Abstract
Initiation factor eIF4G is an essential protein required for initiation of mRNA translation via the 5′ cap-dependent pathway. It interacts with eIF4E (the mRNA 5′ cap-binding protein) and serves as an anchor for the assembly of further initiation factors. With treatment of Saccharomyces cerevisiae with rapamycin or with entry of cells into the diauxic phase, eIF4G is rapidly degraded, whereas initiation factors eIF4E and eIF4A remain stable. We propose that nutritional deprivation or interruption of the TOR signal transduction pathway induces eIF4G degradation.
Keywords: cell cycle, starvation, protein degradation
The availability of nutrients plays a central role in growth regulation of the yeast Saccharomyces cerevisiae. Starvation of cells for carbon or nitrogen, a common condition in natural habitats (1), leads to lowering of transcription and translation of most genes and cessation of growth in the G1 phase of the cell cycle (2). As an exception, expression of genes that allow entry into the stationary or G0 phase is enhanced (for reviews, see refs. 1 and 3). Entry into the G0 phase in response to starvation is accompanied by and depends on reduced expression of the G1 phase cyclins. Cells that express stable cyclin2 (Cln2-1p) enter S phase despite nutritional deprivation (4, 5) and lose viability. G0 phase cells are able to survive for long periods of time under conditions of nutritional deprivation and to resume growth when nutrients become available again (1, 3).
A response similar to starvation is triggered by treatment of yeast cells with the immunosuppressant rapamycin (6, 7). Rapamycin interacts with the FK506-binding protein (FKBP; a proline rotamase) (6), and the resulting complex binds to and inhibits the proteins Tor1p and Tor2p (6, 8), two phosphatidylinositol kinase homologs. This results in G1 phase arrest, inhibition of translation initiation, and acquirement of a G0 phase-like phenotype reminiscent of nutrient-starved cells (7). G1 arrest in rapamycin-treated cells is probably caused by reduced CLN3 (cyclin3) mRNA translation. Cln3p induces the expression of other G1 phase cyclins that promote G1 to S phase transition (for reviews, see refs. 9 and 10). A gene construct carrying the promoter and the 5′ untranslated region of UBI4 (the polyubiquitin-encoding gene) fused to the CLN3 ORF renders translation of CLN3 mRNA less dependent on initiation factor eIF4E (eukaryotic initiation factor 4E), thereby suppressing the rapamycin-induced G1 phase arrest and conferring starvation sensitivity to the cells (7). This result indicates that Tor1p and Tor2p control the activity of eIF4E or eIF4E-associated protein(s).
A similar rapamycin-sensitive transduction pathway signaling through a Tor-like protein (mTOR/FRAP/RAFT1) (11, 12) and regulating the activity of translation factors has been identified in mammalian cells (for a review, see ref. 13). Inhibition of signaling through this pathway blocks the p70(S6)/p85(S6) kinase, inhibits the phosphorylation of ribosomal protein S6 and of eIF4E binding proteins (4E-BPs), and promotes the association of 4E-BPs with eIF4E (14–18). Dephosphorylated 4E-BPs inhibit the interaction of eIF4E with eIF4G through competition for the same binding site on eIF4E (19, 20). Blocking of the signaling pathway with rapamycin affects mostly translation of mRNAs containing a polypyrimidine tract in their 5′ untranslated region; among those, mRNAs encoding ribosomal proteins, translation elongation factors (14, 21), and proteins involved in growth control (22).
eIF4G, which in Saccharomyces cerevisiae is encoded by the genes TIF4631 and TIF4632 (23), is an essential initiation factor required for mRNA translation via the 5′ cap structure (24). Mammalian eIF4G serves as an anchor for the binding of further initiation factors such as eIF3 and eIF4A (25) and the 40S ribosomal subunit (for a review, see ref. 26). Here, we report that initiation factor eIF4G is rapidly degraded in S. cerevisiae on entry in the diauxic growth phase (transition from fermentative to oxidative metabolism) or on treatment with rapamycin.
MATERIALS AND METHODS
Yeast Strains.
Strain CBY12 was obtained by crossing a spontaneous ura3− mutant of strain CBY2.1 (transformed with pCB2) with strain CBY1.2. Haploid spores carrying the desired auxotrophic markers were selected by replica-plating on minimal medium plates (0.67% yeast nitrogen base) containing 2% galactose. The plasmid pCB2 contains the entire TIF4631 cDNA (EcoRI fragment) (23) in the vector p301-URA3 under the GAL1/10 promoter. For further details on the characteristics of yeast strains and relevant plasmids used, see Table 1.
Table 1.
Relevant yeast strains and plasmids
| Strain | Genotype | Source/ref. |
|---|---|---|
| CWO4 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3 | 45 |
| ASZ1 | Isogenic to CWO4, MATa | 45 |
| CBY1.1 | Isogenic to CWO4, MATα tif4631::LEU2 | This study, ref. 23 |
| CBY1.2 | Isogenic to CWO4, MATa tif4631::LEU2 | This study, ref. 23 |
| CBY2.1 | Isogenic to CWO4, MATα tif4632::URA3 | This study, ref. 23 |
| CBY12 | CWO4 tif4631::LEU2 tif4632::ura3 〈pCB2〉 | This study, ref. 23 |
| PDY4 | cdc63-1 trp1 ura3 | This study |
| Plasmid | ||
| pPW2 | 〈tor1-1; URA3〉 | 31 |
| pCB2 | 〈p301-TIF4631cDNA; URA3〉 | This study, ref. 23 |
Growth Conditions.
