Abstract
A large superfamily of transmembrane receptors control cellular responses to diverse extracellular signals by catalyzing activation of specific types of heterotrimeric GTP-binding proteins. How these receptors recognize and promote nucleotide exchange on G protein α subunits to initiate signal amplification is unknown. The three-dimensional structure of the transducin (Gt) α subunit C-terminal undecapeptide Gtα(340–350) IKENLKDCGLF was determined by transferred nuclear Overhauser effect spectroscopy while it was bound to photoexcited rhodopsin. Light activation of rhodopsin causes a dramatic shift from a disordered conformation of Gtα(340–350) to a binding motif with a helical turn followed by an open reverse turn centered at Gly-348, a helix-terminating C capping motif of an αL type. Docking of the NMR structure to the GDP-bound x-ray structure of Gt reveals that photoexcited rhodopsin promotes the formation of a continuous helix over residues 325–346 terminated by the C-terminal helical cap with a unique cluster of crucial hydrophobic side chains. A molecular mechanism by which activated receptors can control G proteins through reversible conformational changes at the receptor–G protein interface is demonstrated.
Visual signals in retinal rod cells are triggered by the interaction of photoexcited rhodopsin (R*) with the heterotrimeric GTP-binding protein transducin (Gt). This prototypical G protein-coupled receptor (GPCR) system has been well characterized with regard to the light activation of R and the resulting nucleotide exchange on Gt that leads to the dissociation of Gt from R*. The GTP-bound form of activated Gt differs markedly from the inactive GDP-bound state (1–4). Molecular details of the mechanism by which R* recognizes Gt and catalyzes nucleotide exchange remain obscure. Several domains on the α and βγ subunits of Gt contribute to its R* recognition site (5–7). One of the best studied is the extreme C-terminal region of Gtα. Pertussis toxin-mediated ADP ribosylation of Cys-347 is well known to uncouple Gt from the receptor (8). Epitope mapping, mutational, and peptide competition studies have established that an 11-residue segment, IKENLKDCGLF, Gtα(340–350), plays a major role in receptor recognition and, potentially, in nucleotide exchange on Gtα (9–12). A synthetic undecapeptide, Gtα(340–350), derived from the C-terminal region of Gtα is a competitor for the Gt site on R* (10). The same peptide mimics the ability of Gt to stabilize an active intermediate metarhodopsin II, MII (10, 13). Sequence similarities in C-terminal regions of Gα subunits suggest that receptor-interacting domains of G proteins bear common structural elements with enough sequence diversity between Gα classes to provide recognition of various receptor subtypes.
Conformational changes at the receptor-interacting surface of G proteins upon binding to activated GPCRs are thought to trigger nucleotide exchange and G protein activation (14). Direct x-ray studies of interactions in receptor–G protein complexes, however, have yet to be realized and the C-terminal segment of Gtα is disordered in all x-ray structures of Gt (1–4). Transferred nuclear Overhauser effect (TRNOE) spectroscopy (15) is a viable approach to determining conformations of ligands in complexes with their cognate receptors. Previous application of TRNOE spectroscopy to study the R*–Gt complex faced unexpected difficulties (16). The mutant Gtα(340–350)K341R was used because of technical reasons and was reported to be more constrained when bound to the dark-adapted R rather than to MII. Gtα(340–350)K341R appeared to relax upon receptor activation. This contradicts the known property of Gt and Gtα(340–350) to selectively bind and stabilize MII. The experimental data suffered from overall low signal-to-noise and the structural interpretation was compromised by errors in NMR assignments (17). Herein, we elucidate the structural changes of the native Gtα(340–350) undecapeptide upon association with highly purified R* in its natural lipid environment by TRNOE spectroscopy. In accord with biochemical data, Gtα(340–350) is structured only after light activation. We propose a mechanism of specific conformational changes in Gt resulting from association with R*.
MATERIALS AND METHODS
Sample Preparation.
Gtα(340–350) was synthesized on solid phase and purified by reverse-phase FPLC (13). Cattle eyes were dark-adapted overnight and R was purified under dim red light or in the dark. Urea-washed membranes were prepared from rod outer segments (18, 19). Purity of R was estimated at 95–99% by SDS/PAGE and spectroscopy (20). The NMR laboratory and NMR spectrometer were light-proofed. NMR samples contained Gtα(340–350) (5 mg/ml), 20 mM sodium phosphate (pH 7.5), 100 mM KCl, 0.1 mM EDTA, 1 mM DTT, 10% 2H2O. R-containing samples included R at 2 mg/ml, which was activated by 480-nm light produced by 150-W Fiber Lite for 30 s.
NMR.
One-dimensional proton spectra were acquired at 1°C (Varian Unity-500 spectrometer, vnmr 5.1 software), 5,200-Hz spectral width, 16,000 data points, and 5.0-s preacquisition delay. Spectra from TOCSY (total correlation spectroscopy) (21) used the MLEV-17 mixing sequence of 120 ms, flanked by two 2-ms trim pulses, a 0.5-s preaquisition delay, and a 1.0-s presaturation. Phase-sensitive two-dimensional spectra were obtained with the hypercomplex method. Nuclear Overhauser effect spectroscopy (NOESY) (22) spectra used a 2 × 280 × 2,049 data matrix with 16 scans per t1 value (1.9 s per scan; total time for NOESY, 4.7 hr). Conditions for a typical experiment are as follows: 1°C, 7–9 hr of TOCSY–NOESY on dark-adapted samples (phase 1), light exposure for 30 s, and NOESY data acquisition (phase 2). Light-exposed samples were allowed to decay for 18–24 hr followed by a third round of NOESY (phase 3). Similar experiments using a range of mixing times Tmix = 50–400 ms were analyzed for possible spin diffusion effects, as intensities of the NOE cross-peaks most accurately reflect the interproton distances at mixing times during the linear phase of NOE build up. Phase 1 NOEs were at maximum by Tmix = 350 ms; phase 2 was at maximum by Tmix = 100 ms.
