagenesis experiments.
Both bases that we have examined have two major tautomeric states. Incorporation of these bases into oligonucleotides caused some lowering of the Tm for some duplexes. Although only a relatively small number of duplexes were examined, some trends are apparent. Thus when P is substituted for T, there is a negligible effect on the melting temperature, but there is a slight destabilization when substituted for C. Incorporation of K is generally more destabilizing, particularly when replacing G. The superior stability of P as T, rather than as C, and of K as A, rather than G, mirrors the behavior of these modified bases in templates for Taq polymerase.
Both P and K function as template bases for DNA synthesis by Taq DNA polymerase and are copied with the expected specificity: P is copied exclusively as a pyrimidine, and K exclusively as a purine; i.e., no transversion mutations were observed. P shows a slight preference for being copied as T under PCR conditions (T/C ratio = 3:2), whereas the preference of K for being copied as A rather than G was more pronounced (A/G ratio = 7:1), particularly in some positions of the template. Nevertheless, primers containing K paired with C in the sequence to be amplified clearly function efficiently. Earlier work suggested that Taq polymerase in a primer extension assay incorporated only A opposite P and T with only a small amount of C opposite K (31). Our results derive, in each case, from 96 copying events where the analogue has a variety of sequence contexts.
Oligonucleotides containing six residues replaced by a mixture of P and K (M) primed DNA synthesis efficiently, both in standard sequencing reactions using T7 DNA polymerase and in PCRs using Taq polymerase. Three important conditions had to be met for these reactions to work. First, the concentration of the primers had to be raised to 10 pmol in sequencing reactions and to 50 or 200 pmol in PCRs, suggesting that only a small proportion of the primers were being used. In fact, if complete specificity of the primers is assumed, only one primer molecule in each 64 (26 primers) should prime; this would imply that each primer was at an effective concentration of 3 pmol per PCR.
Secondly, all primers containing K needed annealing times of up to 1 min, as opposed to only 5 sec for primers containing six P or six I residues. We attribute this to the methoxyl group of the K base existing in the syn configuration and the consequent inability to form K⋅T or K⋅C pairs of reasonable structures without prior configurational inversion to the anti form. The evidence from NMR hybridization studies of oligomers giving rise to mo4C⋅G base pairs is that a slow step is involved in the hybridization. This is clearly attributable to such an inversion prior to stabilization of the duplex (32, 33). Where several such residues are present, the effect on the hybridization forward rate should be considerable. We assume that these considerations can be extended to K. Thirdly, the optimal annealing temperature of 44°C for primers containing K was noticeably depressed from the temperature of 50°C at which primers not containing K worked. This is consistent with the lower Tm values for duplexes containing K compared with those containing P. Within the relatively small number of assays done, the equimolar mixture of P and K was superior to dI, the most commonly used universal nucleoside analogue. Nevertheless, clearly some modifications can be made to improve the performance of the degenerate bases, now that the principle has been established. Of these, the most obvious is the synthesis of a purine analogue in which, as with P, the methoxyl residue is constrained in the anti conformation (for example, by forming a third ring) and work to this end is in progress.
The polymerase recognition of P and K as template bases is complemented by the analysis of the corresponding nucleoside 5′-triphosphates as mutagenic substrates. In PCR amplifications, dPTP is an excellent mutagenic substrate, giving a ratio of the transitions T → C to C → T of about 5:1 (34). This value results from base recognition both as a triphosphate substrate and as a template base derived from incorporation of the triphosphate. In primer extension experiments, the relative efficiencies of insertion opposite A and G are about 10:1 (34). This is to be compared with our observed A/G relative insertion rates opposite template P of 1.5:1. We may draw the conclusion that there is a strong qualitative agreement between these sets of results and the tautomeric constant of P, which is assumed to be closely similar to that of mo4C at 0.03 (8). Beyond this there appears to be a leveling effect consequent on base pairing because only the imino tautomer of dP is observed in NMR experiments. In this connection, evidence for a shift in the tautomeric ratio (of mo6A) by complementary bases in nonaqueous solution has been given (35).
Similar considerations apply to dK and dKTP; the tautomeric ratio of the nucleoside (9:1 by 1H NMR) and the mutagenic spectrum of the triphosphate are described elsewhere (36). It is evident that where the free energy difference between the tautomers is small, other features, for instance in the enviroment of the polymerase active site, may override the more obvious connection between the tautomeric and the insertion ratios. Further studies of analogues of the kind discussed herein in templates and as substrates are in progress and may help to elucidate mechanistic details of polymerase base recognition.
Acknowledgments
We thank Jan Fogg and Richard Grenfell for oligonucleotide synthesis. We are grateful to Dr. Alan Coulson who supplied the cosmid clone K07A1 and the cDNA clone cm06e3 and to Dr. O. Sundin who provided the recipe for the oligonucleotide hybridization solution. Finally, we thank the Medical Research Council for financial support.
ABBREVIATIONS
- P
6H,8H-3,4-dihydropyrimido[4,5-c][1,2]oxazin-7-one
- K
N6-methoxy-2,6-diaminopurine
- mo
methoxy
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U96695).