Unless indicated otherwise, cells were grown in YPD (1% yeast extract, 2% peptone, 2% glucose) or YPGal (1% yeast extract, 2% peptone, 2% galactose) at 30°C.
Manipulation of Yeast Cells.
Yeast cells were transformed by using the lithium acetate method (27).
DNA Manipulations.
DNA manipulations were carried out according to Sambrook et al. (28) by using Escherichia coli strain XL2B (Stratagene) for subclonings and maintenance of plasmid DNA.
Drug Treatment.
Final concentrations of drugs added to exponentially growing cell cultures were as follows: 0.2 μg/ml rapamycin (dissolved in vehicle: 90% ethanol/10% Tween-20), 10 μg/ml cycloheximide (Sigma), 2 μM α-factor (Sigma), 0.1 M hydroxyurea (Fluka), 25 μg/ml nocodazole (Fluka; dissolved in dimethyl sulfoxide). Samples (10–20 ml cell cultures) were harvested at different time points after addition of drugs, and cells were collected by centrifugation at 5,000 × g for 3 min, washed once, and immediately processed for protein analysis (or frozen at −80°C for later processing).
SDS/PAGE and Western Blot Analysis.
Cell pellets were resuspended in 50–100 μl of buffer A (20 mM Tris⋅HCl, pH 7.5/100 mM KCl/2 mM MgCl2/0.1 mM EDTA/7 mM 2-mercaptoethanol) and mixed with 0.2–0.4 g glass beads rinsed with buffer A. Cells were lysed by vortex mixing eight times for 20 s. After centrifugation at 12,000 × g for 5–10 min, the ribosome content of the supernatants was determined by measuring the absorption at 260 nm. Unless indicated otherwise, equal amounts of proteins were loaded on 12.5% SDS/PAGE and blotted onto nitrocellulose for 45 min at 60 V in a Mini Trans Blot Cell (Bio-Rad). Blots were saturated with 2.5% BSA in TBS (10 mM Tris⋅HCl, pH 7.5/150 mM NaCl) for 5 min at room temperature and incubated overnight with rat polyclonal antibodies (1:1,000 dilutions in TBS containing 0.5% BSA). After washing with TBS blots were decorated for 1 h with peroxidase-conjugated rabbit anti-rat Igs (Dako) and stained with 0.018% chloronaphthol and 0.006% H2O2 in TBS. Equal protein loading was verified by Coomassie blue staining. Amounts corresponding to 0.8 A260 were loaded for the detection of eIF4G1, 1.2 A260 for eIF4G2, 0.2 A260 for eIF4E, and 0.08 A260 for eIF4A.
Flow Cytometry.
Cell culture samples (300 μl culture) were kept in 70% ethanol at 4°C overnight or longer. Washed cells (dH2O) were resuspended in 500 μl of 50 mM sodium citrate (pH 7.4) containing 0.25 μg/ml RNase A, sonicated for 3 min, and incubated for 1 h at 37°C. DNA was stained by addition of 500 μl of citrate buffer containing 16 μg/ml propidium iodide. For each time point, 10,000 events were analyzed for DNA content by using a Becton Dickinson FACScan machine, and data were processed by using cell quest software (Cell Quest, Lincoln Park, NJ).
RNA Preparation and Northern Blot Analysis.
All solutions were prepared by using diethylpyrocarbonate (DEPC)-treated ddH2O. Cell pellets (10 ml cultures) were resuspended in 500 μl TEL buffer (10 mM Tris⋅HCl, pH 7.5/2 mM EDTA/150 mM LiCl) plus 1% lithium dodecyl sulfate. Cell extracts were prepared by vortex mixing with 0.5 g glass beads in the presence of 0.5 ml phenol/chloroform (1:1) and centrifuged at 12,000 × g for 10 min. Phenolic phases were reextracted with 200 μl TEL buffer, and the aqueous phases were reextracted twice with phenol/chloroform (1:1). RNA was precipitated from the combined aqueous phases with 2.5 vol of absolute ethanol in the presence of 0.1 M KAc. RNA pellets were washed once with 70% ethanol, air dried, and resuspended in 10 μl (DEPC-treated) ddH2O. The RNA yield was determined by measuring the optical density at 260 nm (1 A260 = 40 μg total RNA). Equal amounts of RNA (15 μg) were dissolved in FR buffer (100 mM Mops/50 mM sodium acetate, pH 7.0/5 mM EDTA), 6% formaldehyde, and 50% formamide, incubated for 15 min at 65°C, and loaded directly on a 0.7% agarose gel containing FR buffer and 6% formaldehyde. Gel electrophoresis was carried out for 3–4 h at 30 V. The agarose gels were blotted on Zeta-probe membrane (Schleicher & Schüll) overnight. Blotted RNA was fixed for 2 min by UV treatment and baking of the membrane for 1 h at 80°C. Prehybridization of the membranes was done for 1 h at 42°C in 50% formamide, 5× SSC (750 mM sodium chloride/75 mM sodium citrate, pH 7.5), 5× Denhardt’s reagent [0.5 g of Ficoll (type 400, Pharmacia)/0.5 g polyvinylpyrrolidone/0.5 g of BSA (fraction V; Sigma)/0.5% SDS]. The blots were probed with 32P labeled TIF4631 cDNA (23) by incubating overnight at 42°C. They were extensively washed with 5× SSC/0.1% SDS, 2× SSC/0.1% SDS, and 0.5× SSC/0.1% SDS at room temperature, and 0.2× SSC/0.1% SDS at 42°C. They were then exposed to x-ray films.