Structure Calculations.
NMR structures of Gtα(340–350) were calculated with tinker 3.4, http://dasher.wustl.edu/tinker/(23) using the charmm22 (24) force field. Additional NOEs were classified according to their intensities as weak, medium, and strong and translated to distance constraints of 1.9–5.0 Å, 1.9–3.5 Å, and 1.9–2.7 Å, respectively. Appropriate prochiral corrections to the upper distance limits were introduced for the CH2, CH3, and aromatic ring protons (1.8 Å and 4.9 Å, respectively) in those cases where stereospecific assignment was ambiguous because of the proton chemical shift degeneracy. Separate distance constraints for each of the prochiral resonances with a weighting in the penalty function equal to the inverse of its degeneracy were entered. This allows all real atoms to be used in structure calculations. Distance constraints derived from the NOE cross-peaks that allowed alternative assignments were tested in parallel calculations, and constraints resulting in the best agreement with the rest of the distance matrix were kept. Initial models were generated by distance geometry (distgeom) and 20 low-energy structures with a maximum distance violation of no more than 0.1 Å were energy-minimized (newton). Constrained molecular dynamics at 1,000K and simulated annealing (anneal, 2-fs time steps, up to a total of 25–100 ps) led to the final 20 structures that were superimposed, and rmsd values from the average structure were calculated in molmol (25). The stereochemical quality of peptide structures in the ensemble was calculated with procheck 3.5 (26). For docking the NMR structures to the x-ray structure of Gt, the backbone of the last four residues of Gtα was superimposed on the backbone of the first four residues of R* bound Gtα(340–350) with an rmsd of 0.299. The overlapping residues 340–344 were removed from the x-ray structure and Gtα(340–350) was covalently attached to Ile-339 followed by energy minimization to improve the local geometry.
RESULTS AND DISCUSSION
Sequence-specific and stereospecific assignments were established by analyzing appropriate regions of TOCSY and NOESY spectra of free peptide and peptide in the presence of R by using standard procedures (27). Proton chemical shifts in the presence of R did not differ significantly from those of the free peptide, but peptide resonances were slightly broadened by light activation of R and overall intensity was diminished. This is likely to reflect a fast exchange between R*-bound and free peptide, because Gtα(340–350) has a relatively low affinity for MII, EC50 = 100 μM (Fig. 1A). Resonance assignments are summarized in Table 1. Distinguishing δ-methyl protons of Leu-344 and Leu-349 was straightforward, and the difference in chemical shifts for the side-chain protons of Lys-341 and Lys-345 was small but reproducible. Proton resonance assignment is 96% complete, with the ɛNH3 and ɛCH2 of Lys-345 and Lys-341 having degenerate chemical shifts under the experimental conditions. All connectivities between adjacent residues were observed in the NH–αH region of the NOESY spectra. An explicit continuous path through the residues Phe-350 to Ile-340 (numbers 11 to 1) can be suggested (Fig. 2B). A majority of the sequential NH–NH connectivities can also be observed (Fig. 2A). Due to the close position of the corresponding resonances, sequential NH–NH NOEs of Lys-341/Glu-342, Lys-345/Asp-346, and Asp-346/Cys-347 are too close to the main diagonal to be identified unambiguously.
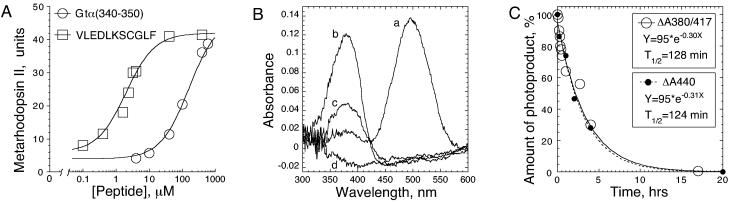
Figure 1.
Light activation of R in the presence of native Gtα(340–350) peptide and the high-affinity analog VLEDLKSCGLF (28). (A) Dose–response results of the MII formation. Extra MII was calculated as the A380/A417 difference (29). (B) UV/visible difference spectra of R alone (trace a), in the presence of 2 mM of Gtα(340–350) (traces b–d), 1 min (trace b), 3 hr (trace c), and 18 hr (trace d) after light activation were recorded at 1°C as described (29). Identical samples bleached for 3 hr at 25°C were used as a light-scattering blank. All spectra are normalized to the A800 values. Note that MII stabilization proceeds at the expense of MI and no photoproducts other than MII are being generated. (C) Decay of the MII–Gtα(340–350) complex (open symbols). Amount of extra MII generated in the presence of 2 mM Gtα(340–350) peptide at 1°C 1 min after light activation was calculated as the A380/A417 difference and was taken as 100%. The authenticity of the MII photoproduct, which contains all-trans-retinal covalently attached to the Lys-296 via a deprotonated Schiff base, was confirmed in acid-trapping experiments with final concentration of HCl at 1% (vol/vol). Acid denaturation generates the product with Amax = 440 nm only if retinal is covalently bound (30). Solid symbols show the amount of the 440-nm product generated after addition of HCl at indicated times during the decay of the 380-nm species; A440 in the control fully bleached sample plus HCl is subtracted; ΔA440 at 1 min is 100%. At 1°C and pH 7.5, direct decay of the extra MII stabilized by the Gtα(340–350) to form opsin and all-trans-retinal (bypassing MIII, Amax = 465 nm) is consistent with previous reports that when bound to Gt, alternative pathway of MII decay via MIII is suppressed (for review, see ref. 31).