References
- 1.Ohtsuka E, Matsuki S, Ikehara M, Takahashi Y, Matsubara K. J Biol Chem. 1985;260:2605–2608. [PubMed] [Google Scholar]
- 2.Takahashi Y, Kato K, Hayashizaki Y, Wakabayashi T, Ohtsuka E, Matsuki S, Ikehara M, Matsubara K. Proc Natl Acad Sci USA. 1985;82:1931–1935. doi: 10.1073/pnas.82.7.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawase Y, Iwai S, Inoue H, Miura K, Ohtsuka E. Nucleic Acids Res. 1986;14:7727–7736. doi: 10.1093/nar/14.19.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin F H, Castro M M, Aboul-ela F, Tinoco I. Nucleic Acids Res. 1985;13:8927–8938. doi: 10.1093/nar/13.24.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aboul-Ela F, Koh D, Tinoco I, Martin F H. Nucleic Acids Res. 1985;13:4811–4827. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drake J W. The Molecular Basis of Mutation. San Francisco: Holden–Day; 1970. [Google Scholar]
- 7.Anand N N, Brown D M, Salisbury S A. Nucleic Acids Res. 1987;15:8167–8176. doi: 10.1093/nar/15.20.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown D M, Hewlins M J E, Schell P. J Chem Soc. 1968;C:1925–1929. doi: 10.1039/j39680001925. [DOI] [PubMed] [Google Scholar]
- 9.Kwiatkowski, J. S. & Pullman, B. (1975) in Adv. Heterocycl. Chem. 18, pp.199–335.
- 10.Shugar D, Huber C P, Birnbaum G I. Biochim Biophys Acta. 1976;447:274–284. doi: 10.1016/0005-2787(76)90050-2. [DOI] [PubMed] [Google Scholar]
- 11.Morozov Y V, Savin F A, Chekhov V O, Budowsky E I, Yakovlev D Y. J Photochem. 1982;20:229–252. [Google Scholar]
- 12.Kong Thoo Lin P, Brown D M. Nucleic Acids Res. 1989;17:10373–10383. doi: 10.1093/nar/17.24.10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Negishi K, Williams D M, Inoue Y, Imura T, Katahira M, Kanazawa H, Uesugi S, Hayatsu H, Brown D M. Nucleic Acids Symp Ser. 1995;34:231–232. [PubMed] [Google Scholar]
- 14.Negishi K, Williams D M, Inoue Y, Moriyama K, Brown D M, Hayatsu H. Nucleic Acids Res. 1997;25:1548–1552. doi: 10.1093/nar/25.8.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujii T, Saito T, Itaya T, Kizu K, Kumazawa Y, Nakajima S. Chem Pharm Bull. 1987;35:4482–4493. [Google Scholar]
- 16.Brown D M, Kong Thoo Lin P. Carbohydr Res. 1991;216:129–139. doi: 10.1016/0008-6215(92)84156-m. [DOI] [PubMed] [Google Scholar]
- 17.Nishio H, Ono A, Matsuda A, Ueda T. Nucleic Acids Res. 1992;20:777–782. doi: 10.1093/nar/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong Thoo Lin P, Brown D M. Nucleic Acids Res. 1992;20:5149–5152. doi: 10.1093/nar/20.19.5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gralla J, Crothers D M. J Mol Biol. 1973;78:301–319. doi: 10.1016/0022-2836(73)90118-6. [DOI] [PubMed] [Google Scholar]
- 20.Marchuk D, Drumm M, Saulino A, Collins F S. Nucleic Acids Res. 1991;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coulson A, Sulston J, Brenner S, Karn J. Proc Natl Acad Sci USA. 1986;83:7821–7825. doi: 10.1073/pnas.83.20.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waterston R, Martin C, Craxton M, Huynh C, Coulson A, Hillier L, Durbin R, Green P, Shownkeen R, Halloran N, et al. Nat Genet. 1992;1:114–123. doi: 10.1038/ng0592-114. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.Sulston J, Hodgkin J. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 587–606. [Google Scholar]
- 25.McGeoch D J. Nucleic Acids Res. 1990;18:4105–4110. doi: 10.1093/nar/18.14.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Aerschot A, Peeters B, Van Derhaeghe H. Nucleosides and Nucleotides. 1987;6:437–439. [Google Scholar]
- 27.Liu H, Nichols R. Biotechniques. 1994;16:24–26. [PubMed] [Google Scholar]
- 28.Kamiya H, Sakaguchi T, Murata N, Fujimuro M, Miura H, Ishikawa K, Shimizu M, Inoue H, Nishimura S, Matsukage A, et al. Chem Pharm Bull. 1992;40:2792–2795. doi: 10.1248/cpb.40.2792. [DOI] [PubMed] [Google Scholar]
- 29.Bergstrom D E, Zhang P, Toma P H, Andrews P C, Nichols R. J Am Chem Soc. 1995;117:1201–1209. [Google Scholar]
- 30.Habener J F, Vo C D, Le D B, Gryan G P, Ercolani L, Wang A H-J. Proc Natl Acad Sci USA. 1988;85:1735–1739. doi: 10.1073/pnas.85.6.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamiya H, Murata-Kamiya N, Kong Thoo Lin P, Brown D M, Ohtsuka E. Nucleosides and Nucleotides. 1994;13:1483–1492. [Google Scholar]
- 32.Stone M J, Nedderman A N R, Williams D H, Kong Thoo Lin P, Brown D M. J Mol Biol. 1991;222:711–723. doi: 10.1016/0022-2836(91)90507-3. [DOI] [PubMed] [Google Scholar]
- 33.Gdaniec Z, Bann B, Sowers L C, Fazakerley G V. Eur J Biochem. 1996;242:271–279. doi: 10.1111/j.1432-1033.1996.0271r.x. [DOI] [PubMed] [Google Scholar]
- 34.Zaccolo M, Williams D M, Brown D M, Gherardi E. J Mol Biol. 1996;255:589–603. doi: 10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- 35.Kierdaszuk B, Stolarski R, Shugar D. FEBS Lett. 1983;158:128–130. doi: 10.1111/j.1432-1033.1983.tb07186.x. [DOI] [PubMed] [Google Scholar]
- 36.Hill F, Williams D M, Loakes D, Brown D M. Nucleic Acids Res. 1998;26:1144–1149. doi: 10.1093/nar/26.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]