RESULTS
Rapamycin Treatment Causes the Disappearance of eIF4G.
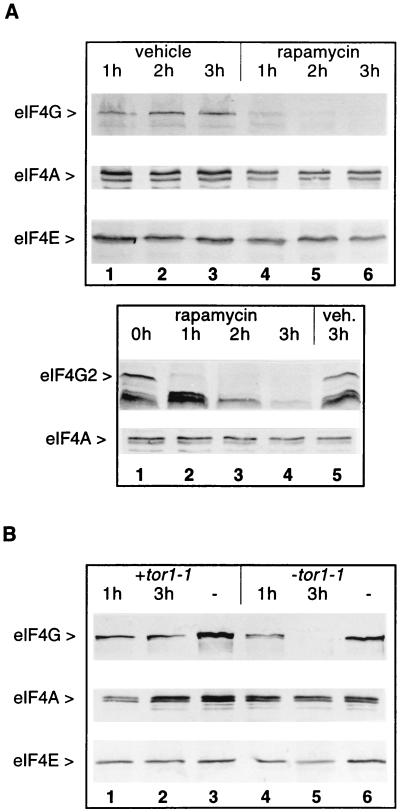
Treatment of S. cerevisiae with the immunosuppressant rapamycin induces inhibition of translation at the level of initiation, arrest of growth in the G1 phase of the cell cycle, and expression of G0 phase-specific genes (6, 7). We asked whether factors involved in 5′ cap-dependent translation such as eIF4E and eIF4G were affected by rapamycin treatment. Exponentially growing wild-type cells (strain CWO4; Table 1) were treated either with rapamycin or drug vehicle as a control. Aliquots of treated cultures were harvested at hourly intervals and analyzed by Western blotting (Fig. 1A). Between 1 and 2 h after rapamycin addition, eIF4G levels were reduced and were hardly detectable after 3 h of drug treatment (Fig. 1A, first row, lanes 4–6). In some experiments, we detected the degradation of most eIF4G only after 3 h of rapamycin treatment (Fig. 1B). In contrast, initiation factors eIF4A (Fig. 1A, second row) and eIF4E (Fig. 1A, third row) as well as eIF4G from cells incubated with vehicle (Fig. 1A, first row, lanes 1–3) were not affected by the treatment. The antibody used to detect eIF4G [rat polyclonal antibody against a glutathione S-transferase (GST)-eIF4G1/160–492 (aa 160–492) fusion protein (29)] recognizes eIF4G1 (encoded by TIF4631) and, with somewhat lower affinity, also eIF4G2 (encoded by TIF4632). Fig. 1A Lower shows that eIF4G also disappears in a strain that only expresses eIF4G2 (strain CBY1.1; Table 1) upon treatment of cells with rapamycin. The two lower bands are probably degradation products of eIF4G2. They accumulate after 1 h of rapamycin treatment (lane 2) and then disappear gradually (lanes 3 and 4). Identical results were obtained when cells were directly boiled in SDS sample buffer. The disappearance of intact eIF4G was also observed when Western blots were decorated with other antibodies: rat polyclonal anti-eIF4F antibody (30), rat polyclonal antibody against GST-eIF4G1/542–883 (aa 542–883), or anti-HA antibody for a strain carrying a construct encoding HA-tagged Tif4632p. We conclude that levels of both eIF4G1 and eIF4G2 are dramatically reduced after 1–3 h of rapamycin treatment, at a time when levels of other initiation factors such as eIF4E and eIF4A are almost unchanged.
Figure 1.
Rapamycin causes the disappearance of eIF4G. (A) Exponentially growing cells from strains CWO4 (Upper) or CBY1.1 (Lower) were treated with rapamycin or vehicle. Aliquots (20 ml) were harvested at the times indicated after addition of the drug (or vehicle). Cell extracts were fractionated by SDS/PAGE, blotted onto nitrocellulose, and decorated with rat polyclonal antibodies against eIF4G (Upper and Lower, first row), eIF4A (Upper and Lower, second row), and eIF4E (Upper, third row). (B) Cell extracts from strain CWO4 transformed with the plasmid pPW2 carrying the dominant tor1-1 allele (lanes 1–3) or a vector without tor1-1 (lanes 4–6) were treated as described in A with rapamycin (lanes 1, 2, 4 and 5) or vehicle (lanes 3 and 6) and analyzed on Western blots for eIF4G (first row), eIF4A (second row), or eIF4E content (third row).
The Disappearance of eIF4G Is Regulated by the TOR Signaling Pathway.
Rapamycin (complexed with FKBP) exerts its effects on the cell cycle through interaction with Tor1p and Tor2p. Mutation of a serine residue in the lipid-kinase domain of TOR1 or TOR2 abolishes binding of the FKBP–rapamycin complex and confers rapamycin resistance to the cells (31, 32). To test whether the disappearance of eIF4G was caused by blockage of the TOR signal transduction pathway, we transformed wild-type yeast cells with the tor1-1 allele (plasmid pPW2; Table 1), which confers partial rapamycin resistance, and analyzed eIF4G levels by Western blotting after treatment of the cells with rapamycin (Fig. 1B). In this strain, rapamycin treatment did not lead to the disappearance of eIF4G (first row, lanes 1 and 2). The levels of eIF4A (second row) and eIF4E (third row) also remained unchanged. Interestingly, the amount of eIF4G was lower in rapamycin-treated cells than in vehicle-treated cells (compare lanes 1 and 2 with lane 3). This may be due to the fact that the tor1-1 allele confers only partial resistance to rapamycin (6). In a control experiment, the same yeast strain was transformed with a plasmid carrying the same auxotrophic marker as pPW2 but without tor1-1. In this case rapamycin treatment led to the disappearance of eIF4G (Fig. 1B, lanes 4 and 5). We conclude from these data that the disappearance of eIF4G in rapamycin-treated cells is caused by the blockage of the TOR signal transduction pathway.