Table 1.
Proton resonance assignments (ppm) for Gtα(340–350) at 1°C and pH 7.5
| Residue | NH | αH | βH | γH | δH | Others |
|---|---|---|---|---|---|---|
| Ile-340 | 3.90 | 1.99 | γCH2, 1.21, γCH3, 1.01, δCH3, 0.96 | |||
| Lys-341 | 8.79 | 4.33 | 1.84, 1.80 | 1.48 | 1.71 | ɛNH3, 7.65, ɛCH2, 3.01 |
| Glu-342 | 8.80 | 4.24 | 2.03, 1.96 | 2.29 | ||
| Asn-343 | 8.76 | 4.66 | 2.87, 2.76 | γNH2, 7.79, 7.13 | ||
| Leu-344 | 8.50 | 4.32 | 1.69 | 1.62 | δCH3, 0.94, 0.89 | |
| Lys-345 | 8.42 | 4.26 | 1.82 | 1.45 | 1.69 | ɛNH3, 7.65 |
| Asp-346 | 8.48 | 4.59 | 2.78, 2.69 | |||
| Cys-347 | 8.44 | 4.49 | 2.99 | |||
| Gly-348 | 8.65 | 3.93 | ||||
| Leu-349 | 8.08 | 4.30 | 1.52 | 1.46 | δCH3, 0.91, 0.84 | |
| Phe-350 | 7.82 | 4.43 | 3.19, 2.97 | δ(2,4), 7.26, ɛ(3,5), 7.37, z(4), 7.32 |
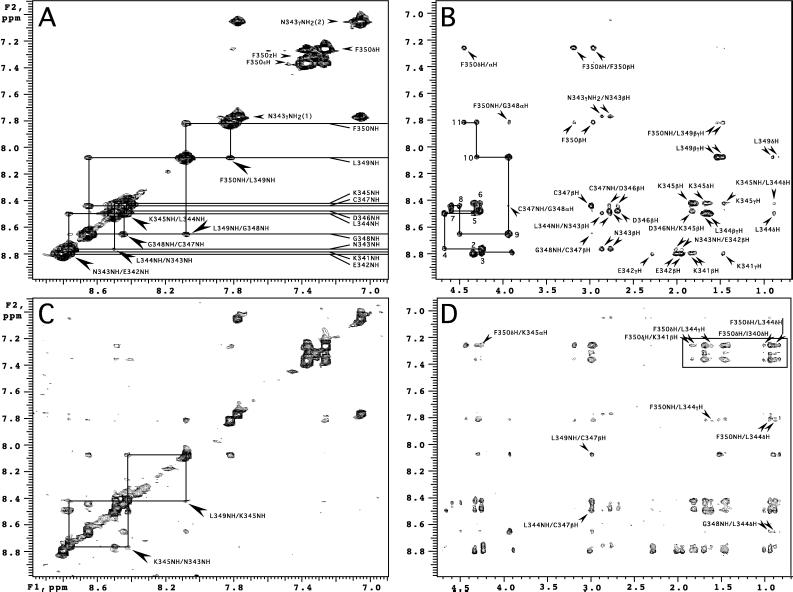
Figure 2.
Representative regions from the NOESY spectra of Gtα(340–350) (Tmix = 200 ms) in the presence of the dark-adapted R (A and B) and R* (C and D). One-letter amino acid code is used for annotations. (A) Amide region of the dark spectrum. Resonances of the amide protons are indicated with horizontal lines. Selected sequential NH–NH cross-peaks are labeled with larger arrows. (B) The path over the sequential αHi, NHi+1 connectivities is shown as a continuous solid line. Positions of the intraresidue αHi, NHi cross-peaks corresponding to the residues 340–350 are numbered 1–11. (C) Light-induced additional cross-peaks in the amide region. Diagonal symmetry of the Leu-349NH/Lys-345NH and Lys-345NH/Asn-343NH NOE connectivities are highlighted with rectangles. (D) Light-induced cross-peaks in the NH-aliphatic and aromatic aliphatic regions. Interactions between Phe-350 aromatic protons and side-chain protons of Leu-349, Leu-344, and Lys-341 are boxed. NOE cross-peaks common in B and D are assigned in B. Selected additional cross-peaks are labeled.
NMR experiments on Gtα(340–350) were performed for the peptide alone and in the presence of R in three phases, (i) dark-adapted R, (ii) photoexcited R*, and (iii) opsin. Spectra of the free peptide contained only sequential connectivities, characteristic of a random conformation in solution (data not shown). Phase 1 and 3 data were identical. They differed from the spectra of the free peptide only by the presence of an additional NOE Gly-348/Phe-350 (αHi, NHi+2), and a weak and somewhat unusual Cys-347/Gly-348 (NHi, αHi+1) (Fig. 2B). However, careful examination of the free peptide spectrum revealed that the Gly-348/Phe-350 (αHi, NHi+2) was in fact present, though of much weaker intensity. Observation of these two NOEs may indicate the propensity of the last four amino acids to form a turn at Gly-348 even in solution. Overall, the lack of additional medium to long-range NOEs in the phase 1 spectra indicates no detectable binding of Gtα(340–350) to dark-adapted R. This observation is in accord with the known weak interaction of Gt with inactive R (32, 33) and in distinct contrast to the previous report (16).