The Disappearance of eIF4G Is Caused by Degradation.
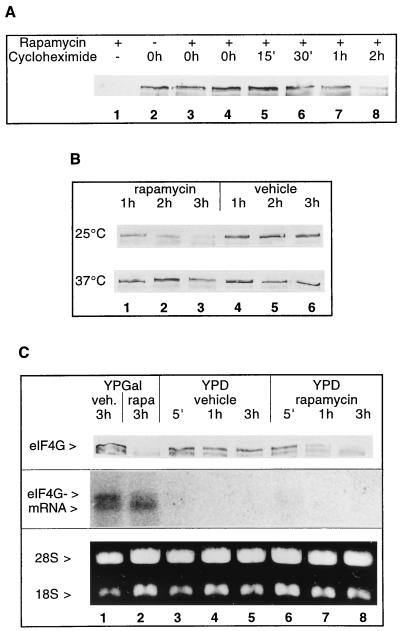
To determine whether the disappearance of eIF4G after rapamycin treatment was caused by enhanced degradation or reduced synthesis of the protein, we attempted to measure its half-life. Because several attempts to label eIF4G with [35S]methionine were not successful, we began to study eIF4G in the presence or absence of rapamycin under conditions where de novo protein synthesis is blocked. Three hours after addition of the translation elongation inhibitor cycloheximide, the eIF4G level was not affected (Fig. 2A, lane 2). Simultaneous treatment of yeast cells with rapamycin and cycloheximide prevented rapamycin-mediated eIF4G degradation (Fig. 2A; compare lane 1 with lanes 3 and 4). When cycloheximide was added 15 min to 1 h (but not 2 h) after the addition of rapamycin, it inhibited degradation of eIF4G (Fig. 2A, lanes 5–8).
Figure 2.
Stability of eIF4G. (A) Effect of rapamycin treatment on eIF4G (strain CWO4) when de novo protein synthesis is blocked with cycloheximide. Rapamycin was added to exponentially growing cells at time point 0 h, except for lane 2 (no rapamycin added) and lane 3 where it was added 15 min after cycloheximide addition. The time points of cycloheximide addition are indicated. The total duration of incubation in the presence of the drugs was 3 h. Shown are Western blots from extracts decorated for eIF4G. (B) Effect of rapamycin (lanes 1–3) or vehicle (lanes 4–6) on eIF4G stability in the mutant strain PDY4 (cdc63-1) grown at 25°C (first row) and after shift from 25°C to 37°C (second row). Time points after addition of the drug (or vehicle) are indicated. Shown are Western blots decorated for eIF4G. (C) Exponentially growing cells in YPGal (strain CBY12) were shifted to fresh medium containing YPGal (lanes 1 and 2) or YPD (lanes 3–8) and incubated with rapamycin or vehicle for the times indicated. First row shows Western blot decorated for eIF4G. Equal volumes of cell cultures were loaded. Second row shows Northern blot of total RNA isolated from cells harvested at the time points indicated and probed with a cDNA corresponding to the whole TIF4631 ORF (23). Third row shows ethidium bromide staining of 28S and 18S rRNAs used for the Northern blot analysis.
In a further experiment protein synthesis was blocked by incubating PDY4 cells carrying the temperature sensitive cdc63-1 allele [CDC63 encodes Prt1p, a subunit of translation initiation factor eIF3 (33–35)] at the nonpermissive temperature (37°C). After 3 h at 37°C, eIF4G was still present (Fig. 2B, second row, lanes 4–6). Although rapamycin (Fig. 2B, first row, lanes 1–3) but not vehicle (Fig. 2B, first row, lanes 4–6) induced eIF4G degradation at 25°C, incubation of cdc63-1 cells at 37°C led to stabilization of eIF4G in the presence of rapamycin (Fig. 2B, second row, lanes 1–3). Taken together the data in Fig. 2 A and B suggest that eIF4G is a stable protein and that the rapamycin-mediated destabilization requires ongoing protein synthesis.
To verify that eIF4G is an intrinsically stable protein we constructed a conditionally lethal strain in which eIF4G is expressed from TIF4631 (as its only eIF4G source) under the control of the galactose-regulatable GAL1/10 promoter (strain CBY12; Table 1). Expression of eIF4G is shut off when cells are transferred from galactose- to glucose-containing medium. Through dilution with fresh medium the cells were kept at low density. Cells at different time points after transfer to fresh glucose-containing medium (or to galactose-containing medium as a control) and addition of rapamycin or vehicle were analyzed for eIF4G protein and eIF4G mRNA content (Fig. 2C). Already 5 min after shift to glucose, hardly any eIF4G1 mRNA could be detected (Fig. 2C, second row, lanes 3–5). Despite the very quick loss of eIF4G mRNA, eIF4G1 protein levels did not decrease significantly (Fig. 2C, first row, lanes 3–5). For the Western blot, equal volumes of cell cultures were loaded (instead of equal A260 units) to compensate for residual growth of the cells during the first hours after the shift to glucose-containing medium despite the shut off of TIF4631 gene expression. Rapamycin treatment of cells maintained in galactose had little effect on eIF4G1 mRNA decay (Fig. 2C, second row, lane 2) but led to eIF4G protein degradation (Fig. 2C, first row, lane 2). In the cells shifted to glucose-containing medium and additionally treated with rapamycin, eIF4G mRNA faded as expected (Fig. 2C, second row, lanes 6–8) and the protein levels decreased rapidly (Fig. 2C, first row, lanes 6–8). It is somewhat difficult to estimate the half-life of eIF4G from these experiments. Nevertheless, in absence of rapamycin, the protein level was only slightly reduced 3 h after the disappearance of eIF4G mRNA (Fig. 2C, first and second rows, lane 5). In contrast, more than 50% of the protein was degraded after 1 h of rapamycin treatment (Fig. 2C, lane 7), suggesting a half-life shorter than 1 h. These results show that in exponentially growing cells eIF4G is a stable protein and strongly suggest that the disappearance of eIF4G is due to its accelerated degradation upon rapamycin treatment.
Degradation of eIF4G at the Diauxic Shift.
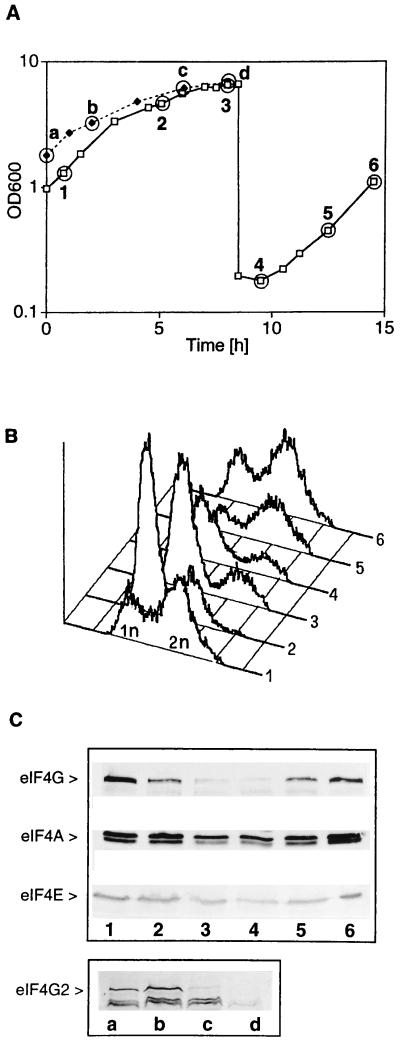
Because rapamycin treatment of yeast cells simulates nutritional deprivation (7) we looked for eIF4G degradation under starvation conditions. Fig. 3A depicts growth curves of wild-type cells (strain CWO4, time points 1–6) and cells deleted for TIF4631 (strain CBY1.1, time points a–d) grown in YPD to high density (a stage at which most of the glucose is metabolized) and then diluted back into fresh medium to replenish glucose. When cells reached high density and slowed down growth in the diauxic phase (Fig. 3A, time points 2 and 3, and c and d) most of them had a 1n DNA content (1n referring to the DNA content of a haploid cell before replication) (Fig. 3B, graphs 2 and 3) and lost eIF4G (Fig. 3C, Upper, lane 3) and eIFG2 (Fig. 3C, Lower, lanes c and d). Under these conditions translation initiation factors eIF4A and eIF4E still persisted (Fig. 3C, Upper, second and third rows). When cells were kept for longer times at high density (e.g., 12 h), eIF4A- and eIF4E-levels also declined [in contrast to the ribosome content which remained constant (results not shown)]. When cells were diluted back into fresh medium they accumulated eIF4G again (Fig. 3C, Upper, lanes 5 and 6), entered S phase, as measured by the appearance of cells with a 2n DNA content (Fig. 3B, graphs 5 and 6), and resumed exponential growth (Fig. 3A, points 5 and 6). Identical results were obtained when Western blots were decorated either with anti-eIF4F or anti-eIF4G1/542–883 antibody. These data show that exponentially growing cells contain eIF4G1 and eIF4G2 and that both are degraded upon entry into the diauxic phase.
Figure 3.
eIF4G degradation is induced upon entry into the diauxic phase. (A) Strain CWO4 was allowed to grow to the diauxic phase (points 2 and 3), diluted into fresh medium, and allowed to resume exponential growth (points 4–6). Strain CBY1.1 (TIF4631 deletion) was allowed to grow to the diauxic phase (points c and d). Time points indicated (1–6) correspond to CWO4 cells harvested at a cell density (OD600) of 1.302 (time point 1), 4.66 (time point 2), 6.6 (time point 3), 0.178 (time point 4), 0.445 (time point 5), and 1.10 (time point 6) and to CBY1.1 cells with an OD600 of 1.81 (time point a), 3.29 (time point b), 6.19 (time point c), and 6.88 (time point d). (B) Flow cytometry analysis (graphs) of CWO4 cells harvested at time points 1–6 in A. (C) Western blots of extracts from CWO4 cells harvested at time points 1–6 (lanes 1–6) and CBY1.1 (lanes a–d) and decorated with antibodies against eIF4G, eIF4A, and eIF4E.