Exposure of the dark-adapted sample to light produced a remarkable increase in the number of longer-range interactions indicative of a discrete conformation in the R*-bound state (Fig. 3). Representative regions of the NOESY contour plots for the peptide in the presence of the dark-adapted and photoexcited R* are compared in Fig. 2. Under the experimental conditions (1°C, pH 7.5), photoexcitation of R alone leads to the equilibrium of two long-lived intermediates, MI (Amax = 490 nm), and MII (Amax = 380 nm), with MI as predominant species. Gtα(340–350) or its analogs mimic Gt and bind specifically to MII shifting MI–MII equilibrium toward MII in a concentration-dependent manner (Fig. 1) (10, 13). Thus, light activation of R in the presence of saturating amounts of the undecapeptide generates a uniform sample of the MII–Gtα(340–350) complex (Fig. 1). The efforts to estimate the stability of this complex under the exact NMR conditions are hampered by the strong light-scattering effects, because the concentration of rod outer segment membranes is at least 10 times higher compared with the typical concentrations used for spectral analysis. Under the normal UV/visible assay conditions, decay of MII in the presence of Gtα(340–350) proceeds slowly (T½ = 128 min; Fig. 1C) without generation of any other photoproducts of R stable on the time scale of NMR experiments. The final product in the sample is opsin/all-trans-retinal and Gtα(340–350) peptide (Fig. 1B). NOESY spectra acquired 18 hr after light activation showed an NOE pattern indistinguishable from that obtained during phase 1. Thus, the undecapeptide becomes structured only when bound to the photoexcited state of R, MII. This observation with Gtα(340–350) parallels the ability of the heterotrimeric Gt to form a transient high-affinity complex with R* (34). It also suggests that the relatively short undecapeptide Gtα(340–350) and Gt share a common mode of binding to R*.
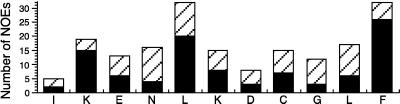
Figure 3.
Summary of the NOEs observed for the R*-bound Gtα(340–350). Each NOE for a particular proton pair is counted twice. Solid bars, medium- and long-range interactions; hatched bars, short range NOEs.
NMR models of Gtα(340–350) bound to R* were determined by a distance geometry/simulated annealing protocol (Table 2). Independent computations gave 20 structures with a high degree of convergence (Table 2 and Fig. 4A) with a helical turn in the middle of the undecapeptide followed by an open reverse turn (Fig. 4). A notable feature of the bound conformation is a hydrophobic cluster formed by the side chains of Phe-350, Leu-349, Leu-344, and Lys-341 (Fig. 4 B and C). Long-range interactions between aromatic-ring protons of Phe-350 and side-chain protons of Leu-344, Lys-341, and Ile-340 constitute a significant proportion of R*-induced NOE cross-peaks (Fig. 2D). The NOE pattern between aromatic-ring protons and side-chain protons of Lys (Fig. 2D, boxed) was assigned as Phe/Lys-341 based on the small but reliable difference in the β-, γ-, and δ-methylene proton resonances of Lys-341 and Lys-345. The choice of Lys-341 was also supported by the identified interactions of the aromatic ring with the side-chain protons of Ile-340 adjacent to Lys-341. Nevertheless, Phe-350 to Lys-341 and Phe-350 to Lys-345 assignments were used in generating the initial set of structures by distance geometry calculations. Selection of Lys-341 constraints consistently produced better agreement for the entire set of distance constraints. Interactions between Phe-350 and Lys-345, however, are not excluded, because final structures show the Lys-345βH protons within the 5.0-Å NOE limit from the Phe-350 aromatic ring.
Table 2.
Statistics of the NMR structure and sterochemical quality by PROCHECK and MOLMOL
| Value for ensemble of 20 structures | Value for average structure | Reference value for x-ray structures (2.0 Å) | |
|---|---|---|---|
| Ramachandran plot statistics | |||
| Residues in most favored regions [A,B,L], % | 43.1 | 62.5 | 75 ± 10 |
| Residues in additional allowed regions [a,b,l,p], % | 50 | 37.5 | |
| Residues in generously allowed regions [∼a,∼b,∼l,∼p], % | 6.9 | 0 | |
| Residues in disallowed regions, % | 0% | 0% | |
| Main-chain statistics | |||
| SD of ω angle, degrees | 6.3 | 7.4 | 6.0 ± 3.0 |
| Bad contacts per 100 residues | N/A* | 0 | 4.2 ± 10 |
| Cα chirality, SD of ζ angle, degrees | 2.4 | 1.5 | 3.1 ± 1.6 |
| H bond energy, kcal/mol | 0.9 | 0.8 | 0.6 ± 0.2 |
| Side-chain statistics | |||
| χ-1 gauche minus, degrees | 11.7 | 10.7 | 18.1 ± 6.5 |
| χ-1 trans, degrees | 9.0 | 8.1 | 19.0 ± 5.3 |
| χ-1 gauche plus, degrees | 9.4 | 6.9 | 17.5 ± 4.9 |
| χ-1 pooled, degrees | 16.3 | 14.8 | 18.2 ± 4.8 |
| χ-2 trans, degrees | 33.6 | 32.5 | 20.4 ± 5.0 |
| Distance constraint violations (>0.3 Å) | 0 | N/A | N/A |
| Intrapeptide CHARMM energy, kcal/mol | −186.7 ± 1.2 | N/A | N/A |
| Rmsd to the mean (20 structures) for backbone atoms, Å | 0.30 ± 0.12 | N/A | N/A |
| Rmsd to the mean (20 structures) for all heavy atoms, Å | 0.63 ± 0.21 | N/A | N/A |
PROCHECK estimated equivalent resolution for the R*-bound Gtα(340–350) is 2.9 Å. *, Single structures only; N/A, not applicable.