Rapamycin-Induced eIF4G Degradation Is Not Limited to the G1 Phase of the Cell Cycle.
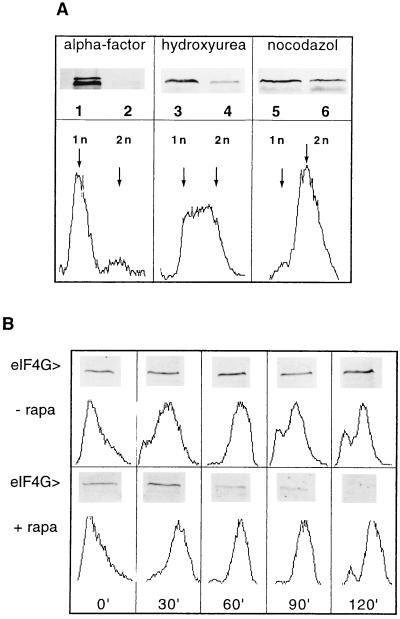
To test whether the degradation of eIF4G in response to rapamycin treatment was dependent on a specific stage of the cell cycle, we arrested cells in different phases and treated them with rapamycin. When cells were arrested in the G1 phase by treatment with α-factor, eIF4G was degraded in the presence of rapamycin (Fig. 4A, lane 2). The same was observed when cells were arrested in the S phase with hydroxyurea, although eIF4G degradation seems to be somewhat less efficient (Fig. 4A, lane 4). To test for degradation of eIF4G in the G2/M (mitosis) phase we arrested cells with the drug nocodazole and subsequently treated them with rapamycin. Because cells could not be arrested for longer periods in the G2/M phase of the cell cycle with nocodazole, they were treated for only 1.5 h with rapamycin. We observe partial eIF4G degradation under these conditions (Fig. 4A, lane 6). Because of this limitation, we do not know whether degradation of eIF4G in the G2/M phase is as efficient as in other phases of the cell cycle. The arrest of the cells in the G1, S phase, or in the G2/M phase was verified by flow cytometry depicting a 1n peak for α-factor-arrested cells (Fig. 4A, Lower Left), a broad peak of fluorescence between 1n and 2n for S phase-arrested cells (Fig. 4A, Lower Center) and predominantly 2n cells for G2/M-phase-arrested cells (Fig. 4A, Lower Right). We extended this analysis by releasing cells synchronized with α-factor from the block in the presence or absence of rapamycin. As presented in Fig. 4B, most cells had duplicated their DNA content after 60 min after release from G1 arrest, both in the absence and presence of rapamycin. Although eIF4G remained intact during progression of the cell cycle in the absence of rapamycin, it was degraded between 60 and 120 min after release in the presence of rapamycin. At this stage most cells have a 2n DNA content confirming that eIF4G degradation also happens at the G2/M stage. We conclude from these data that rapamycin-induced eIF4G degradation can happen at different stages of the cell cycle.
Figure 4.
eIF4G degradation is not confined to a specific stage of the cell cycle. (A Upper) Exponentially growing cells (strain ASZ1) were arrested at an OD600 of 0.2–0.3/ml by treating them either with α-factor for 2 h (lanes 1 and 2), with hydroxyurea for 2 h (lanes 3 and 4), or with nocodazole for 1 h (lanes 5 and 6). Vehicle (lanes 1, 3, and 5) or rapamycin (lanes 2, 4, and 6) was added to the arrested cultures and incubation was continued for 2 h (except nocodazole-treated cells for 1.5 h). Cell extracts were analyzed on Western blots for eIF4G content (lanes 1–6). (A Lower)The graphs show the flow cytometry analysis of cells arrested with the different drugs. (B) Exponentially growing ASZ1 cells were arrested with α-factor (2 h treatment) and subsequently released by incubating with fresh YPD in the absence (Upper) or in the presence of rapamycin (Lower). At the time points indicated (0, 30, 60, 90, and 120 min) cell cultures (10 ml) were harvested, aliquots were prepared for flow cytometry analysis, and cell extracts were analyzed for eIF4G content by Western blotting and immunodecoration.
DISCUSSION
The data presented in this paper show that eIF4G is a stable protein under conditions of exponential growth and becomes unstable with a considerably shorter half-life upon rapamycin treatment or during diauxic phase. The degradation pathway for eIF4G is not known. We show that it is initiated by inhibition of the Tor kinases. The components of the TOR signal transduction pathway upstream and downstream of Tor are unknown in yeast. Because rapamycin induces a response similar to starvation (7), we assume that degradation of eIF4G is mediated by inhibition of the same transduction pathway. The results presented in Fig. 2 demonstrate that ongoing translation is necessary for rapamycin-induced eIF4G degradation. Because both rapamycin and cycloheximide block protein synthesis, it may appear surprising that rapamycin can cause eIF4G degradation. This can be explained by the fact that inhibition of translation by cycloheximide is much faster than by rapamycin (7). In yeast, cellular proteins are degraded either in the vacuole or by the proteasome. Preliminary experiments with phenylmethylsulfonyl fluoride (an inhibitor of vacuolar proteases) or with proteasome mutants suggest that eIF4G degradation happens in the vacuole (Diana Dominguez, personal communication). Ongoing translation may be required for the synthesis of a receptor protein that allows import of eIF4G into the vacuole as it has been proposed for fructose 1,6-bisphosphatase (36). We are currently investigating which determinants on eIF4G target the protein for degradation. Sequence analysis revealed the presence of PEST sequences [proline (P), glutamate (E), serine (S), and threonine (T) rich] in eIF4G (23, 26) which could act as signals for proteolysis.