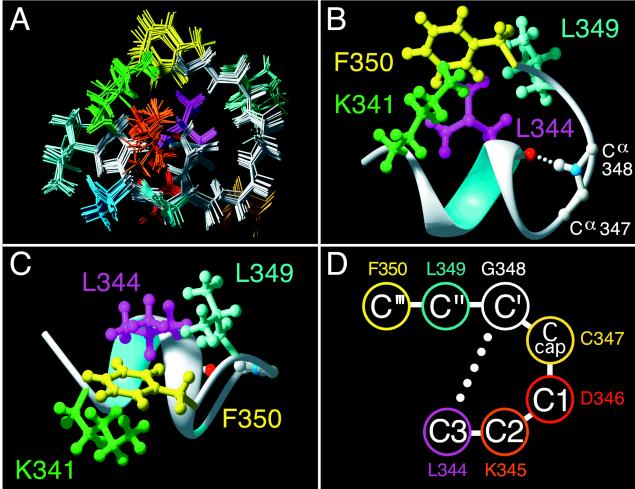
Figure 4.
Model of the Gtα(340–350) undecapeptide bound to R*. (A) Ensemble of 20 R*-bound structures computed independently. Backbone is in white. Side-chains are color coded. (B and C) Ribbon diagram of R*-bound Gtα(340–350) showing the hydrophobic cluster in the bound conformation of Gtα(340–350). Front view (B) and top view (C) of the molecule as likely seen by R* are shown. Side-chains colors: Phe-350, yellow; Leu-344, light blue; Leu-344, magenta; Lys-341, green. The H bond between the carbonyl oxygen of Leu-344 and the amide hydrogen of Gly-348 is shown as a white dotted line. Note the proximity of the charged Lys-341 amino group to the Phe-350 aromatic ring. (D) Amino acid sequence of Gtα(344–350) and nomenclature for the C-terminal capping motif (35). The αL-defining 5 → 1 hydrogen bond is shown as a dotted line. Figures are prepared with molmol (25) and pov-ray ppc.
Structure calculations show that the hydrophobic cluster favors formation of a H-bond (5 → 1 type) between the carbonyl oxygen of Leu-344 and the amide hydrogen of Gly-348, which additionally stabilizes the reverse turn (Fig. 4B) and allows cross-relaxation between the amide protons of Leu-349 to Lys-345 (Fig. 2C). Experimentally determined constraints define the folding of a Glu-342 to Asp-346 stretch into a helical turn. Additionally, each of the 20 models in the ensemble show two main-chain H bonds between Lys-341 and Lys-345 and between Glu-342 and Asp-346, characteristic of an α-helical conformation. The NOE cross-peak Lys-345/Asn-343, (NHi,i+2) also agrees with the presence of a helical turn (Fig. 2C). Main-chain geometry for Ile-340 and Lys-341 are also consistent with the helical conformation. Nonetheless, these residues are not strictly helical as defined by the Kabsch/Sander DPSS algorithm (36) based on the H-bonding pattern. On the basis of the results of molecular modeling, we expected to find at least two additional NOE cross-peaks of an αHi,NHi+4 type: Lys-341/Lys-345 and Glu-342/Asp-346. Unfortunately, the positions of both NOEs, which are commonly rather weak, overlap with two very strong sequential connectivities Lys-345/Leu-344 and Asp-346/Lys-345 (Fig. 2D). Thus, the data argue for a one full helical turn in R*-bound Gtα(340–350).
Examination of the φ/ψ torsional angles of R*-bound Gtα(340–350) revealed that Gly-348 occupied a conformation normally prohibited for l-amino acids, which allows the peptide chain to turn sharply back upon itself. In fact, Gly-348 is highly conserved in G proteins of Gi family and appears invariant at position 348 in 24 Gtα(340–350) analogs selected for R* binding (28). Expression of Gt mutants with substitutions of Gly-348 with Ala or Pro (11, 12) in the Gtα severely impairs interaction with R*. An earlier NMR study suggested that Gly-348 was part of a type II β-turn at the C terminus of the Gtα (16). Our results do not support this hypothesis and suggest an alternative interpretation of the Gly-348 requirement.
Gly residues have been noted to participate in the termination of helices with high frequency (35, 37, 38). The last C-terminal turn of a helix is less stable because its three carbonyls do not participate in H bonds due to helix termination (39). By allowing the peptide chain to turn back upon itself, Gly provides an opportunity for an alternative H bonding pattern and concomitant stabilization of the C-terminal portion of the helix. Comparison of the R*-bound structure of Gtα(340–350) with known types of helix-terminating motifs revealed a high degree of similarity with the αL type of helix capping (Gly-348 is C′, adjacent residues toward the N terminus are named Ccap, C1, C2, and C3; residues toward the C terminus are named C′′ and C′′′) (35) (Fig. 4D). This motif is characterized by an H bond between the amide hydrogen at C′ and carbonyl oxygen at C3 (5 → 1 type), a criterion met by the R*-bound Gtα(340–350) (Fig. 4B). The lack of a β-turn H bonding pattern between Asp-346 and Leu-349 and φ/ψ values for Cys-347 are inconsistent with a type II reverse turn, as proposed (16). Our data strongly argue for a different type of reverse turn at this position, namely, a variant of a classical αL-type, the C-terminal capping motif (37).
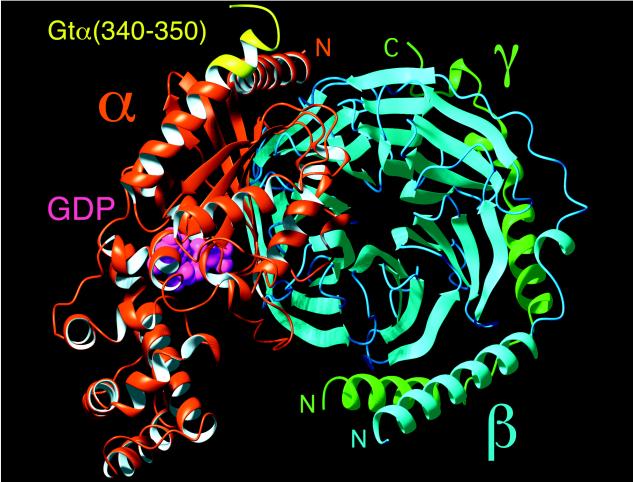
Interestingly, helix-terminating reverse turns are generally found to be solvent exposed (35). In αL type motifs, the conformation of the residue that follows the C′(glycine) requires the C′′ side chain to point outward, as for R*-bound Gtα(340–350). Sequence comparison revealed that in the helices terminated by the αL motif, the residue at C′′ is invariably polar. Gtα(340–350), however, contains the hydrophobic Leu-349 at this C′′ position. If this segment were solvent-exposed, an apolar residue at C′′ combined with a C′-to-C3 hydrogen bond would make helix capping extremely unfavorable. This agrees with the fact that in all x-ray structures of Gt obtained so far, residues 344–350 are disordered (1–4). We suggest that it is the interaction of Gtα(340–350) with the hydrophobic core of the light-induced Gt-binding site on R* that counteracts the hydrophobicity of Leu-349 and makes this motif stable. With the highly hydrophobic patch formed by the side chains of Phe-350, Leu-344, and Lys-341, the Leu-349 side chain likely makes up a significant portion of the specific R*-interacting surface (Fig. 4C). In fact, when docked to the GDP-liganded crystal structure of Gt that is known to be the form of G protein ready for interaction with R*, the NMR structure of Gtα(340–350) provides a perfect continuation of the C-terminal helix α5, Gtα(325–340), terminated by the C-cap and hydrophobic cluster (Fig. 5). The suggested hydrophobic patch (Lys-341/Leu-344/Leu-349/Phe-350) points away from Gt in the direction where R* would be found in a putative ternary complex with Gt. Hydrophobic shielding of this motif from water clearly requires a specific binding site on R*. Light-induced exposure of a potential complementary binding site on the second (CD) and third (EF) intracellular loops (40) is consistent with the movement of transmembrane helices C and F recently shown to be required for light activation (41, 42). Dissociation from the receptor would be expected to expose Leu-349 to solvent, leading to transition of the Gtα(340–350) segment back into a disordered state and providing a mechanism for reversible changes in Gt.
Figure 5.
R*-bound conformation of Gtα(340–350) (yellow) docked to the structure of a GDP-bound form of heterotrimeric Gt (1GOT) showing the predicted effect of R* on Gt conformation. At the top of Gt is the putative R*-interacting surface of the heterotrimer. GDP in the nucleotide binding site of the α subunit is shown in magenta as a space-filling model.
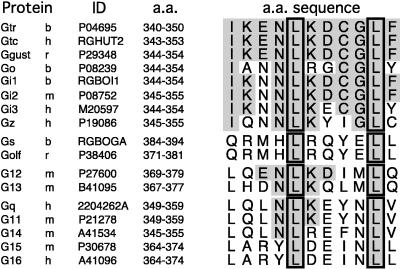
Two Leu residues, Leu-344 and Leu-349, appear remarkably conserved among members of functionally distinct G protein classes (Fig. 6). The pivotal roles of Leu-344 and Leu-349 in R* binding have been established with specific mutations at these sites in Gt (11, 12, 43) and by studies of Gtα(340–350) peptide analogs (28, 40). Substitutions of Leu-344 and Leu-349 by any other amino acid severely affect R* binding. The role for these two residues appears different based on the structure of the R*-bound Gtα(340–350). Leu-344 is crucial to the overall folding of the C-terminal domain. It contributes a central hydrophobic side chain that is in the core of a Lys-341/Leu-344/Leu-349/Phe-350 hydrophobic patch (Fig. 4C). This is in agreement with the previous observation that Leu344Ala mutation not only has a dramatic effect on the local stability of Gtα(340–350) but also can affect global folding of Gt (12, 43). In contrast, according to the structure presented in this article, Leu-349 has a more prominent regulatory role in R* recognition. R* appears to require the hydrophobic side chain of Leu-349 that causes reversibility in initiation and C-capping of the long C-terminal α5 helix of the Gt α subunit.
Figure 6.
Amino acid sequence comparison of the C-terminal regions of G protein α subunits. Identical amino acids are shaded. Conserved Leu-344 and Leu-349 are outlined. Protein sources: b, bovine; h, human; m, mouse; r, rat. Full protein sequences are available from the National Center for Biotechnology Information at http://www.ncbi.nlm.nih.gov/Entrez/protein.html.
Several factors contribute to the credibility of our results. The conformation of the bound undecapeptide has features suggesting that it occupies a hydrophobic pocket. The hydrophobic cluster on the exterior of the peptide is unexpected in water; the cation–aromatic interaction between the Lys-341 amine and the Phe-350 phenyl ring (Fig. 4B) is commonly seen in the interior of proteins (44) and is often exploited in specific ligand–receptor recognition (45). Helical turns at the terminals of small peptides in solution are also somewhat unusual. Relatively higher magnitudes of the sequential αH–NH and NH–NH NOEs of Phe-350, Leu-349, and Leu-344 in phase 2 spectra are likely to reflect less conformational averaging in these regions due to constraints imposed by the complementary binding site on rhodopsin. The bound conformation reflects NOEs seen only with R* and is consistent with the lack of structure observed with the peptide alone or in the presence of dark-adapted R or opsin. Similar TRNOE experiments on an analog of Gtα(340–350), VLEDLKSCGLF, with an affinity approximately 100-fold greater (Fig. 1), show diminished NOE peaks (data not shown), in accord with requirements for an appropriate exchange rate in ligand–receptor complexes (15). An unexpected confirmation for the R*-induced conformational changes in Gtα(340–350) that we propose comes from the analysis of the x-ray structure of Giα–AlF4 complexed with RGS4 protein in the transition state for GTP hydrolysis (46). The extreme C-terminal region of Giα resembles the overall fold of the R*-bound Gtα(340–350) with rmsd for the backbone of only 0.51 Å. Remarkably, the C-terminal segment is stabilized by the same mechanism of Leu-344 side chain (Leu-343 in Giα) being shielded from solvent. For the Giα–AlF4–RGS4 complex, such shielding is achieved by burying Leu-344 in the intramolecular interface with an N-terminal helix. Thus, helix initiation and capping at the C termini of Gα is a likely event provided a complementary hydrophobic environment is presented.