We speculated that a check point in G1 would control eIF4G degradation and therefore that rapamycin could exert its effect only during a specific stage of the cell cycle. The results presented in Fig. 4 show that this is not the case. It seems that the machinery used for the degradation of eIF4G by rapamycin is present during all phases of the cell cycle. However, the efficiency of rapamycin-induced eIF4G degradation may vary somewhat with the different phases of the cell cycle.
Is degradation of eIF4G the primary cause or a secondary effect of the translational down-regulation caused by starvation? Dissociation of eIF4G from eIF4E might precede degradation of eIF4G. In mammalian cells, rapamycin leads to the dephosphorylation of 4E-BPs and the enhancement of their affinity for eIF4E (37, 38). Unfortunately, it was not tested and we therefore do not know whether this leads to the dissociation of eIF4E from eIF4G. In temperature-sensitive yeast strains exhibiting a weakened eIF4E–eIF4G interaction caused by mutations in the eIF4E gene, we observed degradation of eIF4G caused by shift to the nonpermissive temperature (results not shown). This result indicates that free eIF4G is more susceptible to degradation than eIF4G in a complex with eIF4E. This may also be true for eIF4G in mammalian cells where overexpression of eIF4E antisense RNA caused reduced eIF4E expression and consequently a decrease in the cellular levels of eIF4G (39). In S. cerevisiae p20, an eIF4E-BP (40, 41) has been shown to compete with eIF4G for binding to eIF4E (29). However, we have no evidence that rapamycin induces dissociation of eIF4G from eIF4E in yeast cells. It remains to be elucidated whether p20 is modified after rapamycin treatment of yeast cells and whether it is involved in the dissociation of the eIF4E–eIF4G complex in vivo. Preliminary results indicate that a mutant which does not express p20 (29) shows very similar kinetics of eIF4G degradation like wild-type cells.
At present, we can only speculate on the physiological role of eIF4G degradation upon glucose starvation of yeast cells in the diauxic phase. eIF4G is most likely the limiting factor in cap-dependent translation in yeast, as we detected only ≈0.1 mol eIF4G per mol ribosomes, whereas the concentration of other initiation factors such as eIF4E and eIF4A is at least in the range of 1 mol factor per mol ribosomes (unpublished data). Degradation of eIF4G may serve to down-regulate translation and arrest growth in the G1 phase of the cell cycle. Indeed, strain CBY12 (whose only eIF4G gene copy is under the control of the galactose-regulatable GAL1/10 promoter; Table 1) continued growing and dividing normally for approximately four generations after transfer from galactose- to glucose-containing medium until most eIF4G significantly decreased because of factor dilution and turnover. The cells arrested in the G1 phase (even at low cell density) indicating that eIF4G activity is required for G1 to S phase transition and that its down-regulation causes G1 arrest (results not shown). In addition, degradation of this factor could be required for proper entry of the cells into the stationary phase. It would be interesting to check whether yeast cells containing a degradation-resistant form of eIF4G are unable to correctly enter the G0 phase and would consequently lose viability upon starvation.
An intriguing question is how cells synthesize eIF4G when the conditions are favorable to resume growth after a period of starvation. Cap-dependent translation requires intact eIF4G and eIF4E–eIF4G interaction (29, 42). In contrast, internal initiation (42, 43), translation of uncapped mRNAs (42), and translation of certain capped mRNAs (7, 29) do not. It is possible that translation of eIF4G mRNA itself does not need intact eIF4G. However, it could require a fragment of eIF4G. In mammalian cells, the translation of eIF4G mRNA is initiated internally (44) and is therefore expected to depend only on a C-terminal fragment of eIF4G. In yeast, we have not yet been able to detect a specific fragment of eIF4G that would persist after degradation of the protein. It is also possible that residual levels of eIF4G (undetected in our assays) are protected from degradation and initiate eIF4G mRNA translation when nutritional conditions are benign.
Acknowledgments
We thank Elisabeth Kislig and Barbara Fischli-Wittmer for excellent technical assistance, Parisa Danaie for strain PDY4, Patrick Linder for strains CWO4 and ASZ1, and Mike Hall and Stephen Helliwell for plasmid pPW2 and helpful discussions. This work was supported by Grants 31-42374.94 and 31-45528.95 from the Swiss National Science Foundation.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: eIF, eukaryotic initiation factor; GST, glutathione S-transferase; BP, binding protein.