Our results are in striking contrast with the published data and conclusions (16). Low signal-to-noise and assignment errors in the previous work (17) hinder comparison of the NMR models and conformational changes in Gtα(340–350) upon binding to R*. More importantly, the lack of binding of native Gtα(340–350) to the dark-adapted R in our experiments contradicts the paradoxical interaction of mutant Gtα(340–350)K341R with dark-adapted R previously suggested. It is likely that the existence of a “precoupled” state for the Gtα(340–350)K341R was artificially exaggerated by the “dark” data acquired by using significantly longer time periods and then directly comparing “dark” and “light” spectra (16). It is also possible that the use of a native not a mutant peptide, higher purity of R, and special attention to preventing premature light activation of our samples account for the experimental differences.
This study resolves a part of the puzzle of how conformational changes in activated GPCRs control the GDP–GTP exchange in the nucleotide-binding site of G proteins that is some 35 Å away from the receptor–G protein interface. Conceptually, the Gα C terminus and switch regions I, II, and III that are affected most upon nucleotide exchange appear structurally coupled by reciprocal order–disorder transitions (47). Our results provide direct evidence for the reversible ordering of the C-terminal 11-residue segment of Gtα induced by R*. If structurally coupled to the conformation of the nucleotide binding site, R*-catalyzed C capping of Gtα(340–350) could trigger GDP release. Mutational studies point to the long C-terminal α5 helix as the possible bridge involved in communication of the distant R*-interacting interface and the nucleotide-binding site (11, 43). However, recently identified contact sites of GPCRs and βγ subunits (13, 20, 29, 48–50) provide additional means for transmitting structural changes. A more detailed analysis of any concerted movements of G protein subunits in the R*-bound state is required to understand G protein activation. Application of similar TRNOE studies to the interaction of the C-terminal Gtγ peptide with R* (13, 20, 29) should provide additional insights.
Acknowledgments
We thank Dr. E. M. Ross and Dr. K. P. Hofmann for valuable help and comments. This work has been supported by grants from National Institute of Health. N.G. is an Established Investigator of the American Heart Association.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: TRNOE, transferred nuclear Overhauser effect; Gt, transducin; GPCR, G protein-coupled receptor; M, metarhodopsin; NOESY, nuclear Overhauser effect spectroscopy; rmsd, root mean square deviation.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (reference 1aqg).
References
- 1.Noel J P, Hamm H E, Sigler P B. Nature (London) 1993;366:654–663. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- 2.Lambright D G, Noel J P, Hamm H E, Sigler P B. Nature (London) 1994;369:621–628. doi: 10.1038/369621a0. [DOI] [PubMed] [Google Scholar]
- 3.Sondek J, Lambright D G, Noel J P, Hamm H E, Sigler P B. Nature (London) 1994;372:276–279. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- 4.Lambright D G, Sondek J, Bohm A, Skiba N P, Hamm H E, Sigler P B. Nature (London) 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 5.Hamm H E, Gilchrist A. Curr Opin Cell Biol. 1996;8:189–196. doi: 10.1016/s0955-0674(96)80065-2. [DOI] [PubMed] [Google Scholar]
- 6.Helmreich E J, Hofmann K P. Biochim Biophys Acta. 1996;1286:285–322. doi: 10.1016/s0304-4157(96)00013-5. [DOI] [PubMed] [Google Scholar]
- 7.Gudermann T, Schonberg T, Schultz G. Annu Rev Neurosci. 1997;20:399–427. doi: 10.1146/annurev.neuro.20.1.399. [DOI] [PubMed] [Google Scholar]
- 8.West R E, Jr, Moss J, Vaughan M, Liu T, Liu T Y. J Biol Chem. 1985;260:14428–14430. [PubMed] [Google Scholar]
- 9.Deretic D, Hamm H E. J Biol Chem. 1987;262:10839–10847. [PubMed] [Google Scholar]
- 10.Hamm H E, Deretic D, Arendt A, Hargrave P A, Koenig B, Hofmann K P. Science. 1988;241:832–835. doi: 10.1126/science.3136547. [DOI] [PubMed] [Google Scholar]
- 11.Garcia P D, Onrust R, Bell S M, Sakmar T P, Bourne H R. EMBO J. 1995;14:4460–4469. doi: 10.1002/j.1460-2075.1995.tb00125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osawa S, Weiss E R. J Biol Chem. 1995;270:31052–31058. doi: 10.1074/jbc.270.52.31052. [DOI] [PubMed] [Google Scholar]
- 13.Kisselev O, Pronin A, Ermolaeva M, Gautam N. Proc Natl Acad Sci USA. 1995;92:9102–9106. doi: 10.1073/pnas.92.20.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilman A G. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 15.Clore G M, Gronenborn A M. J Magn Reson. 1983;53:423–442. [Google Scholar]
- 16.Dratz E A, Furstenau J E, Lambert C G, Thireault D L, Rarick H, Schepers T, Pakhlevaniants S, Hamm H E. Nature (London) 1993;363:276–281. doi: 10.1038/363276a0. [DOI] [PubMed] [Google Scholar]
- 17.Dratz E A, Furstenau J E, Lambert C G, Thireault D L, Rarick H, Schepers T, Pakhlevaniants S, Hamm H E. Nature (London) 1997;390:424. doi: 10.1038/363276a0. [DOI] [PubMed] [Google Scholar]
- 18.Papermaster D S, Dreyer W J. Biochemistry. 1974;13:2438–2444. doi: 10.1021/bi00708a031. [DOI] [PubMed] [Google Scholar]
- 19.Yamazaki A, Bartucca F, Ting A, Bitensky M W. Proc Natl Acad Sci USA. 1982;79:3702–3706. doi: 10.1073/pnas.79.12.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kisselev O G, Ermolaeva M V, Gautam N. J Biol Chem. 1994;269:21399–21402. [PubMed] [Google Scholar]
- 21.Braunschweiler L, Ernst R R. J Magn Reson. 1983;53:521–528. [Google Scholar]
- 22.Kumar A, Ernst R R, Wüthrich K. Biochem Biophys Res Comm. 1980;95:1–6. doi: 10.1016/0006-291x(80)90695-6. [DOI] [PubMed] [Google Scholar]
- 23.Hodsdon M E, Ponder J W, Cistola D P. J Mol Biol. 1996;264:585–602. doi: 10.1006/jmbi.1996.0663. [DOI] [PubMed] [Google Scholar]
- 24.Brooks B R, Bruccoleri R E, Olafson B D, States D J, Swaminathan S, Karplus M. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 25.Koradi R, Billeter M, Wüthrich K. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 26.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 27.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 28.Martin E L, Rens-Domiano S, Schatz P J, Hamm H E. J Biol Chem. 1996;271:361–366. doi: 10.1074/jbc.271.1.361. [DOI] [PubMed] [Google Scholar]
- 29.Kisselev O G, Ermolaeva M V, Gautam N. J Biol Chem. 1995;270:25356–25358. doi: 10.1074/jbc.270.43.25356. [DOI] [PubMed] [Google Scholar]
- 30.Kito Y, Suzuki T, Azuma M, Sekoguti Y. Nature (London) 1968;218:955–957. doi: 10.1038/218955a0. [DOI] [PubMed] [Google Scholar]
- 31.Hofmann K P. Photobiochem Photobiophys. 1986;13:309–327. [Google Scholar]
- 32.Kuhn H. Nature (London) 1980;283:587–589. doi: 10.1038/283587a0. [DOI] [PubMed] [Google Scholar]
- 33.Fung B K, Hurley J B, Stryer L. Proc Natl Acad Sci USA. 1981;78:152–156. doi: 10.1073/pnas.78.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bornancin F, Pfister C, Chabre M. Eur J Biochem. 1989;184:687–698. doi: 10.1111/j.1432-1033.1989.tb15068.x. [DOI] [PubMed] [Google Scholar]
- 35.Aurora R, Srinivasan R, Rose G D. Science. 1994;264:1126–1130. doi: 10.1126/science.8178170. [DOI] [PubMed] [Google Scholar]
- 36.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 37.Schellman C. In: Protein Folding, The αL Conformation at the End of Helices. Jaenicke R, editor. New York: Elsevier/North Holland; 1980. pp. 53–61. [Google Scholar]
- 38.Richardson J S, Richardson D C. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- 39.Presta L G, Rose G D. Science. 1988;240:1632–1641. doi: 10.1126/science.2837824. [DOI] [PubMed] [Google Scholar]
- 40.Acharya S, Saad Y, Karnik S S. J Biol Chem. 1997;272:6519–6524. doi: 10.1074/jbc.272.10.6519. [DOI] [PubMed] [Google Scholar]
- 41.Farrens D L, Altenbach C, Yang K, Hubbell W L, Khorana H G. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 42.Sheikh S P, Zvyaga T A, Lichtarge O, Sakmar T P, Bourne H R. Nature (London) 1996;383:347–350. doi: 10.1038/383347a0. [DOI] [PubMed] [Google Scholar]
- 43.Onrust R, Herzmark P, Chi P, Garcia P, Lichtarge O, Kingsley C, Bourne H. Science. 1997;275:381–384. doi: 10.1126/science.275.5298.381. [DOI] [PubMed] [Google Scholar]
- 44.Burley S K, Petsko G A. FEBS Lett. 1986;203:139–143. doi: 10.1016/0014-5793(86)80730-x. [DOI] [PubMed] [Google Scholar]
- 45.Dougherty D A. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 46.Tesmer J J, Berman D M, Gilman A G, Sprang S R. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 47.Mixon M B, Lee E, Coleman D E, Berghuis A M, Gilman A G, Sprang S R. Science. 1995;270:954–960. doi: 10.1126/science.270.5238.954. [DOI] [PubMed] [Google Scholar]
- 48.Fukada Y, Takao T, Ohguro H, Yoshizawa T, Akino T, Shimonishi Y. Nature (London) 1990;346:658–660. doi: 10.1038/346658a0. [DOI] [PubMed] [Google Scholar]
- 49.Taylor J M, Jacob-Mosier G G, Lawton R G, VanDort M, Neubig R R. J Biol Chem. 1996;271:3336–3339. doi: 10.1074/jbc.271.7.3336. [DOI] [PubMed] [Google Scholar]
- 50.Yasuda H, Lindorfer M A, Woodfork K A, Fletcher J E, Garrison J C. J Biol Chem. 1996;271:18588–18595. doi: 10.1074/jbc.271.31.18588. [DOI] [PubMed] [Google Scholar]