References
- 1.Werner-Washburne M, Braun E L, Crawford M E, Peck V M. Mol Microbiol. 1996;19:1159–1166. doi: 10.1111/j.1365-2958.1996.tb02461.x. [DOI] [PubMed] [Google Scholar]
- 2.Pringle J R, Hartwell L H. In: The Saccharomyces Cerevisiae Life Cycle. Strathern J N, Jones E W, Broach J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 97–142. [Google Scholar]
- 3.Werner-Washburne M, Braun E, Johnston G C, Singer R A. Microbiol Rev. 1993;57:383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reed S I. Genetics. 1980;95:561–577. doi: 10.1093/genetics/95.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hadwiger J A, Wittenberg C, Richardson H E, de Barros Lopes M, Reed S I. Proc Natl Acad Sci USA. 1989;86:6255–6259. doi: 10.1073/pnas.86.16.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heitman J, Movva N R, Hall M N. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 7.Barbet N C, Schneider U, Helliwell S B, Stansfield I, Tuite M F, Hall M N. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stan R, McLaughlin M M, Cafferkey R, Johnson R K, Rosenberg M, Livi G P. J Biol Chem. 1994;269:32027–32030. [PubMed] [Google Scholar]
- 9.Futcher B. Yeast. 1996;12:1635–1646. doi: 10.1002/(SICI)1097-0061(199612)12:16%3C1635::AID-YEA83%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 10.Nasmyth K. Trends Genet. 1996;12:405–412. doi: 10.1016/0168-9525(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 11.Chiu M I, Katz H, Berlin V. Proc Natl Acad Sci USA. 1994;91:12574–12578. doi: 10.1073/pnas.91.26.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabatini D M, Erdjument-Bromage H, Lui M, Tempst P, Snyder S H. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 13.Brown E J, Schreiber S L. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- 14.Jefferies H B J, Reinhard C, Kozma S C, Thomas G. Proc Natl Acad Sci USA. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ming X F, Burgering B M Th, Wennström S, Claesson-Welsh L, Heldin C H, Bos J L, Kozma S C, Thomas G. Nature (London) 1994;371:426–429. doi: 10.1038/371426a0. [DOI] [PubMed] [Google Scholar]
- 16.Lin T A, Kong X, Haystead T A J, Pause A, Belsham G, Sonenberg N, Lawrence J C. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 17.Pause A, Belsham G J, Gingras A C, Donzé O, Lin T A, Lawrence J C, Sonenberg N. Nature (London) 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 18.von Manteuffel S R, Dennis P B, Pullen N, Gingras A C, Sonenberg N, Thomas G. Mol Cell Biol. 1997;17:5426–5436. doi: 10.1128/mcb.17.9.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haghighat A, Mader S, Pause A, Sonenberg N. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mader S, Lee H, Pause A, Sonenberg N. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terada N, Patel H R, Takase K, Kohno K, Nairn A C, Gelfand E W. Proc Natl Acad Sci USA. 1994;91:11477–11481. doi: 10.1073/pnas.91.24.11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sonenberg N. In: mRNA 5′ Cap-Binding Protein eIF4E and Control of Cell Growth. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 245–269. [Google Scholar]
- 23.Goyer C, Altmann M, Lee H S, Blanc A, Deshmukh M, Woolford J L, Trachsel H, Sonenberg N. Mol Cell Biol. 1993;13:4860–4874. doi: 10.1128/mcb.13.8.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hentze M W. Science. 1997;275:500–501. doi: 10.1126/science.275.5299.500. [DOI] [PubMed] [Google Scholar]
- 25.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 26.Morley S J, Curtis P S, Pain V M. RNA. 1997;3:1085–1104. [PMC free article] [PubMed] [Google Scholar]
- 27.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 29.Altmann M, Schmitz N, Berset C, Trachsel H. EMBO J. 1997;16:1114–1121. doi: 10.1093/emboj/16.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goyer C, Altmann M, Trachsel H, Sonenberg N. J Biol Chem. 1989;264:7603–7610. [PubMed] [Google Scholar]
- 31.Helliwell S B, Wagner P, Kunz J, Deuter-Reinhard M, Henriquez R, Hall M N. Mol Biol Cell. 1994;5:105–118. doi: 10.1091/mbc.5.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cardenas M E, Heitman J. EMBO J. 1995;14:5892–5907. doi: 10.1002/j.1460-2075.1995.tb00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danaie P, Wittmer B, Altmann M, Trachsel H. J Biol Chem. 1995;270:4288–4292. doi: 10.1074/jbc.270.9.4288. [DOI] [PubMed] [Google Scholar]
- 34.Hanic-Joyce P J, Singer R A, Johnston G C. J Biol Chem. 1987;262:2845–2851. [PubMed] [Google Scholar]
- 35.Naranda T, MacMillan S E, Hershey J W B. J Biol Chem. 1994;269:32286–32292. [PubMed] [Google Scholar]
- 36.Chiang H L, Schekman R. Nature (London) 1991;350:313–318. doi: 10.1038/350313a0. [DOI] [PubMed] [Google Scholar]
- 37.Beretta L, Gingras A C, Svitkin Y V, Hall M N, Sonenberg N. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 38.von Manteuffel S R, Gingras A C, Ming X F, Sonenberg N, Thomas G. Proc Natl Acad Sci USA. 1996;93:4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Benedetti A, Joshi-Barve S, Rinker-Schaeffer C, Rhoads R E. Mol Cell Biol. 1991;11:5435–5445. doi: 10.1128/mcb.11.11.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altmann M, Krieger M, Trachsel H. Nucleic Acids Res. 1989;17:7520. doi: 10.1093/nar/17.18.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanker S, Müller P P, Altmann M, Goyer C, Sonenberg N, Trachsel H. J Biol Chem. 1992;267:21167–21171. [PubMed] [Google Scholar]
- 42.Ohlmann T, Rau M, Morley S J, Pain V M. Nucleic Acids Res. 1995;23:334–340. doi: 10.1093/nar/23.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohlmann T, Rau M, Pain V M, Morley S J. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 44.Gan W, Rhoads R E. J Biol Chem. 1996;271:623–626. doi: 10.1074/jbc.271.2.623. [DOI] [PubMed] [Google Scholar]
- 45.Coppolecchia R, Buser P, Stotz A, Linder P. EMBO J. 1993;12:4005–4011. doi: 10.1002/j.1460-2075.1993.tb06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]